SB0099 : myocilin
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | myocilin precursor [Homo sapiens]; myocilin precursor; mutated trabecular meshwork-induced glucocorticoid response protein; myocilin 55 kDa subunit; Myocilin; Myocilin 55 kDa subunit; Trabecular meshwork-induced glucocorticoid response protein; Myocilin, N-terminal fragment; Myocilin 20 kDa N-terminal fragment; Myocilin, C-terminal fragment; Myocilin 35 kDa N-terminal fragment |
| Gene Names | MYOC; GLC1A, TIGR; GLC1A; TIGR; myocilin, trabecular meshwork inducible glucocorticoid response |
| Gene Locus | 1q23-q24; chromosome 1 |
| GO Function | Not available |
* Information From OMIM
Function: Stone et al. (1997) speculated that the TIGR gene product may cause increased intraocular pressure by obstruction of aqueous outflow. Its expression in trabecular meshwork (TM) and ciliary body (structures of the eye involved in the regulation of intraocular pressure) was consistent with this hypothesis. Obstruction of aqueous outflow is, however, not the only mechanism. Stone (1997) stated that because myocilin is expressed in large amounts in various types of muscle, ciliary body, papillary sphincter, skeletal muscle, heart, and other tissues, it is possible that some muscle-related ciliary body mechanism may be involved in the elevated intraocular pressure. Pilocarpine lowers pressure by constricting the ciliary body chronically; one might then expect a ciliary muscle abnormality to be associated with elevated intraocular pressure.
* Structure Information
1. Primary Information
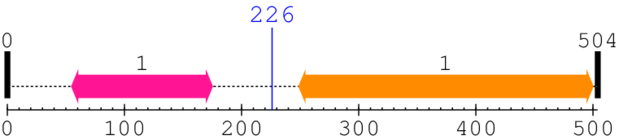
Length: 504 aa
Average Mass: 56.971 kDa
Monoisotopic Mass: 56.937 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Filament-like plant protein, long coiled-coil 1. | 55 | 175 | 695.0 | 9.1 |
| --- cleavage 226 --- | ||||
| Olfactomedin-like domain 1. | 249 | 500 | 2.0 | 0.3 |
3. Sequence Information
Fasta Sequence: SB0099.fasta
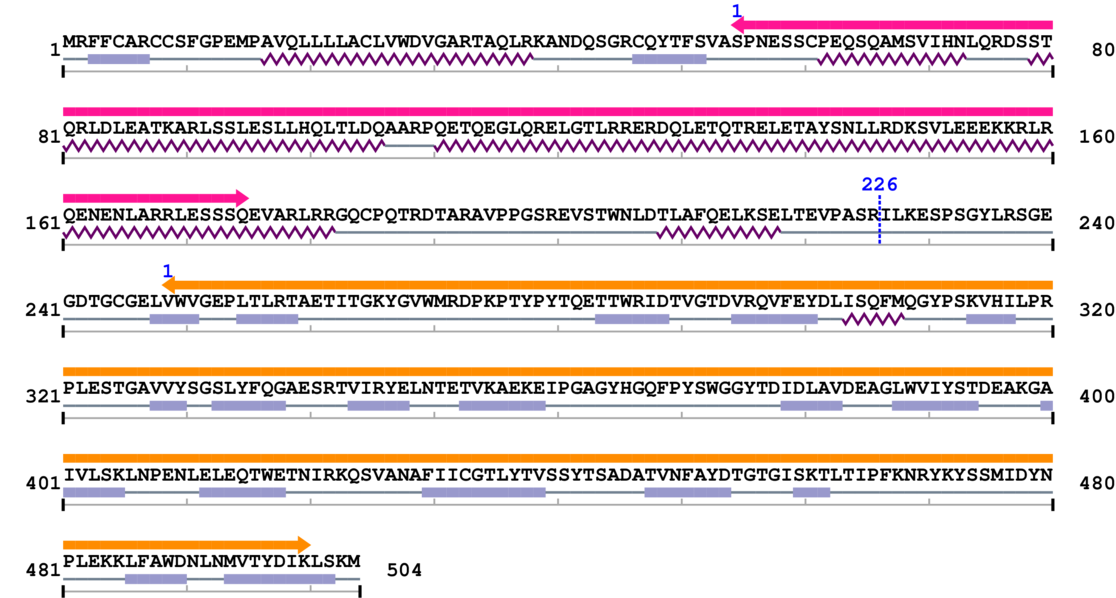
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
1 [sites] cleaved by Calpain 1 and 2
Source Reference: [PubMed ID: 17650508] Sanchez-Sanchez F, Martinez-Redondo F, Aroca-Aguilar JD, Coca-Prados M, Escribano J, Characterization of the intracellular proteolytic cleavage of myocilin and identification of calpain II as a myocilin-processing protease. J Biol Chem. 2007 Sep 21;282(38):27810-24. Epub 2007 Jul 24.
Cleavage sites (±10aa)
[Site 1] SELTEVPASR226-ILKESPSGYL
Arg226  Ile
Ile
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser217 | Glu218 | Leu219 | Thr220 | Glu221 | Val222 | Pro223 | Ala224 | Ser225 | Arg226 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ile227 | Leu228 | Lys229 | Glu230 | Ser231 | Pro232 | Ser233 | Gly234 | Tyr235 | Leu236 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 25450062] Faralli JA, Clark RW, Filla MS, Peters DM, NFATc1 activity regulates the expression of myocilin induced by dexamethasone. Exp Eye Res. 2015 Jan;130:9-16. doi: 10.1016/j.exer.2014.11.009. Epub 2014 Nov