SB0111 : DNA topoisomerase I
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | DNA topoisomerase 1 [Homo sapiens]; DNA topoisomerase 1; type I DNA topoisomerase; 5.99.1.2; DNA topoisomerase I |
| Gene Names | TOP1; topoisomerase (DNA) I |
| Gene Locus | 20q12-q13.1; chromosome 20 |
| GO Function | Not available |
* Information From OMIM
Description: DNA topoisomerases I and II (OMIM:126430) catalyze the breaking and rejoining of DNA strands in a way that allows the strands to pass through one another, thus altering the topology of DNA. Type I topoisomerases (EC 5.99.1.2) break a single DNA strand, whereas the type II enzymes break 2 strands of duplex DNA. Several lines of evidence suggest that topoisomerase I normally functions during transcription (summary by D'Arpa et al., 1988).
* Structure Information
1. Primary Information
Length: 765 aa
Average Mass: 90.725 kDa
Monoisotopic Mass: 90.669 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
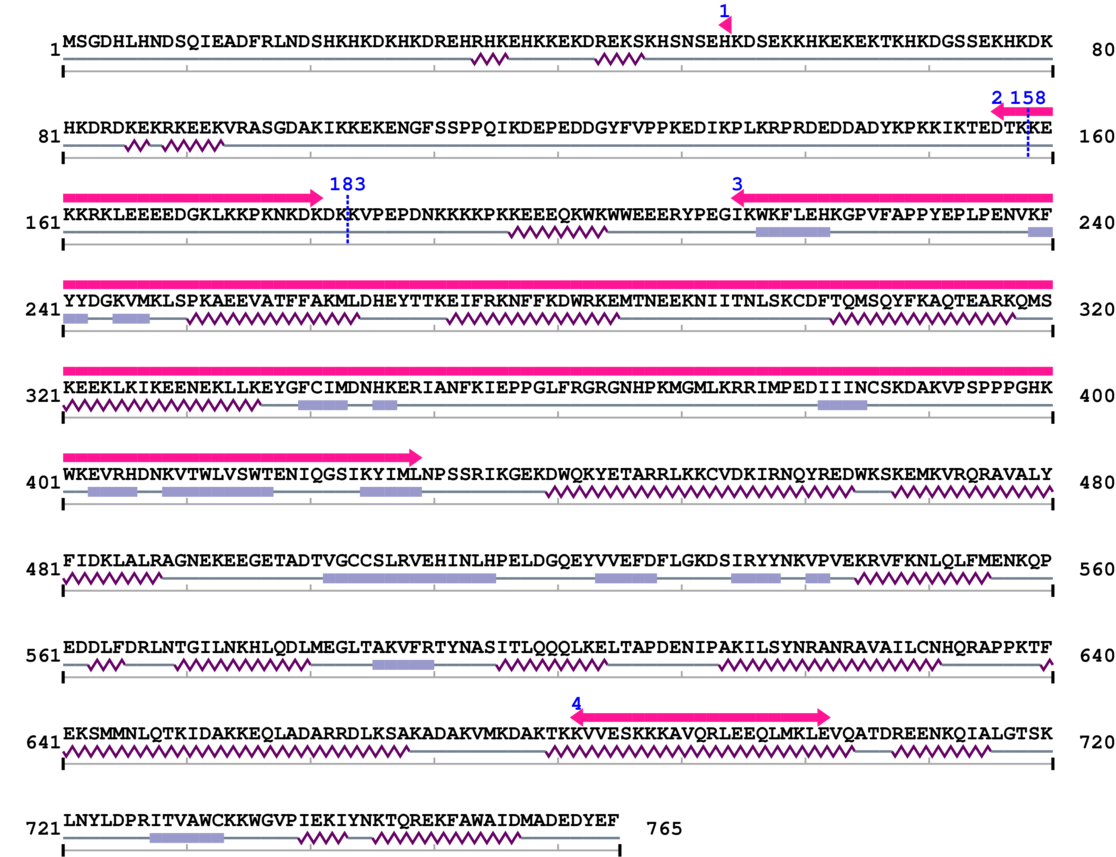
| Eukaryotic DNA topoisomerase I, catalytic core 1. | 54 | 54 | 197.0 | 5.4 |
| Eukaryotic DNA topoisomerase I, DNA binding fragment 1. | 156 | 181 | 94.0 | 13.1 |
| --- cleavage 158 (inside Eukaryotic DNA topoisomerase I, DNA binding fragment 156..181) --- | ||||
| --- cleavage 183 --- | ||||
| Eukaryotic DNA topoisomerase I, DNA binding fragment 2. | 215 | 429 | 1.0 | 8.4 |
| Eukaryotic DNA topoisomerase I, catalytic core 2. | 682 | 702 | 200.0 | 0.7 |
3. Sequence Information
Fasta Sequence: SB0111.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1A31 (X-ray; 210 A; A=175-765), 1A35 (X-ray; 250 A; A=175-765), 1A36 (X-ray; 280 A; A=175-765), 1EJ9 (X-ray; 260 A; A=203-765), 1K4S (X-ray; 320 A; A=174-765), 1K4T (X-ray; 210 A; A=174-765), 1LPQ (X-ray; 314 A; A=202-765), 1NH3 (X-ray; 310 A; A=203-765), 1R49 (X-ray; 313 A; A=174-765), 1RR8 (X-ray; 260 A; C=203-765), 1RRJ (X-ray; 230 A; A=201-765), 1SC7 (X-ray; 300 A; A=174-765), 1SEU (X-ray; 300 A; A=174-765), 1T8I (X-ray; 300 A; A=174-765), 1TL8 (X-ray; 310 A; A=174-765)
* Cleavage Information
2 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 21086148] Chou SM, Huang TH, Chen HC, Li TK, Calcium-induced cleavage of DNA topoisomerase I involves the cytoplasmic-nuclear shuttling of calpain 2. Cell Mol Life Sci. 2011 Aug;68(16):2769-84. doi: 10.1007/s00018-010-0591-4. Epub
Cleavage sites (±10aa)
[Site 1] PKKIKTEDTK158-KEKKRKLEEE
Lys158  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro149 | Lys150 | Lys151 | Ile152 | Lys153 | Thr154 | Glu155 | Asp156 | Thr157 | Lys158 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys159 | Glu160 | Lys161 | Lys162 | Arg163 | Lys164 | Leu165 | Glu166 | Glu167 | Glu168 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
[Site 2] KKPKNKDKDK183-KVPEPDNKKK
Lys183  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys174 | Lys175 | Pro176 | Lys177 | Asn178 | Lys179 | Asp180 | Lys181 | Asp182 | Lys183 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys184 | Val185 | Pro186 | Glu187 | Pro188 | Asp189 | Asn190 | Lys191 | Lys192 | Lys193 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24096022] D'Annessa I, Tesauro C, Wang Z, Arno B, Zuccaro L, Fiorani P, Desideri A, The human topoisomerase 1B Arg634Ala mutation results in camptothecin resistance and loss of inter-domain motion correlation. Biochim Biophys Acta. 2013 Dec;1834(12):2712-21. doi: