SB0097 : Insulin-like growth factor binding protein 2
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | insulin-like growth factor-binding protein 2 precursor [Homo sapiens]; insulin-like growth factor-binding protein 2 precursor; insulin-like growth factor-binding protein 2; IBP-2; IGFBP-2; IGF-binding protein 2; Insulin-like growth factor-binding protein 2 |
| Gene Names | IGFBP2; BP2, IBP2; BP2; IBP2; insulin-like growth factor binding protein 2, 36kDa |
| Gene Locus | 2q35; chromosome 2 |
| GO Function | Not available |
* Information From OMIM
Function: Png et al. (2012) demonstrated that endogenous miR126 (OMIM:611767), a miRNA silenced in a variety of human cancers, non-cell-autonomously regulates endothelial cell recruitment to metastatic breast cancer cells, in vitro and in vivo. It suppresses metastatic endothelial recruitment, metastatic angiogenesis, and metastatic colonization through coordinate targeting of IGFBP2, PITPNC1 (OMIM:605134), and MERTK (OMIM:604705), novel proangiogenic genes and biomarkers of human metastasis. Insulin-like growth factor binding protein-2 (IGFBP2) secreted by metastatic cells recruits endothelia by modulating IGF1 (OMIM:147440)-mediated activation of the IGF type-I receptor (OMIM:147370) on endothelial cells, whereas c-Mer tyrosine kinase (MERTK) receptor cleaved from metastatic cells promotes endothelial recruitment by competitively antagonizing the binding of its ligand GAS6 (OMIM:600441) to endothelial MERTK receptors. Coinjection of endothelial cells with breast cancer cells non-cell-autonomously rescued their miR126-induced metastatic defect, revealing a novel and important role for endothelial interactions in metastatic initiation. Through loss-of-function and epistasis experiments, Png et al. (2012) delineated a miRNA regulator network's individual components as novel and cell-extrinsic regulators of endothelial recruitment, angiogenesis, and metastatic colonization. The authors also identified the IGFBP2/IGF1/IGF1R and GAS6/MERTK signaling pathways as regulators of cancer-mediated endothelial recruitment.
* Structure Information
1. Primary Information
Length: 328 aa
Average Mass: 35.137 kDa
Monoisotopic Mass: 35.114 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Cysteine-rich CWC 1. | 86 | 93 | 20.0 | 4.7 |
| Thyroglobulin type-1 repeat 1. | 101 | 109 | 21.0 | 0.0 |
| --- cleavage 202 --- | ||||
| Insulin-like growth factor binding protein 1. | 271 | 288 | 11.0 | 0.7 |
3. Sequence Information
Fasta Sequence: SB0097.fasta
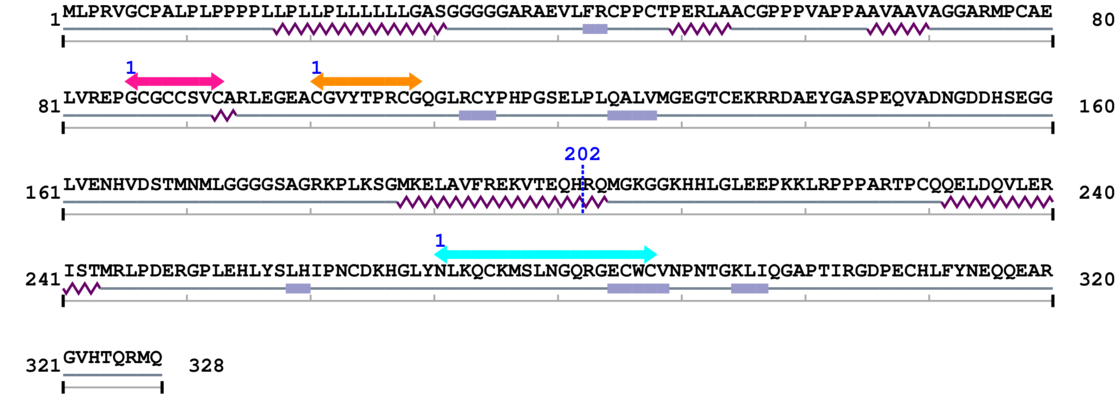
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 2H7T (NMR; -; A=220-325)
* Cleavage Information
1 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 17655506] Berg U, Bang P, Carlsson-Skwirut C, Calpain proteolysis of insulin-like growth factor binding protein (IGFBP) -2 and -3, but not of IGFBP-1. Biol Chem. 2007 Aug;388(8):859-63.
Cleavage sites (±10aa)
[Site 1] VFREKVTEQH202-RQMGKGGKHH
His202  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Val193 | Phe194 | Arg195 | Glu196 | Lys197 | Val198 | Thr199 | Glu200 | Gln201 | His202 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg203 | Gln204 | Met205 | Gly206 | Lys207 | Gly208 | Gly209 | Lys210 | His211 | His212 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24682597] Hu Q, Huang L, Kuang X, Zhang H, Ling G, Chen X, Li K, Deng Z, Zhou J, Is insulin-like growth factor binding protein 2 associated with metastasis in lung cancer? Clin Exp Metastasis. 2014 Jun;31(5):535-41. doi: 10.1007/s10585-014-9647-4. Epub