* Structure Information
1. Primary Information
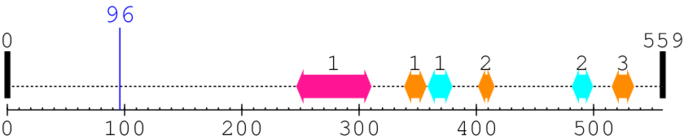
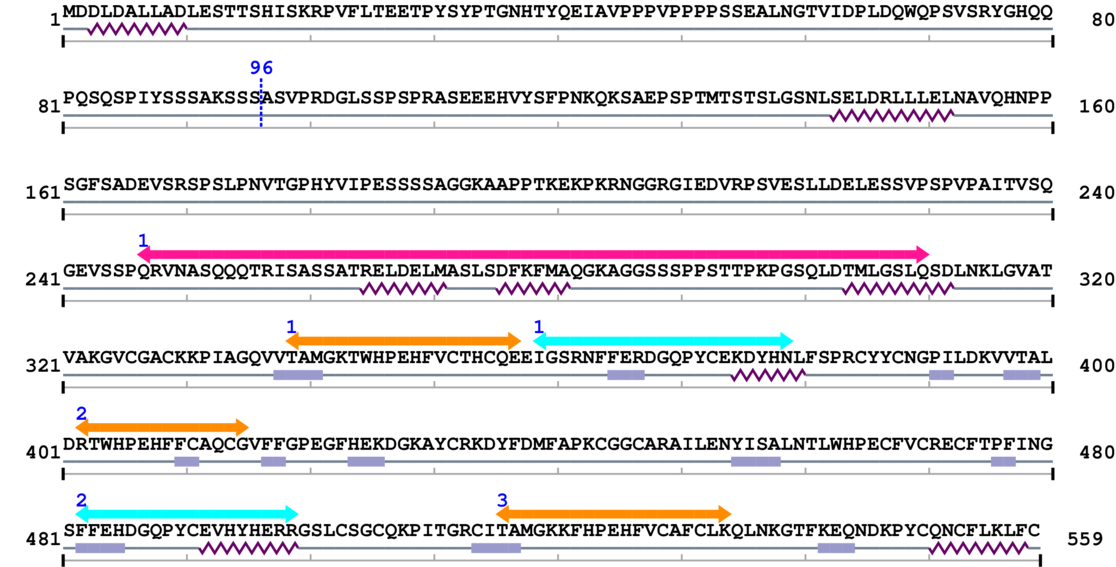
Length: 559 aa
Average Mass: 61.242 kDa
Monoisotopic Mass: 61.203 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
| --- cleavage 96 --- |
| EAP30/Vps36 family 1. | 247 | 310 | 8.0 | 0.0 |
| Ribosomal protein L37e 1. | 339 | 357 | 5.0 | 0.1 |
| Protein of unknown function (DUF1797) 1. | 359 | 379 | 16.0 | 0.0 |
| Ribosomal protein L37e 2. | 402 | 415 | 25.0 | 0.0 |
| Protein of unknown function (DUF1797) 2. | 482 | 499 | 20.0 | 0.0 |
| Ribosomal protein L37e 3. | 516 | 534 | 5.0 | 1.7 |
3. Sequence Information
Fasta Sequence: SB0113.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB:
2L6F (NMR; -; A=140-161, A=262-275), 2L6G (NMR; -; A=140-161), 2L6H (NMR; -; A=263-276), 4R32 (X-ray; 350 A; B/C=139-162)
* Cleavage Information
1 [sites] cleaved by Calpain 1 or 2
Source Reference: [PubMed ID: 21270128] Cortesio CL, Boateng LR, Piazza TM, Bennin DA, Huttenlocher A, Calpain-mediated proteolysis of paxillin negatively regulates focal adhesion dynamics and cell migration. J Biol Chem. 2011 Mar 25;286(12):9998-10006. doi: 10.1074/jbc.M110.187294. Epub
Cleavage sites (±10aa)
[Site 1] IYSSSAKSSS96-ASVPRDGLSS
Ser96  Ala
Ala
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| Ile87 | Tyr88 | Ser89 | Ser90 | Ser91 | Ala92 | Lys93 | Ser94 | Ser95 | Ser96 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Ala97 | Ser98 | Val99 | Pro100 | Arg101 | Asp102 | Gly103 | Leu104 | Ser105 | Ser106 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
DQWQPSVSRYGHQQPQSQSPIYSSSAKSSSASVPRDGLSSPSPRASEEEHVYSFPNKQKS
|
Summary
| # |
organism |
max score |
hits |
top seq |
| 1 |
Gallus gallus |
119.00 |
1 |
paxillin |
| 2 |
Pongo pygmaeus |
91.30 |
1 |
PAXI_PONPY Paxillin gi |
| 3 |
Canis familiaris |
89.40 |
1 |
PREDICTED: similar to Paxillin |
| 4 |
Homo sapiens |
88.20 |
4 |
paxillin gi |
| 5 |
Mus musculus |
72.40 |
5 |
paxillin isoform alpha |
| 6 |
Rattus norvegicus |
61.20 |
1 |
paxillin |
| 7 |
Xenopus laevis |
37.00 |
2 |
paxillin |
| 8 |
Tetraodon nigroviridis |
35.80 |
1 |
unnamed protein product |
| 9 |
Apis mellifera |
32.00 |
1 |
PREDICTED: similar to CG32226-PA |
Top-ranked sequences
| organism | matching |
|---|
| Gallus gallus |
Query 67 DQWQPSVSRYGHQQPQSQSPIYSSSAKSSS#ASVPRDGLSSPSPRASEEEHVYSFPNKQKS 126
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 67 DQWQPSVSRYGHQQPQSQSPIYSSSAKSSS#ASVPRDGLSSPSPRASEEEHVYSFPNKQKS 126
|
| Pongo pygmaeus |
Query 67 DQWQPSVSRYGHQQPQSQSPIYSSSAKSSS#ASVPRDGLSSPSPRASEEEHVYSFPNKQKS 126
|||||| ||+ |||||| ||+| ||||+||# | |+||+ || | ||||||||||||||
Sbjct 67 DQWQPSGSRFIHQQPQSSSPVYGSSAKTSS#VSNPQDGVGSPCSRVGEEEHVYSFPNKQKS 126
|
| Canis familiaris |
Query 67 DQWQPSVSRYGHQQPQSQSPIYSSSAKSSS#ASVPRDGLSSPSPRASEEEHVYSFPNKQKS 126
|||||| ||+ ||||| ||+|||| |+||#| ||+| | ||| ||||||||||||||
Sbjct 78 DQWQPSASRFTHQQPQPPSPVYSSSTKTSS#APVPQDSSGPPCPRAGEEEHVYSFPNKQKS 137
|
| Homo sapiens |
Query 67 DQWQPSVSRYGHQQPQSQSPIYSSSAKSSS#ASVPRDGLSSPSPRASEEEHVYSFPNKQKS 126
|||||| ||+ |||||| ||+| ||||+||# | |+| + || | ||||||||||||||
Sbjct 67 DQWQPSGSRFIHQQPQSSSPVYGSSAKTSS#VSNPQDSVGSPCSRVGEEEHVYSFPNKQKS 126
|
| Mus musculus |
Query 67 DQWQPSVSRYGHQQPQSQSPIYSSSAKSSS#ASVPRDGLSSPSPRASEEEHVYSFPNKQKS 126
|||||| ||| |||| | |+||||||+||#|| +||+ | || ||||||||||||||
Sbjct 67 DQWQPSGSRYAHQQPPSPLPVYSSSAKNSS#ASNTQDGVGSLCSRAGEEEHVYSFPNKQKS 126
|
| Rattus norvegicus |
Query 67 DQWQPSVSRYGHQQPQSQSPIYSSSAKSSS#ASVPRDGLSSPSPRASEEEHVYSFPNKQKS 126
|||||| ||| |||| | ||||||| |+||#|| |+| + | || ||||||||||||||
Sbjct 67 DQWQPSGSRYAHQQPPSPSPIYSSSTKNSS#ASNPQDSVGSLCSRAGEEEHVYSFPNKQKS 126
|
| Xenopus laevis |
Query 82 QSQSPIYSSSAKSSS#ASVPRDGLSSPSPRASEEEHVYSFPNKQKS 126
| ++|+| || # | || ||||||||||||+|
Sbjct 72 QQENPLYCSSGAKDI#---------SGSPSFGEEEHVYSFPNKQRS 107
|
| Tetraodon nigroviridis |
Query 105 SSPSPRASEEEHVYSFPNKQKS 126
|||| ||+|||||||||||
Sbjct 45 SSPSQPVCEEDHVYSFPNKQKS 66
|
| Apis mellifera |
Query 73 VSRYGHQQPQSQSPIYSSSAKSSS#ASVPRDGLSSPSPRASEEEHV 117
|| | +| | +|+| + |+ # ++ | |+| | ||+
Sbjct 1133 VSLYATDKPGSDTPVYIWAVASNG#EALFRRGVSESCPMGVSWEHI 1177
|
* References
[PubMed ID: 18092823] Bertolucci CM, Guibao CD, Zheng JJ, Phosphorylation of paxillin LD4 destabilizes helix formation and inhibits binding to focal adhesion kinase. Biochemistry. 2008 Jan 15;47(2):548-54. Epub 2007 Dec 20.
» More (6) « Hide Below
[PubMed ID: 18171471] ... Scheswohl DM, Harrell JR, Rajfur Z, Gao G, Campbell SL, Schaller MD, Multiple paxillin binding sites regulate FAK function. J Mol Signal. 2008 Jan 2;3:1. doi: 10.1186/1750-2187-3-1.
[PubMed ID: 16717130] ... Nayal A, Webb DJ, Brown CM, Schaefer EM, Vicente-Manzanares M, Horwitz AR, Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J Cell Biol. 2006 May 22;173(4):587-9.
[PubMed ID: 7615549] ... Bellis SL, Miller JT, Turner CE, Characterization of tyrosine phosphorylation of paxillin in vitro by focal adhesion kinase. J Biol Chem. 1995 Jul 21;270(29):17437-41.
[PubMed ID: 7534286] ... Salgia R, Li JL, Lo SH, Brunkhorst B, Kansas GS, Sobhany ES, Sun Y, Pisick E, Hallek M, Ernst T, et al., Molecular cloning of human paxillin, a focal adhesion protein phosphorylated by P210BCR/ABL. J Biol Chem. 1995 Mar 10;270(10):5039-47.
[PubMed ID: 7525621] ... Turner CE, Miller JT, Primary sequence of paxillin contains putative SH2 and SH3 domain binding motifs and multiple LIM domains: identification of a vinculin and pp125Fak-binding region. J Cell Sci. 1994 Jun;107 ( Pt 6):1583-91.
[PubMed ID: 2118142] ... Turner CE, Glenney JR Jr, Burridge K, Paxillin: a new vinculin-binding protein present in focal adhesions. J Cell Biol. 1990 Sep;111(3):1059-68.
 Ala
Ala

 Sequence conservation (by blast)
Sequence conservation (by blast)