SB0118 : Rad21
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | double-strand-break repair protein rad21 homolog [Homo sapiens]; double-strand-break repair protein rad21 homolog; protein involved in DNA double-strand break repair; nuclear matrix protein 1; NXP-1; SCC1 homolog; sister chromatid cohesion 1; Double-strand-break repair protein rad21 homolog; hHR21; Nuclear matrix protein 1 |
| Gene Names | RAD21; HR21, KIAA0078, NXP1; HR21; KIAA0078; NXP1; RAD21 homolog (S. pombe) |
| Gene Locus | 8q24; chromosome 8 |
| GO Function | Not available |
* Information From OMIM
Description: Eukaryotic sister chromatids remain connected from the time of synthesis until they are separated in anaphase. This cohesion depends on a complex of proteins known as cohesins. In vertebrates, unlike in yeast, the cohesins dissociate from chromosome arms earlier in M phase, during prophase. Small amounts of cohesin remain near the centromere until metaphase, with complete removal at the beginning of anaphase. Cohesin complexes contain SCC1 (RAD21), SMC1 (OMIM:300040), SMC3 (CSPG6; OMIM:606062), and either SA1 (STAG1; OMIM:604358) or SA2 (STAG2; OMIM:300826). The complexes, in turn, interact with PDS5 (see OMIM:613200), a protein implicated in chromosome cohesion, condensation, and recombination in yeast (summary by Sumara et al., 2000).
* Structure Information
1. Primary Information
Length: 631 aa
Average Mass: 71.689 kDa
Monoisotopic Mass: 71.645 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Conserved region of Rad21 / Rec8 like protein 1. | 1 | 12 | 23.0 | 0.0 |
| --- cleavage 192 --- | ||||
| DNA polymerase phi 1. | 526 | 577 | 646.0 | 6.4 |
3. Sequence Information
Fasta Sequence: SB0118.fasta
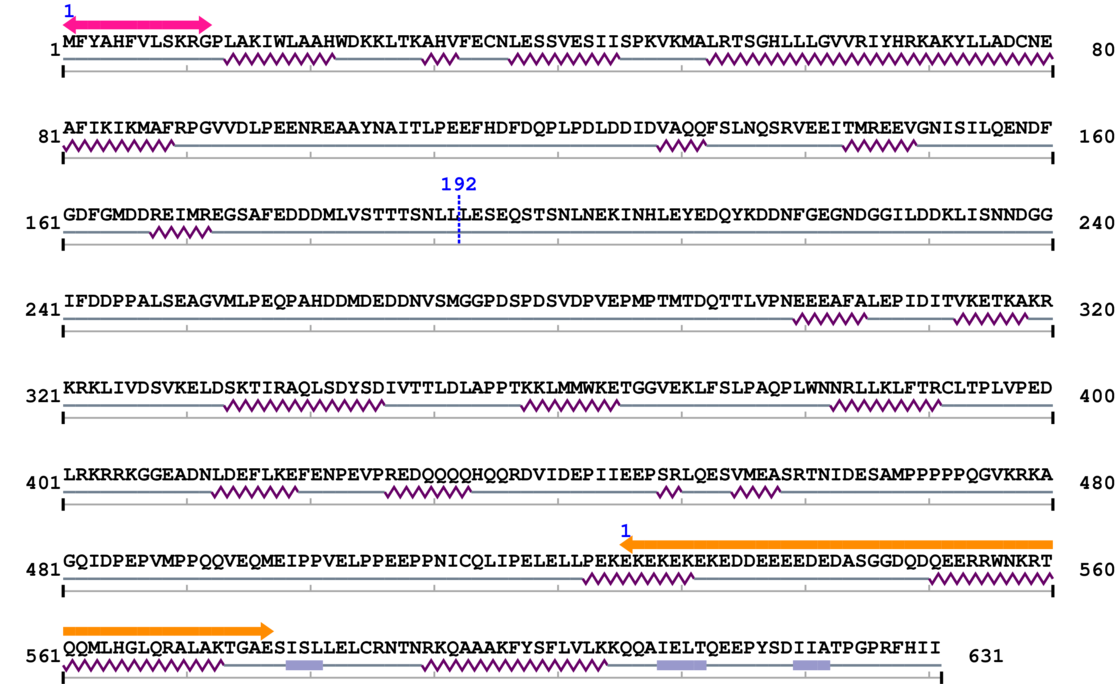
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
1 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 21876002] Panigrahi AK, Zhang N, Mao Q, Pati D, Calpain-1 cleaves Rad21 to promote sister chromatid separation. Mol Cell Biol. 2011 Nov;31(21):4335-47. doi: 10.1128/MCB.06075-11. Epub 2011 Aug
Cleavage sites (±10aa)
[Site 1] LVSTTTSNLL192-LESEQSTSNL
Leu192  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu183 | Val184 | Ser185 | Thr186 | Thr187 | Thr188 | Ser189 | Asn190 | Leu191 | Leu192 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu193 | Glu194 | Ser195 | Glu196 | Gln197 | Ser198 | Thr199 | Ser200 | Asn201 | Leu202 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24148822] Mahmood SF, Gruel N, Chapeaublanc E, Lescure A, Jones T, Reyal F, Vincent-Salomon A, Raynal V, Pierron G, Perez F, Camonis J, Del Nery E, Delattre O, Radvanyi F, Bernard-Pierrot I, A siRNA screen identifies RAD21, EIF3H, CHRAC1 and TANC2 as driver genes within the 8q23, 8q24.3 and 17q23 amplicons in breast cancer with effects on cell growth, survival and transformation. Carcinogenesis. 2014 Mar;35(3):670-82. doi: 10.1093/carcin/bgt351. Epub 2013 Oct