SB0112 : Survival motor neuron protein 1, SMN1
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | survival motor neuron protein isoform d [Homo sapiens]; survival motor neuron protein isoform d; gemin-1; component of gems 1; survival motor neuron 1 protein; tudor domain containing 16A; Survival motor neuron protein; Component of gems 1; Gemin-1 |
| Gene Names | SMN1; SMNC; SMN; SMNT; survival of motor neuron 1, telomeric |
| Gene Locus | 5q13.2; chromosome 5 |
| GO Function | Not available |
* Information From OMIM
Description: The SMN1 and SMN2 (OMIM:601627) genes lie within the telomeric and centromeric halves, respectively, of a large, inverted duplication on chromosome 5q13. These genes share more than 99% nucleotide identity, and both are capable of encoding a 294-amino acid RNA-binding protein, SMN, that is required for efficient assembly of small nuclear ribonucleoprotein (snRNP) complexes. Homozygous loss of the SMN1 gene causes spinal muscular atrophy (SMA; OMIM:253300). Absence of SMN1 is partially compensated for by SMN2, which produces enough SMN protein to allow for relatively normal development in cell types other than motor neurons. However, SMN2 cannot fully compensate for loss of SMN1 because, although SMN2 is transcribed at a level comparable to that of SMN1, a large majority of SMN2 transcripts lack exon 7, resulting in production of a truncated, less stable SMN protein (Lefebvre et al., 1995; Kashima et al., 2007). A small proportion of SMN1 associates with polyribosomes and represses translation of target mRNAs (Sanchez et al., 2013).
* Structure Information
1. Primary Information
Length: 294 aa
Average Mass: 31.848 kDa
Monoisotopic Mass: 31.828 kDa
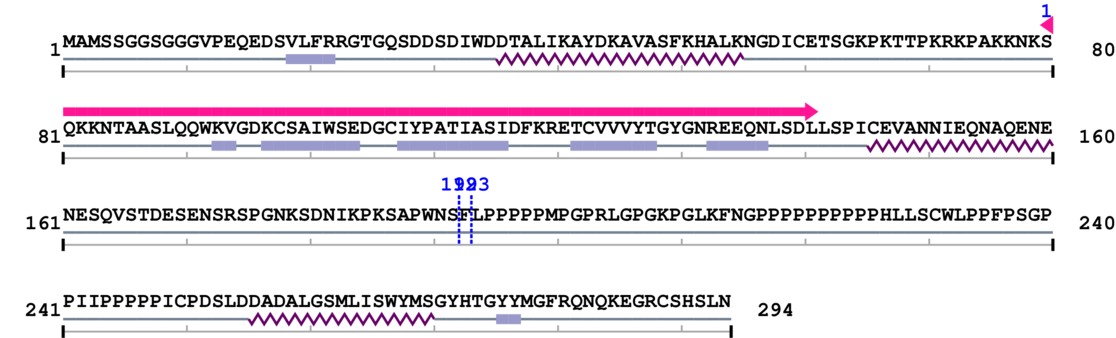
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Tudor domain 1. | 80 | 141 | 43.0 | 0.1 |
3. Sequence Information
Fasta Sequence: SB0112.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1G5V (NMR; -; A=82-169), 1MHN (X-ray; 180 A; A=89-147), 2LEH (NMR; -; B=26-51), 3S6N (X-ray; 250 A; M=26-62), 4A4E (NMR; -; A=84-147), 4A4G (NMR; -; A=84-147), 4GLI (X-ray; 190 A; A=263-294), 4NL6 (X-ray; 550 A; A/B/C=1-294), 4NL7 (X-ray; 300 A; A=11-278), 4QQ6 (X-ray; 175 A; A=82-147)
* Cleavage Information
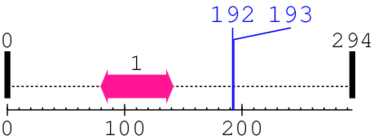
2 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 21209906] Fuentes JL, Strayer MS, Matera AG, Molecular determinants of survival motor neuron (SMN) protein cleavage by the calcium-activated protease, calpain. PLoS One. 2010 Dec 30;5(12):e15769. doi: 10.1371/journal.pone.0015769.
Cleavage sites (±10aa)
[Site 1] IKPKSAPWNS192-FLPPPPPMPG
Ser192  Phe
Phe
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile183 | Lys184 | Pro185 | Lys186 | Ser187 | Ala188 | Pro189 | Trp190 | Asn191 | Ser192 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Phe193 | Leu194 | Pro195 | Pro196 | Pro197 | Pro198 | Pro199 | Met200 | Pro201 | Gly202 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
[Site 2] KPKSAPWNSF193-LPPPPPMPGP
Phe193  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys184 | Pro185 | Lys186 | Ser187 | Ala188 | Pro189 | Trp190 | Asn191 | Ser192 | Phe193 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu194 | Pro195 | Pro196 | Pro197 | Pro198 | Pro199 | Met200 | Pro201 | Gly202 | Pro203 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24711022] Zeng G, Zheng H, Cheng J, Chen R, Lin H, Yang J, Zhang D, [Analysis and carrier screening for copy numbers of SMN and NAIP genes in children with spinal muscular atrophy]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2014 Apr;31(2):152-5. doi: