SB0093 : Amphiphysin I
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | RecName: Full=Amphiphysin; Amphiphysin |
| Gene Names | AMPH1 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 695 aa
Average Mass: 76.256 kDa
Monoisotopic Mass: 76.210 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Protein of unknown function (DUF3584) 1. | 39 | 193 | 903.0 | 9.2 |
| --- cleavage 333 --- | ||||
| --- cleavage 377 --- | ||||
| --- cleavage 392 --- | ||||
| --- cleavage 454 --- | ||||
| --- cleavage 478 --- | ||||
| --- cleavage 527 --- | ||||
| --- cleavage 531 --- | ||||
| --- cleavage 593 --- | ||||
| --- cleavage 609 --- | ||||
| Variant SH3 domain 1. | 629 | 689 | 4.0 | 0.0 |
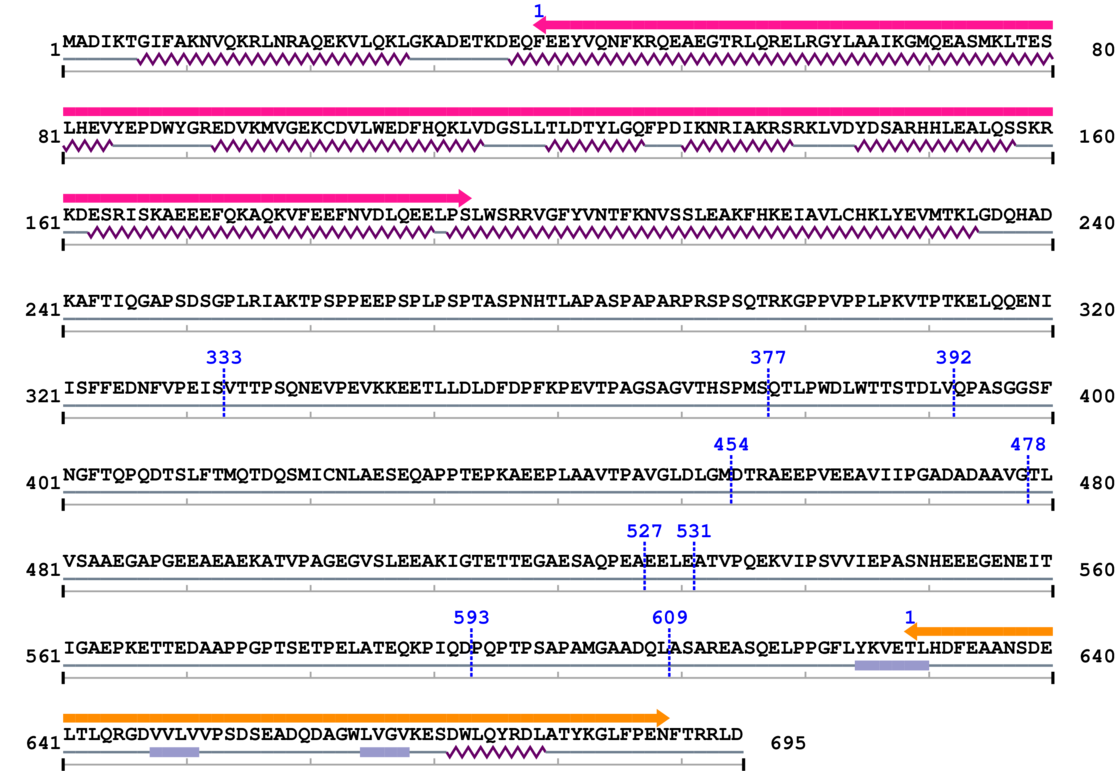
3. Sequence Information
Fasta Sequence: SB0093.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
9 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 17541403] Wu Y, Liang S, Oda Y, Ohmori I, Nishiki T, Takei K, Matsui H, Tomizawa K, Truncations of amphiphysin I by calpain inhibit vesicle endocytosis during neural hyperexcitation. EMBO J. 2007 Jun 20;26(12):2981-90. Epub 2007 May 31.
Cleavage sites (±10aa)
[Site 1] FEDNFVPEIS333-VTTPSQNEVP
Ser333  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe324 | Glu325 | Asp326 | Asn327 | Phe328 | Val329 | Pro330 | Glu331 | Ile332 | Ser333 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val334 | Thr335 | Thr336 | Pro337 | Ser338 | Gln339 | Asn340 | Glu341 | Val342 | Pro343 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PPLPKVTPTKELQQENIISFFEDNFVPEISVTTPSQNEVPEVKKEETLLDLDFDPFKPEV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 120.00 | 14 | amphiphysin isoform 1 |
| 2 | Canis familiaris | 120.00 | 2 | PREDICTED: similar to Amphiphysin |
| 3 | synthetic construct | 120.00 | 1 | amphiphysin |
| 4 | Rattus norvegicus | 112.00 | 2 | amphiphysin 1 |
| 5 | Mus musculus | 111.00 | 2 | amphiphysin |
| 6 | Gallus gallus | 107.00 | 1 | amphiphysin |
| 7 | Mus musculus | 64.30 | 1 | bridging integrator 1 isoform 1 |
| 8 | Danio rerio | 56.20 | 1 | amphiphysin |
| 9 | Homo sapiens | 51.60 | 3 | bridging integrator protein-1 |
| 10 | N/A | 51.20 | 1 | A Chain A, Nmr Structure Of The Tumor Suppressor |
| 11 | Xenopus tropicalis | 46.20 | 1 | bridging integrator 1 |
| 12 | Tetraodon nigroviridis | 38.10 | 1 | unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 |
| Canis familiaris | Query 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 |
| synthetic construct | Query 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 |
| Rattus norvegicus | Query 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||||||||+||||||||||+#||||||||| ||||||||||||||||||+| Sbjct 304 PPLPKVTPTKELQQENIINFFEDNFVPEIN#VTTPSQNEVLEVKKEETLLDLDFDPFKPDV 363 |
| Mus musculus | Query 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||||||||||||+|||||+||||||||||+#||||||||| ||||||||||||||||||+| Sbjct 304 PPLPKVTPTKELKQENIINFFEDNFVPEIN#VTTPSQNEVLEVKKEETLLDLDFDPFKPDV 363 |
| Gallus gallus | Query 304 PPLPKVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 |||||+||||||||||||+ |+|||||||+#||||||||+|| || |+||||||||||||| Sbjct 304 PPLPKLTPTKELQQENIINLFDDNFVPEIN#VTTPSQNEIPETKKVESLLDLDFDPFKPEV 363 |
| Mus musculus | Query 308 KVTPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPE-VKKEETLLDLDFDPFKP 361 | ||+||++|| |+| |+| ||||||#|||||| | | ++ +||||||+| | Sbjct 347 KHTPSKEMKQEQILSLFDDAFVPEIS#VTTPSQFEAPGPFSEQASLLDLDFEPLPP 401 |
| Danio rerio | Query 313 KELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 +||||| || |+ | ||||#||+| || | |+|||||||||||+ Sbjct 313 RELQQEQIIDLFDGGF-PEIS#VTSPQPNERP----GESLLDLDFDPFKPDT 358 |
| Homo sapiens | Query 308 KVTPTKELQQENIISFFEDNFVPEIS#VTTPSQ 339 | ||+||++|| |+| ||| ||||||#|||||| Sbjct 330 KHTPSKEVKQEQILSLFEDTFVPEIS#VTTPSQ 361 |
| N/A | Query 308 KVTPTKELQQENIISFFEDNFVPEIS#VTTPSQ 339 | ||+||++|| |+| ||| ||||||#|||||| Sbjct 46 KHTPSKEVKQEQILSLFEDTFVPEIS#VTTPSQ 77 |
| Xenopus tropicalis | Query 312 TKELQQENIISFFEDNFVPEIS#VTTPSQN 340 +|+ +||||+| |+|+||||||#||||||+ Sbjct 357 SKDFKQENILSLFDDSFVPEIS#VTTPSQS 385 |
| Tetraodon nigroviridis | Query 310 TPTKELQQENIISFFEDNFVPEIS#VTTPSQNEVPEVKKEETLLDLDFDPFKPEV 363 ||+|+++ |||++ |+ |+||#||+|++ + | | +|||+| | | | Sbjct 218 TPSKDMKAENIVNLFDAAAAPDIS#VTSPTEFDRPAV---SSLLDVDLDSFTSTV 268 |
[Site 2] SAGVTHSPMS377-QTLPWDLWTT
Ser377  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser368 | Ala369 | Gly370 | Val371 | Thr372 | His373 | Ser374 | Pro375 | Met376 | Ser377 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln378 | Thr379 | Leu380 | Pro381 | Trp382 | Asp383 | Leu384 | Trp385 | Thr386 | Thr387 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EETLLDLDFDPFKPEVTPAGSAGVTHSPMSQTLPWDLWTTSTDLVQPASGGSFNGFTQPQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 127.00 | 14 | Amphiphysin |
| 2 | synthetic construct | 127.00 | 1 | amphiphysin |
| 3 | Rattus norvegicus | 118.00 | 2 | amphiphysin 1 |
| 4 | Mus musculus | 114.00 | 2 | amphiphysin |
| 5 | Gallus gallus | 94.00 | 1 | amphiphysin |
| 6 | Danio rerio | 60.50 | 1 | amphiphysin |
| 7 | Canis familiaris | 38.10 | 2 | PREDICTED: similar to Myc box dependent interacti |
| 8 | Mus musculus | 36.20 | 1 | bridging integrator 1 isoform 1 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 407 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 407 |
| synthetic construct | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 407 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 407 |
| Rattus norvegicus | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 407 ||||||||||||||+||||||| ||||||#|||||||||||||||||||||||| ||||| Sbjct 348 EETLLDLDFDPFKPDVTPAGSAAATHSPMS#QTLPWDLWTTSTDLVQPASGGSFNDFTQPQ 407 |
| Mus musculus | Query 348 EETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQPQ 407 ||||||||||||||+| ||||| ||||||#|||||||||||||||||||||||| ||| | Sbjct 348 EETLLDLDFDPFKPDVAPAGSAAATHSPMS#QTLPWDLWTTSTDLVQPASGGSFNDFTQAQ 407 |
| Gallus gallus | Query 349 ETLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFTQ 405 |+||||||||||||| | ||||||||#||||||||||+++|||||| +|||| | Sbjct 349 ESLLDLDFDPFKPEVV---STGVTHSPMS#QTLPWDLWTTTSELVQPASSTAFNGFAQ 402 |
| Danio rerio | Query 349 ETLLDLDFDPFKPEV-TPAGSAGVTHSPMS#QTLPWDLWTTSTDLVQPASGGSFNGFT 404 |+|||||||||||+ || | | ||+|#||||||||| | ||| + ||| Sbjct 344 ESLLDLDFDPFKPDTSTPIGQ---TQSPVS#QTLPWDLWT--GDAAQPAQPATDAGFT 395 |
| Canis familiaris | Query 350 TLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLW 385 +|||||||| | +| + +| #|++||||| Sbjct 491 SLLDLDFDPLPPVASPVKAP----TPSG#QSIPWDLW 522 |
| Mus musculus | Query 350 TLLDLDFDPFKPEVTPAGSAGVTHSPMS#QTLPWDLW 385 +||||||+| | +| + +| #|++||||| Sbjct 390 SLLDLDFEPLPPVASPVKAP----TPSG#QSIPWDLW 421 |
[Site 3] DLWTTSTDLV392-QPASGGSFNG
Val392  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp383 | Leu384 | Trp385 | Thr386 | Thr387 | Ser388 | Thr389 | Asp390 | Leu391 | Val392 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln393 | Pro394 | Ala395 | Ser396 | Gly397 | Gly398 | Ser399 | Phe400 | Asn401 | Gly402 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| VTPAGSAGVTHSPMSQTLPWDLWTTSTDLVQPASGGSFNGFTQPQDTSLFTMQTDQSMIC |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 121.00 | 10 | amphiphysin isoform 1 |
| 2 | synthetic construct | 121.00 | 1 | amphiphysin |
| 3 | Rattus norvegicus | 110.00 | 1 | amphiphysin 1 |
| 4 | Mus musculus | 108.00 | 2 | Amph protein |
| 5 | Gallus gallus | 74.30 | 1 | amphiphysin |
| 6 | Canis familiaris | 61.20 | 1 | PREDICTED: similar to Amphiphysin |
| 7 | Danio rerio | 40.80 | 1 | amphiphysin |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 422 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 422 |
| synthetic construct | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 422 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 422 |
| Rattus norvegicus | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSM 420 ||||||| |||||||||||||||||||||#||||||||| ||||||||||||||||+| Sbjct 363 VTPAGSAAATHSPMSQTLPWDLWTTSTDLV#QPASGGSFNDFTQPQDTSLFTMQTDQNM 420 |
| Mus musculus | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSM 420 | ||||| |||||||||||||||||||||#||||||||| ||| ||||||||||||+| Sbjct 50 VAPAGSAAATHSPMSQTLPWDLWTTSTDLV#QPASGGSFNDFTQAQDTSLFTMQTDQNM 107 |
| Gallus gallus | Query 368 SAGVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFTQPQDTSLFTMQTDQSM 420 | ||||||||||||||||||+++||#|||| +|||| |||+ | +|+++++ Sbjct 365 STGVTHSPMSQTLPWDLWTTTSELV#QPASSTAFNGFA--QDTTAFAVQSNENV 415 |
| Canis familiaris | Query 363 VTPAGSAGVTHSPMSQTLPWDLWTTSTDLV#QP 394 | | || |||||||||||||||||||||||#|| Sbjct 382 VAPVGSVGVTHSPMSQTLPWDLWTTSTDLV#QP 413 Query 393 QPASGGSFNGFTQPQDTSLFTMQTDQSMIC 422 + ||||+||||||||||||||||||||||| Sbjct 586 EKASGGTFNGFTQPQDTSLFTMQTDQSMIC 615 |
| Danio rerio | Query 370 GVTHSPMSQTLPWDLWTTSTDLV#QPASGGSFNGFT 404 | | ||+|||||||||| | #||| + ||| Sbjct 363 GQTQSPVSQTLPWDLWT--GDAA#QPAQPATDAGFT 395 |
[Site 4] TPAVGLDLGM454-DTRAEEPVEE
Met454  Asp
Asp
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Thr445 | Pro446 | Ala447 | Val448 | Gly449 | Leu450 | Asp451 | Leu452 | Gly453 | Met454 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asp455 | Thr456 | Arg457 | Ala458 | Glu459 | Glu460 | Pro461 | Val462 | Glu463 | Glu464 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| AESEQAPPTEPKAEEPLAAVTPAVGLDLGMDTRAEEPVEEAVIIPGADADAAVGTLVSAA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 113.00 | 8 | amphiphysin isoform 1 |
| 2 | synthetic construct | 113.00 | 1 | amphiphysin |
| 3 | Rattus norvegicus | 40.80 | 1 | amphiphysin 1 |
| 4 | Mus musculus | 36.20 | 2 | amphiphysin |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 425 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 484 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 425 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 484 |
| synthetic construct | Query 425 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 484 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 425 AESEQAPPTEPKAEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLVSAA 484 |
| Rattus norvegicus | Query 425 AESEQAPPTEPKAEEPLAAVTPAV-GLDLGM#DTRAEEPVEEAVIIPGADADAAVGT 479 ||+||| ||||+|||| |||||+#+ ||| ||| | || || ||| Sbjct 421 AETEQALPTEPQAEEPPTTAAAPTAGLDLGL#EM--EEPKEEAAIPPGTDAGETVGT 474 |
| Mus musculus | Query 425 AESEQAPPTEPK-AEEPLAAVTPAVGLDLGM#DTRAEEPVEEAVIIPGADADAAVGTLV 481 ||+||| ||||+ | | | | |||||+#+ ||| ||||| | | | | | Sbjct 421 AETEQALPTEPQAEEPPATAAAPTAGLDLGL#EM--EEPKEEAVIPPATDTGETVETAV 476 |
[Site 5] PGADADAAVG478-TLVSAAEGAP
Gly478  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro469 | Gly470 | Ala471 | Asp472 | Ala473 | Asp474 | Ala475 | Ala476 | Val477 | Gly478 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr479 | Leu480 | Val481 | Ser482 | Ala483 | Ala484 | Glu485 | Gly486 | Ala487 | Pro488 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GLDLGMDTRAEEPVEEAVIIPGADADAAVGTLVSAAEGAPGEEAEAEKATVPAGEGVSLE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 113.00 | 8 | amphiphysin isoform 1 |
| 2 | synthetic construct | 110.00 | 1 | amphiphysin |
| 3 | Rattus norvegicus | 40.40 | 1 | amphiphysin 1 |
| 4 | Xanthobacter autotrophicus Py2 | 36.20 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 5 | Bradyrhizobium japonicum USDA 110 | 35.80 | 2 | malonic semialdehyde oxidative decarboxylase |
| 6 | Canis familiaris | 35.00 | 1 | PREDICTED: similar to Amphiphysin |
| 7 | Acidiphilium cryptum JF-5 | 34.30 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 8 | Agrobacterium tumefaciens str. C58 | 33.90 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 9 | Mycobacterium gilvum PYR-GCK | 33.50 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 10 | Nitrobacter winogradskyi Nb-255 | 33.50 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 11 | Rhizobium leguminosarum bv. viciae 3841 | 33.50 | 1 | malonic semialdehyde oxidative decarboxylase |
| 12 | Bradyrhizobium sp. BTAi1 | 33.10 | 1 | Methylmalonate-semialdehyde dehydrogenase |
| 13 | Nitrobacter sp. Nb-311A | 32.70 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 14 | Caulobacter crescentus CB15 | 32.70 | 1 | methylmalonate-semialdehyde dehydrogenase, putati |
| 15 | Colwellia psychrerythraea 34H | 32.30 | 3 | methylmalonate-semialdehyde dehydrogenase |
| 16 | Rhizobium etli CFN 42 | 32.30 | 1 | methylmalonate-semialdehyde dehydrogenase (acylat |
| 17 | Sinorhizobium meliloti 1021 | 32.30 | 1 | PUTATIVE MALONIC SEMIALDEHYDE OXIDATIVE DECARBOXY |
| 18 | Pseudomonas aeruginosa 2192 | 32.30 | 1 | COG1012: NAD-dependent aldehyde dehydrogenases |
| 19 | Rhodopseudomonas palustris BisA53 | 32.30 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 20 | Alteromonas macleodii 'Deep ecotype' | 32.00 | 2 | methylmalonate-semialdehyde dehydrogenase |
| 21 | Rhodopseudomonas palustris CGA009 | 32.00 | 1 | putative malonic semialdehyde oxidative decarboxy |
| 22 | Pseudomonas fluorescens Pf-5 | 32.00 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 23 | Pseudomonas fluorescens PfO-1 | 32.00 | 1 | Methylmalonate-semialdehyde dehydrogenase |
| 24 | Pseudomonas putida KT2440 | 32.00 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 25 | Rhodopseudomonas palustris BisB5 | 32.00 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 26 | Rhodopseudomonas palustris BisB18 | 32.00 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 27 | Pseudomonas aeruginosa PAO1 | 32.00 | 1 | probable aldehyde dehydrogenase |
| 28 | Pseudomonas syringae pv. phaseolicola 1448A | 32.00 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 29 | Rhodospirillum rubrum ATCC 11170 | 32.00 | 1 | Methylmalonate-semialdehyde dehydrogenase |
| 30 | Rhodopseudomonas palustris HaA2 | 32.00 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 31 | Pseudomonas syringae pv. tomato str. DC3000 | 32.00 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 32 | Pseudomonas syringae pv. syringae B728a | 32.00 | 1 | Methylmalonate-semialdehyde dehydrogenase |
| 33 | Mycobacterium bovis AF2122/97 | 31.60 | 1 | PROBABLE METHYLMALONATE-SEMIALDEHYDE DEHYDROGENAS |
| 34 | Brucella suis 1330 | 31.60 | 1 | methylmalonic acid semialdehyde dehydrogenase |
| 35 | Sphingopyxis alaskensis RB2256 | 31.60 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 36 | Arthrobacter sp. FB24 | 31.60 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 37 | Bordetella parapertussis 12822 | 31.60 | 1 | putative oxidoreductase |
| 38 | Oceanospirillum sp. MED92 | 31.60 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 39 | Mycobacterium tuberculosis CDC1551 | 31.60 | 1 | methylmalonic acid semialdehyde dehydrogenase |
| 40 | Mycobacterium tuberculosis H37Rv | 31.60 | 1 | PROBABLE METHYLMALONATE-SEMIALDEHYDE DEHYDROGENAS |
| 41 | Chromohalobacter salexigens DSM 3043 | 31.60 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 42 | Bordetella pertussis Tohama I | 31.60 | 1 | putative oxidoreductase |
| 43 | Oceanicaulis alexandrii HTCC2633 | 31.20 | 1 | methylmalonate-semialdehyde dehydrogenase, putati |
| 44 | Geobacillus kaustophilus HTA426 | 31.20 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 45 | Marinomonas sp. MED121 | 31.20 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 46 | Mycobacterium avium subsp. paratuberculosis K-10 | 31.20 | 1 | MmsA |
| 47 | Mycobacterium tuberculosis C | 31.20 | 1 | COG1012: NAD-dependent aldehyde dehydrogenases |
| 48 | Erythrobacter litoralis HTCC2594 | 30.80 | 1 | methylmalonate-semialdehyde dehydrogenase, putati |
| 49 | Triticum aestivum | 30.80 | 1 | methylmalonate semialdehyde dehydrogenase |
| 50 | Hahella chejuensis KCTC 2396 | 30.80 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 51 | Mesorhizobium loti MAFF303099 | 30.80 | 1 | malonic semialdehyde oxidative decarboxylase |
| 52 | Oryza sativa (japonica cultivar-group) | 30.80 | 1 | Os07g0188800 |
| 53 | Mesorhizobium sp. BNC1 | 30.80 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 54 | Oryza sativa | 30.80 | 1 | methylmalonate semi-aldehyde dehydrogenase |
| 55 | Nitrobacter hamburgensis X14 | 30.80 | 1 | methylmalonate-semialdehyde dehydrogenase |
| 56 | Azotobacter vinelandii AvOP | 30.40 | 1 | Aldehyde dehydrogenase |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 449 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVSLE 508 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 449 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVSLE 508 |
| synthetic construct | Query 449 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVSLE 508 ||||||||||||||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 449 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAETATVPAGEGVSLE 508 |
| Rattus norvegicus | Query 449 GLDLGMDTRAEEPVEEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVSLE 508 |||||++ ||| ||| | || || ||#| ||+ ||||||||| +||||| | | Sbjct 446 GLDLGLEM--EEPKEEAAIPPGTDAGETVG#T-----EGSTGEEAEAEKAALPAGEGESPE 498 |
| Xanthobacter autotrophicus Py2 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 +|+| || | || # |+ || |+ || | || ||| + Sbjct 263 NHMIIMPDADMDQAVD#ALMGAAYGSAGERCMAVSVAVPVGEGTA 306 |
| Bradyrhizobium japonicum USDA 110 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 +++| || | || # |+ || |+ || | || ||| + Sbjct 68 NHMIVMPDADMDQAVD#ALMGAAYGSAGERCMAISVAVPVGEGTA 111 |
| Canis familiaris | Query 466 VIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVSLE 508 ++||| ||| ||#| ||||| || |||||+|| +| ||| +| Sbjct 617 LVIPGTDADVTVG#TSVSAAEEAPVEEAEADKAVLPPGEGAGVE 659 |
| Acidiphilium cryptum JF-5 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | || # |+ || |+ || | || || Sbjct 262 NHMIIMPDADMDQAVD#ALMGAAYGSAGERCMAISVAVPVGE 302 |
| Agrobacterium tumefaciens str. C58 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | || # |+ | |+ || | || || Sbjct 272 NHMIIMPDADLDQAVN#ALMGAGYGSAGERCMAVSVAVPVGE 312 |
| Mycobacterium gilvum PYR-GCK | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | || # |+ | |+ || | || || Sbjct 253 NHMIIMPDADLDQAVD#ALIGAGYGSAGERCMAISVAVPVGE 293 |
| Nitrobacter winogradskyi Nb-255 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | +| # || | || || | || || Sbjct 265 NHMIIMPDADMDQSVE#ALVGAGYGAAGERCMAVSVAVPVGE 305 |
| Rhizobium leguminosarum bv. viciae 3841 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | || # |+ | |+ || | || || Sbjct 252 NHMIIMPDADLDQAVN#ALMGAGYGSAGERCMAISVAVPVGE 292 |
| Bradyrhizobium sp. BTAi1 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 |+++| || | || # |+ | |+ || | || |+ Sbjct 252 NHAIVMPDADLDQAVD#ALIGAGYGSAGERCMAVSVAVPVGK 292 |
| Nitrobacter sp. Nb-311A | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | +| # || | || || | || |+ Sbjct 252 NHMIIMPDADMDQSVD#ALVGAGYGAAGERCMAVSVAVPVGK 292 |
| Caulobacter crescentus CB15 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +++| || | || # ++ || |+ || | || || Sbjct 252 NHGLVMPDADLDQAVA#DIIGAAYGSAGERCMALPVVVPVGE 292 |
| Colwellia psychrerythraea 34H | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 |+|+| || | || # |+ || |+ || | | |+ Sbjct 254 NHAIIMPDADMDNAVN#QLLGAAFGSSGERCMALSVAVAVGD 294 |
| Rhizobium etli CFN 42 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | || # |+ | |+ || | || |+ Sbjct 252 NHMIIMPDADMDQAVN#ALMGAGYGSAGERCMAISVAVPVGK 292 |
| Sinorhizobium meliloti 1021 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | | # |+ | |+ || | || || Sbjct 252 NHMIIMPDADLDQAAN#ALIGAGYGSAGERCMAISVAVPVGE 292 |
| Pseudomonas aeruginosa 2192 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 ||++| || | || # |+ || |+ || | | |+ |+ Sbjct 55 NHAVLMPDADLDNAVS#ALMGAAYGSCGERCMAISVAVCVGDQVA 98 |
| Rhodopseudomonas palustris BisA53 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 |+|+| || | | # |+ | |+ || | || |+ Sbjct 252 NHAIIMPDADMDQTVD#ALIGAGYGSAGERCMAISVAVPVGK 292 |
| Alteromonas macleodii 'Deep ecotype' | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 |+++| || | || # |+ || |+ || | | |+ |+ Sbjct 251 NHAIVMPDADIDNAVN#QLLGAAFGSSGERCMALSVVVAVGDEVA 294 |
| Rhodopseudomonas palustris CGA009 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 |+|+| || | | # |+ | |+ || | || |+ Sbjct 252 NHAIIMPDADIDQTVD#ALIGAGYGSAGERCMAISVAVPVGK 292 |
| Pseudomonas fluorescens Pf-5 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 ||++| || | || # |+ || |+ || | | |+ |+ Sbjct 251 NHAVLMPDADLDNAVS#ALMGAAYGSCGERCMAISVAVCVGDQVA 294 |
| Pseudomonas fluorescens PfO-1 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 ||++| || | || # |+ || |+ || | | |+ |+ Sbjct 251 NHAVLMPDADLDNAVS#ALMGAAYGSCGERCMAISVAVCVGDQVA 294 |
| Pseudomonas putida KT2440 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 ||++| || | || # |+ || |+ || | | |+ |+ Sbjct 251 NHAVLMPDADLDNAVS#ALMGAAYGSCGERCMAISVAVCVGDQVA 294 |
| Rhodopseudomonas palustris BisB5 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 |+|+| || | | # |+ | |+ || | || |+ Sbjct 291 NHAIIMPDADIDQTVD#ALIGAGFGSAGERCMAISVAVPVGK 331 |
| Rhodopseudomonas palustris BisB18 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 |+|+| || | | # |+ | |+ || | || |+ Sbjct 252 NHAIIMPDADLDHTVD#ALIGAGYGSAGERCMAISVAVPVGK 292 |
| Pseudomonas aeruginosa PAO1 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 ||++| || | || # |+ || |+ || | | |+ |+ Sbjct 251 NHAVLMPDADLDNAVS#ALMGAAYGSCGERCMAISVAVCVGDQVA 294 |
| Pseudomonas syringae pv. phaseolicola 1448A | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 ||++| || | || # |+ || |+ || | | |+ ++ Sbjct 251 NHAVLMPDADLDNAVS#ALMGAAYGSCGERCMAISVAVCVGDQIA 294 |
| Rhodospirillum rubrum ATCC 11170 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | | # |+ | |+ || | || || Sbjct 255 NHMIIMPDADLDQTVD#ALIGAGYGSAGERCMAISVAVPVGE 295 |
| Rhodopseudomonas palustris HaA2 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 |+|+| || | | # |+ | |+ || | || |+ Sbjct 252 NHAIIMPDADLDQTVD#ALIGAGYGSAGERCMAISVAVPVGK 292 |
| Pseudomonas syringae pv. tomato str. DC3000 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 ||++| || | || # |+ || |+ || | | |+ ++ Sbjct 251 NHAVLMPDADLDNAVS#ALMGAAYGSCGERCMAISVAVCVGDQIA 294 |
| Pseudomonas syringae pv. syringae B728a | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 ||++| || | || # |+ || |+ || | | |+ ++ Sbjct 251 NHAVLMPDADLDNAVS#ALMGAAYGSCGERCMAISVAVCVGDQIA 294 |
| Mycobacterium bovis AF2122/97 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +++| || | || # |+ | |+ || | || |+ Sbjct 253 NHMIVMPDADLDQAVD#ALIGAGYGSAGERCMAISVAVPVGD 293 |
| Brucella suis 1330 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | | # |+ | |+ || | || || Sbjct 252 NHLLIMPDADMDQTVD#ALIGAGYGSAGERCMAISVAVPVGE 292 |
| Sphingopyxis alaskensis RB2256 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +++| || | | # | || |+ || | || || Sbjct 250 NHGIVMPDADLDQVVN#DLTGAAFGSAGERCMALPVVVPVGE 290 |
| Arthrobacter sp. FB24 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 ||+| || | || # |+ | |+ || | || |+ Sbjct 253 NHMVIMPDADLDMAVD#ALMGAGYGSAGERCMAISVAVPVGK 293 |
| Bordetella parapertussis 12822 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 ||+| || + |+ # || || || |+ | | || Sbjct 255 NHCVILPDADPEVALN#QLVGAAFGAAGQRCMATSVAVMVGE 295 |
| Oceanospirillum sp. MED92 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 |+++| || | || # |+ || |+ || | | |+ Sbjct 252 NHAIVMPDADMDNAVN#QLLGAAFGSSGERCMALSVAVAVGD 292 |
| Mycobacterium tuberculosis CDC1551 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +++| || | || # |+ | |+ || | || |+ Sbjct 253 NHMIVMPDADLDQAVD#ALIGAGYGSAGERCMAISVAVPVGD 293 |
| Mycobacterium tuberculosis H37Rv | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +++| || | || # |+ | |+ || | || |+ Sbjct 253 NHMIVMPDADLDQAVD#ALIGAGYGSAGERCMAISVAVPVGD 293 |
| Chromohalobacter salexigens DSM 3043 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 ||++| || | | #||+ || |+ || | | |+ Sbjct 250 NHAVVLPDADLDNAAS#TLIGAAFGSAGERCMAISVAVCVGD 290 |
| Bordetella pertussis Tohama I | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 ||+| || + |+ # || || || |+ | | || Sbjct 246 NHCVILPDADPEVALN#QLVGAAFGAAGQRCMATSVAVMVGE 286 |
| Oceanicaulis alexandrii HTCC2633 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | | # || +| |+ || | || |+ Sbjct 255 NHGIILPDADLDQVVQ#DLVGSAYGSAGERCMALPVAVPVGD 295 |
| Geobacillus kaustophilus HTA426 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 ++++| || | || # +++|| |+ || | | |+ Sbjct 258 NHSIVMPDADLDMAVT#NIINAAFGSAGERCMACSVVVAVGD 298 |
| Marinomonas sp. MED121 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEA 494 |+++| || | || # || || |+ || | Sbjct 251 NHAIVMPDADMDNAVN#QLVGAAFGSSGERCMA 282 |
| Mycobacterium avium subsp. paratuberculosis K-10 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +++| || | || # |+ | |+ || | || |+ Sbjct 253 NHMIVMPDADLDQAVD#ALIGAGYGSAGERCMAISVAVPVGD 293 |
| Mycobacterium tuberculosis C | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +++| || | || # |+ | |+ || | || |+ Sbjct 253 NHMIVMPDADLDQAVD#ALIGAGYGSAGERCMAISVAVPVGD 293 |
| Erythrobacter litoralis HTCC2594 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +++| || | | # | || |+ || | || || Sbjct 251 NHGIVMPDADLDQVVN#DLAGAAFGSAGERCMALPVVVPVGE 291 |
| Triticum aestivum | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAG 502 |+|+| || || + # |++| || |+ | | | Sbjct 140 NHAIILPDADRDATLN#ALIAAGFGAAGQRCMALSTAVFVG 179 |
| Hahella chejuensis KCTC 2396 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGEGVS 506 |++| || + ||#+|| || |+ || | | |+ |+ Sbjct 252 NHMVVMPDADPEQVVG#SLVGAAYGSAGERCMAISVAVCVGDEVA 295 |
| Mesorhizobium loti MAFF303099 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | | # |+ | |+ || | || | Sbjct 252 NHMIIMPDADMDQTVD#ALIGAGYGSAGERCMAISVAVPVGN 292 |
| Oryza sativa (japonica cultivar-group) | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAG 502 |+|+| || || + # |++| || |+ | | | Sbjct 289 NHAIILPDADRDATLN#ALIAAGFGAAGQRCMALSTAVFVG 328 |
| Mesorhizobium sp. BNC1 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || | | # |+ | |+ || | || |+ Sbjct 252 NHLIIMPDADLDQTVD#ALIGAGYGSAGERCMAVSVAVPVGK 292 |
| Oryza sativa | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAG 502 |+|+| || || + # |++| || |+ | | | Sbjct 288 NHAIILPDADRDATLN#ALIAAGFGAAGQRCMALSTAVFVG 327 |
| Nitrobacter hamburgensis X14 | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEAEKATVPAGE 503 +|+| || + +| # || | || || | || |+ Sbjct 252 NHMIIMPDADMEQSVD#ALVGAGYGAAGERCMAVSVAVPVGK 292 |
| Azotobacter vinelandii AvOP | Query 463 EEAVIIPGADADAAVG#TLVSAAEGAPGEEAEA 494 |+++| || | || #+|+ || |+ || | Sbjct 251 NHAILMPDADLDNAVN#SLLGAAFGSSGERCMA 282 |
[Site 6] EGAESAQPEA527-EELEATVPQE
Ala527  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu518 | Gly519 | Ala520 | Glu521 | Ser522 | Ala523 | Gln524 | Pro525 | Glu526 | Ala527 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu528 | Glu529 | Leu530 | Glu531 | Ala532 | Thr533 | Val534 | Pro535 | Gln536 | Glu537 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TVPAGEGVSLEEAKIGTETTEGAESAQPEAEELEATVPQEKVIPSVVIEPASNHEEEGEN |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 113.00 | 7 | Amphiphysin |
| 2 | synthetic construct | 113.00 | 1 | amphiphysin |
| 3 | Rattus norvegicus | 69.30 | 1 | amphiphysin 1 |
| 4 | Canis familiaris | 59.30 | 1 | PREDICTED: similar to Amphiphysin |
| 5 | Mus musculus | 57.00 | 3 | amphiphysin |
| 6 | Gallus gallus | 44.70 | 1 | amphiphysin |
| 7 | Theileria annulata strain Ankara | 31.20 | 1 | SfiI-subtelomeric fragment related protein family |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 498 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 557 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 498 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 557 |
| synthetic construct | Query 498 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 557 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 498 TVPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 557 |
| Rattus norvegicus | Query 499 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHE 552 +||||| | | ||| |+|| | | |+|# |||| |||||||||||||||||| Sbjct 489 LPAGEGESPEGAKIDVESTELASSESPQA#AELEAGAPQEKVIPSVVIEPASNHE 542 |
| Canis familiaris | Query 499 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 557 +| ||| +|||+ || |+ + + + #|| || | ||||||||||||||+| ||++ Sbjct 650 LPPGEGAGVEEARPDTEATQAVDGDRSQL#EEAEAVAPLEKVIPSVVIEPASNNEGEGDH 708 |
| Mus musculus | Query 499 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPASNHEEEGEN 557 +||||| | | ||| |+|| | | |+ # | || || |||||||||||||| |||+ Sbjct 493 LPAGEGGSPEGAKIDGESTELAISESPQP#VEPEAGAPQ--VIPSVVIEPASNHEGEGEH 549 |
| Gallus gallus | Query 499 VPAGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKV--IPSVVIEPASNHEEEGE 556 | || | | + || || +|+ #|| + | |||| |||||||||||+| ||| Sbjct 494 VAAGAG----EGAVRTEQEAAAEGDKPQG#EEKDVDVSQEKVSSIPSVVIEPASNNEGEGE 549 |
| Theileria annulata strain Ankara | Query 501 AGEGVSLEEAKIGTETTEGAESAQPEA#EELEATVPQEKVIPSVVIEPA-SNHEEEGEN 557 || | | + | ||| || | # | | | | +|| || || ++ Sbjct 2052 AGHCKKLPEPAVPTSTTEAEESTDPAQ#PAQPAGQPAESAETSTSAQPAESNQPEEHDS 2109 |
[Site 7] SAQPEAEELE531-ATVPQEKVIP
Glu531  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser522 | Ala523 | Gln524 | Pro525 | Glu526 | Ala527 | Glu528 | Glu529 | Leu530 | Glu531 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala532 | Thr533 | Val534 | Pro535 | Gln536 | Glu537 | Lys538 | Val539 | Ile540 | Pro541 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GEGVSLEEAKIGTETTEGAESAQPEAEELEATVPQEKVIPSVVIEPASNHEEEGENEITI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 113.00 | 8 | Amphiphysin |
| 2 | synthetic construct | 113.00 | 1 | amphiphysin |
| 3 | Rattus norvegicus | 64.70 | 1 | amphiphysin 1 |
| 4 | Canis familiaris | 58.20 | 1 | PREDICTED: similar to Amphiphysin |
| 5 | Mus musculus | 53.10 | 3 | amphiphysin |
| 6 | Gallus gallus | 43.90 | 1 | amphiphysin |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 502 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 561 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 502 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 561 |
| synthetic construct | Query 502 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 561 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 502 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENEITI 561 |
| Rattus norvegicus | Query 502 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHE 552 ||| | | ||| |+|| | | |+| |||#| |||||||||||||||||| Sbjct 492 GEGESPEGAKIDVESTELASSESPQAAELE#AGAPQEKVIPSVVIEPASNHE 542 |
| Canis familiaris | Query 502 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENE 558 ||| +|||+ || |+ + + + || |#| | ||||||||||||||+| ||++| Sbjct 653 GEGAGVEEARPDTEATQAVDGDRSQLEEAE#AVAPLEKVIPSVVIEPASNNEGEGDHE 709 |
| Mus musculus | Query 502 GEGVSLEEAKIGTETTEGAESAQPEAEELE#ATVPQEKVIPSVVIEPASNHEEEGENE 558 ||| | | ||| |+|| | | |+ | |#| || |||||||||||||| |||++ Sbjct 496 GEGGSPEGAKIDGESTELAISESPQPVEPE#AGAPQ--VIPSVVIEPASNHEGEGEHQ 550 |
| Gallus gallus | Query 509 EAKIGTETTEGAESAQPEAEELE#ATVPQEKV--IPSVVIEPASNHEEEGE 556 | + || || +|+ || +# | |||| |||||||||||+| ||| Sbjct 500 EGAVRTEQEAAAEGDKPQGEEKD#VDVSQEKVSSIPSVVIEPASNNEGEGE 549 |
[Site 8] LATEQKPIQD593-PQPTPSAPAM
Asp593  Pro
Pro
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu584 | Ala585 | Thr586 | Glu587 | Gln588 | Lys589 | Pro590 | Ile591 | Gln592 | Asp593 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Pro594 | Gln595 | Pro596 | Thr597 | Pro598 | Ser599 | Ala600 | Pro601 | Ala602 | Met603 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EPKETTEDAAPPGPTSETPELATEQKPIQDPQPTPSAPAMGAADQLASAREASQELPPGF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 117.00 | 6 | amphiphysin isoform 2 |
| 2 | synthetic construct | 114.00 | 1 | amphiphysin |
| 3 | Mus musculus | 44.30 | 3 | amphiphysin |
| 4 | Rattus norvegicus | 43.50 | 1 | amphiphysin 1 |
| 5 | Canis familiaris | 40.40 | 1 | PREDICTED: similar to Amphiphysin |
| 6 | Gallus gallus | 33.90 | 1 | amphiphysin |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 564 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 623 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 564 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 623 |
| synthetic construct | Query 564 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 623 || |||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 564 EPMETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 623 |
| Mus musculus | Query 564 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 623 ||+| || | | | |+||| |+ |# | | | | || | || +|+||||||| Sbjct 557 EPREAAEDVAAQGSAGEKQEVATEPTPL-D#SQATLPASA-GAVDASLSAGDATQELPPGF 614 |
| Rattus norvegicus | Query 571 DAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 623 | || || | ||||| |+ # || ||| || | || +|+||||||| Sbjct 561 DVAPQGPAGEKQELATEPTPLDS#QAATP-APA-GAVDASLSAGDAAQELPPGF 611 |
| Canis familiaris | Query 572 AAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 623 || ||||+| |||| | | +|# || ||| | | | | |+|||| Sbjct 722 AAAPGPTTELPELAPEPKAGED#SQPLPSALA---------ASEESLEVPPGF 764 |
| Gallus gallus | Query 564 EPKETTEDAAPPGPTSETPELATEQKPIQD#PQPTPSAPAMGAADQLASAREASQELPPGF 623 | |+ + | ||| ++ +||| ++# | ||| || ||| + + ++|||| Sbjct 557 ESKDAAAEMGTQGTDSETSQIGSEQKATEE#IQTTPS------QDQPASAGDTASDMPPGF 610 |
[Site 9] APAMGAADQL609-ASAREASQEL
Leu609  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala600 | Pro601 | Ala602 | Met603 | Gly604 | Ala605 | Ala606 | Asp607 | Gln608 | Leu609 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala610 | Ser611 | Ala612 | Arg613 | Glu614 | Ala615 | Ser616 | Gln617 | Glu618 | Leu619 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ETPELATEQKPIQDPQPTPSAPAMGAADQLASAREASQELPPGFLYKVETLHDFEAANSD |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 122.00 | 22 | AF498092_1 amphiphysin I variant CT2 |
| 2 | synthetic construct | 121.00 | 2 | amphiphysin |
| 3 | Rattus norvegicus | 68.60 | 3 | amphiphysin 1 |
| 4 | Mus musculus | 66.60 | 4 | amphiphysin |
| 5 | Canis familiaris | 63.90 | 2 | PREDICTED: similar to Amphiphysin |
| 6 | Gallus gallus | 62.00 | 1 | amphiphysin |
| 7 | Tetraodon nigroviridis | 42.40 | 2 | unnamed protein product |
| 8 | Danio rerio | 41.20 | 1 | amphiphysin |
| 9 | N/A | 36.20 | 4 | SH3P9 |
| 10 | Mus musculus | 36.20 | 1 | bridging integrator 1 isoform 1 |
| 11 | Xenopus tropicalis | 35.80 | 1 | bridging integrator 1 |
| 12 | Xenopus laevis | 35.00 | 1 | bridging integrator 1 |
| 13 | Homo sapiens | 33.50 | 4 | bridging integrator protein-1 |
| 14 | Bos taurus | 33.50 | 1 | bridging integrator 1 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 580 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 639 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 580 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 639 |
| synthetic construct | Query 580 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 639 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 580 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 639 |
| Rattus norvegicus | Query 580 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 639 | ||||| |+ || ||| || | # || +|+||||||||||||||||||||||| Sbjct 570 EKQELATEPTPLDSQAATP-APA-GAVDAS#LSAGDAAQELPPGFLYKVETLHDFEAANSD 627 |
| Mus musculus | Query 580 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 639 | |+||| |+ | | | | | || | # || +|+||||||||||||||||||||||| Sbjct 573 EKQEVATEPTPL-DSQATLPASA-GAVDAS#LSAGDATQELPPGFLYKVETLHDFEAANSD 630 |
| Canis familiaris | Query 580 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 639 | |||| | | +| || ||| | # | | | |+|||||||||||||||||||| Sbjct 730 ELPELAPEPKAGEDSQPLPSALA-------#--ASEESLEVPPGFLYKVETLHDFEAANSD 780 |
| Gallus gallus | Query 580 ETPELATEQKPIQDPQPTPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 639 || ++ +||| ++ | ||| || #||| + + ++|||||+||| |||||||||| Sbjct 573 ETSQIGSEQKATEEIQTTPSQ------DQP#ASAGDTASDMPPGFLFKVEVLHDFEAANSD 626 |
| Tetraodon nigroviridis | Query 618 ELPPGFLYKVETLHDFEAANSD 639 +|| ||+||||+||||||||| Sbjct 761 QLPEDFLFKVETMHDFEAANSD 782 |
| Danio rerio | Query 616 SQELPPGFLYKVETLHDFEAANSD 639 | +|| ||+||||+||||||| | Sbjct 706 SSGMPPRFLFKVETMHDFEAANPD 729 |
| N/A | Query 597 TPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 639 | || || + # || +|||||++||+ ||+ | ++| Sbjct 338 TFSATVNGAVE--#GSAGTGRLDLPPGFMFKVQAQHDYTATDTD 378 |
| Mus musculus | Query 597 TPSAPAMGAADQL#ASAREASQELPPGFLYKVETLHDFEAANSD 639 | || || + # || +|||||++||+ ||+ | ++| Sbjct 492 TFSATVNGAVE--#GSAGTGRLDLPPGFMFKVQAQHDYTATDTD 532 |
| Xenopus tropicalis | Query 618 ELPPGFLYKVETLHDFEAANSD 639 |+|||||||| ||+ + +|| Sbjct 481 EMPPGFLYKVRAQHDYASTDSD 502 |
| Xenopus laevis | Query 618 ELPPGFLYKVETLHDFEAANSD 639 |+|||||||| ||+ | + | Sbjct 401 EMPPGFLYKVRAQHDYTATDGD 422 |
| Homo sapiens | Query 618 ELPPGFLYKVETLHDFEAANSD 639 +|||||++||+ ||+ | ++| Sbjct 420 DLPPGFMFKVQAHHDYTATDTD 441 |
| Bos taurus | Query 618 ELPPGFLYKVETLHDFEAANSD 639 +|||||++||+ ||+ | ++| Sbjct 356 DLPPGFMFKVQAQHDYTATDTD 377 |
* References
[PubMed ID: 8076697] David C, Solimena M, De Camilli P, Autoimmunity in stiff-Man syndrome with breast cancer is targeted to the C-terminal region of human amphiphysin, a protein similar to the yeast proteins, Rvs167 and Rvs161. FEBS Lett. 1994 Aug 29;351(1):73-9.
[PubMed ID: 7757077] ... Yamamoto R, Li X, Winter S, Francke U, Kilimann MW, Primary structure of human amphiphysin, the dominant autoantigen of paraneoplastic stiff-man syndrome, and mapping of its gene (AMPH) to chromosome 7p13-p14. Hum Mol Genet. 1995 Feb;4(2):265-8.
[PubMed ID: 9513187] ... Floyd S, Butler MH, Cremona O, David C, Freyberg Z, Zhang X, Solimena M, Tokunaga A, Ishizu H, Tsutsui K, De Camilli P, Expression of amphiphysin I, an autoantigen of paraneoplastic neurological syndromes, in breast cancer. Mol Med. 1998 Jan;4(1):29-39.
[PubMed ID: 12690205] ... Scherer SW, Cheung J, MacDonald JR, Osborne LR, Nakabayashi K, Herbrick JA, Carson AR, Parker-Katiraee L, Skaug J, Khaja R, Zhang J, Hudek AK, Li M, Haddad M, Duggan GE, Fernandez BA, Kanematsu E, Gentles S, Christopoulos CC, Choufani S, Kwasnicka D, Zheng XH, Lai Z, Nusskern D, Zhang Q, Gu Z, Lu F, Zeesman S, Nowaczyk MJ, Teshima I, Chitayat D, Shuman C, Weksberg R, Zackai EH, Grebe TA, Cox SR, Kirkpatrick SJ, Rahman N, Friedman JM, Heng HH, Pelicci PG, Lo-Coco F, Belloni E, Shaffer LG, Pober B, Morton CC, Gusella JF, Bruns GA, Korf BR, Quade BJ, Ligon AH, Ferguson H, Higgins AW, Leach NT, Herrick SR, Lemyre E, Farra CG, Kim HG, Summers AM, Gripp KW, Roberts W, Szatmari P, Winsor EJ, Grzeschik KH, Teebi A, Minassian BA, Kere J, Armengol L, Pujana MA, Estivill X, Wilson MD, Koop BF, Tosi S, Moore GE, Boright AP, Zlotorynski E, Kerem B, Kroisel PM, Petek E, Oscier DG, Mould SJ, Dohner H, Dohner K, Rommens JM, Vincent JB, Venter JC, Li PW, Mural RJ, Adams MD, Tsui LC, Human chromosome 7: DNA sequence and biology. Science. 2003 May 2;300(5620):767-72. Epub 2003 Apr 10.
[PubMed ID: 12853948] ... Hillier LW, Fulton RS, Fulton LA, Graves TA, Pepin KH, Wagner-McPherson C, Layman D, Maas J, Jaeger S, Walker R, Wylie K, Sekhon M, Becker MC, O'Laughlin MD, Schaller ME, Fewell GA, Delehaunty KD, Miner TL, Nash WE, Cordes M, Du H, Sun H, Edwards J, Bradshaw-Cordum H, Ali J, Andrews S, Isak A, Vanbrunt A, Nguyen C, Du F, Lamar B, Courtney L, Kalicki J, Ozersky P, Bielicki L, Scott K, Holmes A, Harkins R, Harris A, Strong CM, Hou S, Tomlinson C, Dauphin-Kohlberg S, Kozlowicz-Reilly A, Leonard S, Rohlfing T, Rock SM, Tin-Wollam AM, Abbott A, Minx P, Maupin R, Strowmatt C, Latreille P, Miller N, Johnson D, Murray J, Woessner JP, Wendl MC, Yang SP, Schultz BR, Wallis JW, Spieth J, Bieri TA, Nelson JO, Berkowicz N, Wohldmann PE, Cook LL, Hickenbotham MT, Eldred J, Williams D, Bedell JA, Mardis ER, Clifton SW, Chissoe SL, Marra MA, Raymond C, Haugen E, Gillett W, Zhou Y, James R, Phelps K, Iadanoto S, Bubb K, Simms E, Levy R, Clendenning J, Kaul R, Kent WJ, Furey TS, Baertsch RA, Brent MR, Keibler E, Flicek P, Bork P, Suyama M, Bailey JA, Portnoy ME, Torrents D, Chinwalla AT, Gish WR, Eddy SR, McPherson JD, Olson MV, Eichler EE, Green ED, Waterston RH, Wilson RK, The DNA sequence of human chromosome 7. Nature. 2003 Jul 10;424(6945):157-64.
[PubMed ID: 15489334] ... Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P, Guyer M, Peck AM, Derge JG, Lipman D, Collins FS, Jang W, Sherry S, Feolo M, Misquitta L, Lee E, Rotmistrovsky K, Greenhut SF, Schaefer CF, Buetow K, Bonner TI, Haussler D, Kent J, Kiekhaus M, Furey T, Brent M, Prange C, Schreiber K, Shapiro N, Bhat NK, Hopkins RF, Hsie F, Driscoll T, Soares MB, Casavant TL, Scheetz TE, Brown-stein MJ, Usdin TB, Toshiyuki S, Carninci P, Piao Y, Dudekula DB, Ko MS, Kawakami K, Suzuki Y, Sugano S, Gruber CE, Smith MR, Simmons B, Moore T, Waterman R, Johnson SL, Ruan Y, Wei CL, Mathavan S, Gunaratne PH, Wu J, Garcia AM, Hulyk SW, Fuh E, Yuan Y, Sneed A, Kowis C, Hodgson A, Muzny DM, McPherson J, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madari A, Young AC, Wetherby KD, Granite SJ, Kwong PN, Brinkley CP, Pearson RL, Bouffard GG, Blakesly RW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Griffith M, Griffith OL, Krzywinski MI, Liao N, Morin R, Palmquist D, Petrescu AS, Skalska U, Smailus DE, Stott JM, Schnerch A, Schein JE, Jones SJ, Holt RA, Baross A, Marra MA, Clifton S, Makowski KA, Bosak S, Malek J, The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC). Genome Res. 2004 Oct;14(10B):2121-7.
[PubMed ID: 16903783] ... Schmid EM, Ford MG, Burtey A, Praefcke GJ, Peak-Chew SY, Mills IG, Benmerah A, McMahon HT, Role of the AP2 beta-appendage hub in recruiting partners for clathrin-coated vesicle assembly. PLoS Biol. 2006 Sep;4(9):e262.
[PubMed ID: 20946875] ... Dergai O, Novokhatska O, Dergai M, Skrypkina I, Tsyba L, Moreau J, Rynditch A, Intersectin 1 forms complexes with SGIP1 and Reps1 in clathrin-coated pits. Biochem Biophys Res Commun. 2010 Nov 12;402(2):408-13. doi:
[PubMed ID: 12057195] ... Brett TJ, Traub LM, Fremont DH, Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure. 2002 Jun;10(6):797-809.