SB0101 : C-terminal binding protein 1
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | C-terminal-binding protein 1 isoform 1 [Homo sapiens]; C-terminal-binding protein 1 isoform 1; brefeldin A-ribosylated substrate; C-terminal-binding protein 1; CtBP1; 1.1.1.- |
| Gene Names | CTBP1; CTBP; C-terminal binding protein 1 |
| Gene Locus | 4p16; chromosome 4 |
| GO Function | Not available |
* Information From OMIM
Function: Polycomb (Pc) is part of a Pc group (PcG) protein complex that is involved in repression of gene activity during Drosophila and vertebrate development. Using a yeast 2-hybrid assay, Sewalt et al. (1999) found that Xenopus Ctbp1 interacts with Xenopus Pc and that human CTBP2 interacts with PC2 (OMIM:603079), a human Pc homolog. Immunofluorescence studies indicated that CTBP1 and CTBP2 partially colocalize with PC2 in large PcG domains in interphase nuclei. As with PC2, chimeric LexA-CTBP2 and LexA-CTBP1 proteins repressed gene activity when targeted to a reporter gene. Sewalt et al. (1999) suggested that PC2-mediated repression of gene expression involves an association with corepressors such as the CTBPs. They speculated that the interference of the adenoviral E1A protein with the transcription machinery of the infected cell may involve interference with PcG-mediated repression through disruption of the CTBP-PcG interaction. Northern blot analysis revealed that the CTBP1 gene was expressed as a 2.4-kb mRNA in all human tissues tested.
* Structure Information
1. Primary Information
Length: 440 aa
Average Mass: 47.535 kDa
Monoisotopic Mass: 47.505 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Cellulose synthase subunit D 1. | 170 | 198 | 88.0 | 0.0 |
| XdhC Rossmann domain 1. | 337 | 374 | 44.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0101.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
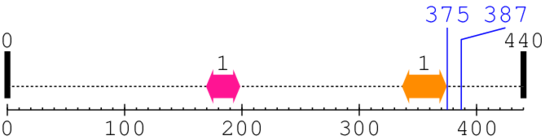
2 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 23707407] Ono Y, Iemura S, Novak SM, Doi N, Kitamura F, Natsume T, Gregorio CC, Sorimachi H, PLEIAD/SIMC1/C5orf25, a novel autolysis regulator for a skeletal-muscle-specific calpain, CAPN3, scaffolds a CAPN3 substrate, CTBP1. J Mol Biol. 2013 Aug 23;425(16):2955-72. doi: 10.1016/j.jmb.2013.05.009. Epub
Cleavage sites (±10aa)
[Site 1] DPAVVHPELN375-GAAYRYPPGV
Asn375  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp366 | Pro367 | Ala368 | Val369 | Val370 | His371 | Pro372 | Glu373 | Leu374 | Asn375 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly376 | Ala377 | Ala378 | Tyr379 | Arg380 | Tyr381 | Pro382 | Pro383 | Gly384 | Val385 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
[Site 2] AYRYPPGVVG387-VAPTGIPAAV
Gly387  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala378 | Tyr379 | Arg380 | Tyr381 | Pro382 | Pro383 | Gly384 | Val385 | Val386 | Gly387 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val388 | Ala389 | Pro390 | Thr391 | Gly392 | Ile393 | Pro394 | Ala395 | Ala396 | Val397 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 23907728] Bizama C, Benavente F, Salvatierra E, Gutierrez-Moraga A, Espinoza JA, Fernandez EA, Roa I, Mazzolini G, Sagredo EA, Gidekel M, Podhajcer OL, The low-abundance transcriptome reveals novel biomarkers, specific intracellular pathways and targetable genes associated with advanced gastric cancer. Int J Cancer. 2014 Feb 15;134(4):755-64. doi: 10.1002/ijc.28405. Epub 2013 Aug