SB0103 : [beta]2-Adaptin, AP2B1
[ CaMP Format ]
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | AP-2 complex subunit beta [Rattus norvegicus]; AP-2 complex subunit beta; AP105B; beta-adaptin; beta2-adaptin; adaptor protein complex AP-2 subunit beta; adapter-related protein complex 2 beta subunit; clathrin assembly protein complex 2 beta large chain; plasma membrane adaptor HA2/AP2 adaptin beta subunit; beta-2-adaptin; beta adaptin; adapter-related protein complex 2 subunit beta; adaptor-related protein complex 2 subunit beta; AP complex subunit beta {ECO:0000256|PIRNR:PIRNR002291} |
| Gene Names | Ap2b1; rCG_33026 {ECO:0000313|EMBL:EDM05477.1}; adaptor-related protein complex 2, beta 1 subunit |
| Gene Locus | 10q26; chromosome 10 |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| NP_542150 | NM_080583 | 140670 | Q3ZB97 | N/A | N/A | N/A | rno:140670 |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 951 aa
Average Mass: 105.691 kDa
Monoisotopic Mass: 105.625 kDa
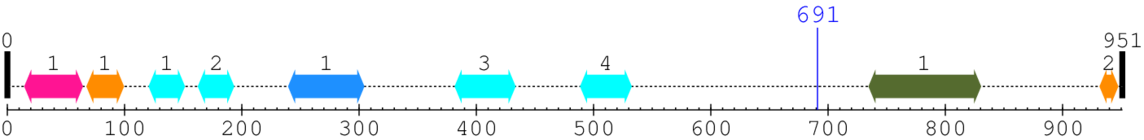
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| non-SMC mitotic condensation complex subunit 1 1. | 15 | 64 | 105.0 | 0.3 |
| Guanine nucleotide exchange factor in Golgi transport N-terminal 1. | 68 | 99 | 109.0 | 0.0 |
| CLASP N terminal 1. | 121 | 151 | 177.0 | 0.2 |
| CLASP N terminal 2. | 163 | 193 | 180.0 | 0.0 |
| Armadillo-like 1. | 240 | 304 | 98.0 | 0.1 |
| CLASP N terminal 3. | 382 | 433 | 86.0 | 0.1 |
| CLASP N terminal 4. | 489 | 532 | 118.0 | 0.0 |
| --- cleavage 691 --- | ||||
| Adaptin C-terminal domain 1. | 735 | 830 | 9.0 | 0.4 |
| Guanine nucleotide exchange factor in Golgi transport N-terminal 2. | 932 | 947 | 135.0 | 0.0 |
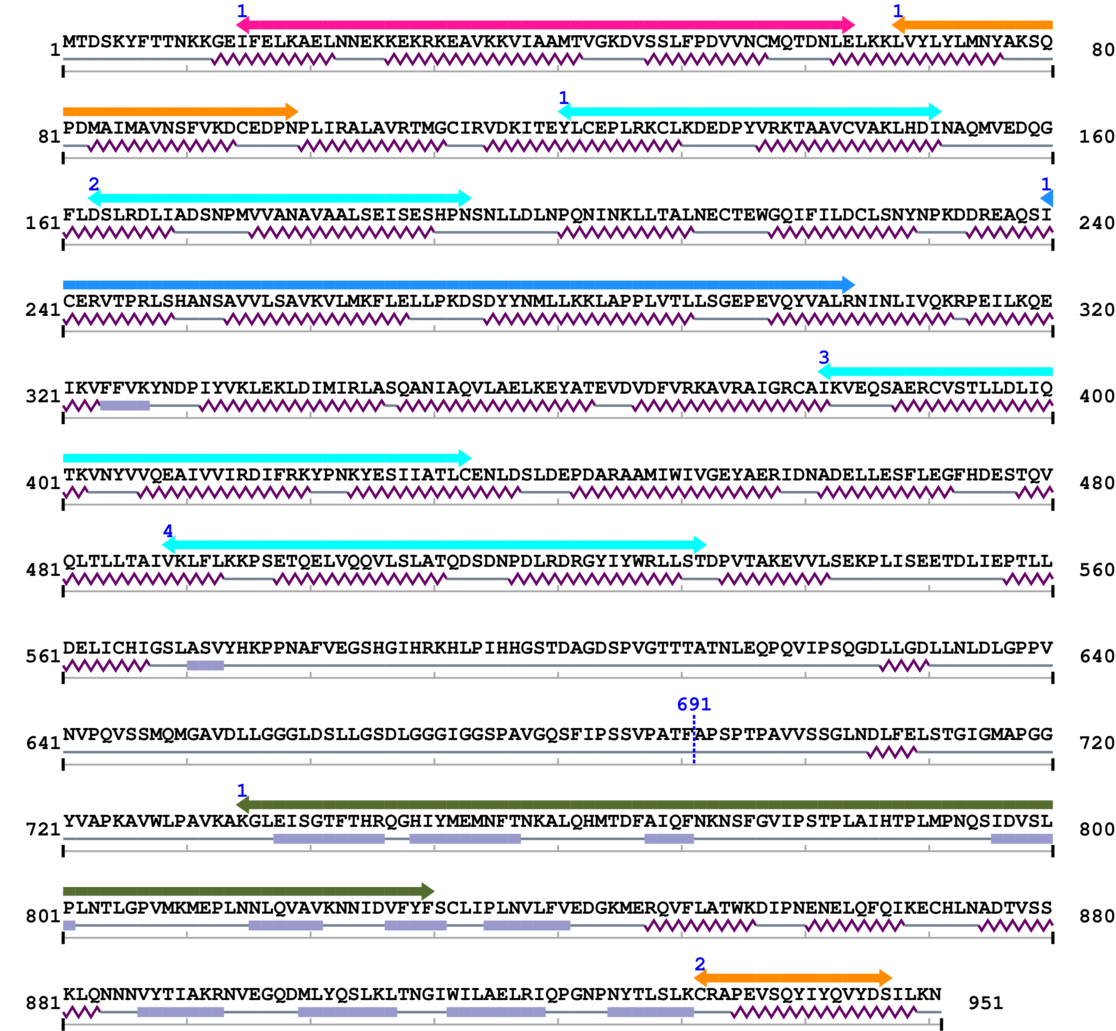
3. Sequence Information
Fasta Sequence: SB0103.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites] cleaved by Calpain 1 and 2
Source Reference: [PubMed ID: 19240038] Rudinskiy N, Grishchuk Y, Vaslin A, Puyal J, Delacourte A, Hirling H, Clarke PG, Luthi-Carter R, Calpain hydrolysis of alpha- and beta2-adaptins decreases clathrin-dependent endocytosis and may promote neurodegeneration. J Biol Chem. 2009 May 1;284(18):12447-58. doi: 10.1074/jbc.M804740200. Epub 2009
Cleavage sites (±10aa)
[Site 1] FIPSSVPATF691-APSPTPAVVS
Phe691  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe682 | Ile683 | Pro684 | Ser685 | Ser686 | Val687 | Pro688 | Ala689 | Thr690 | Phe691 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala692 | Pro693 | Ser694 | Pro695 | Thr696 | Pro697 | Ala698 | Val699 | Val700 | Ser701 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 17022975] Tokumitsu H, Hatano N, Yokokura S, Sueyoshi Y, Nozaki N, Kobayashi R, Phosphorylation of Numb regulates its interaction with the clathrin-associated adaptor AP-2. FEBS Lett. 2006 Oct 16;580(24):5797-801. Epub 2006 Sep 27.