SB0119 : Caspase-3
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | caspase-3 preproprotein [Homo sapiens]; caspase-3 preproprotein; caspase 3, apoptosis-related cysteine protease; PARP cleavage protease; cysteine protease CPP32; SREBP cleavage activity 1; caspase-3; procaspase3; apopain; CASP-3; CPP-32; protein Yama; Caspase-3; 3.4.22.56; Apopain; Cysteine protease CPP32; Protein Yama; SCA-1; Caspase-3 subunit p17; Caspase-3 subunit p12 |
| Gene Names | CASP3; CPP32; caspase 3, apoptosis-related cysteine peptidase |
| Gene Locus | 4q34; chromosome 4 |
| GO Function | Not available |
* Information From OMIM
Description: Cysteinyl aspartate-specific proteases, or caspases, such as CASP3, cleave substrates directly after an aspartic acid residue and play essential roles in programmed cell death. Caspases are synthesized in a dormant form with an N-terminal prodomain followed by a large subunit and a small subunit. Proteolytic processing releases the caspase large and small subunits, resulting in activation (summary by Parker et al., 2010).
* Structure Information
1. Primary Information
Length: 277 aa
Average Mass: 31.608 kDa
Monoisotopic Mass: 31.587 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 7 --- | ||||
| Caspase domain 1. | 45 | 275 | 1.0 | 0.1 |
3. Sequence Information
Fasta Sequence: SB0119.fasta
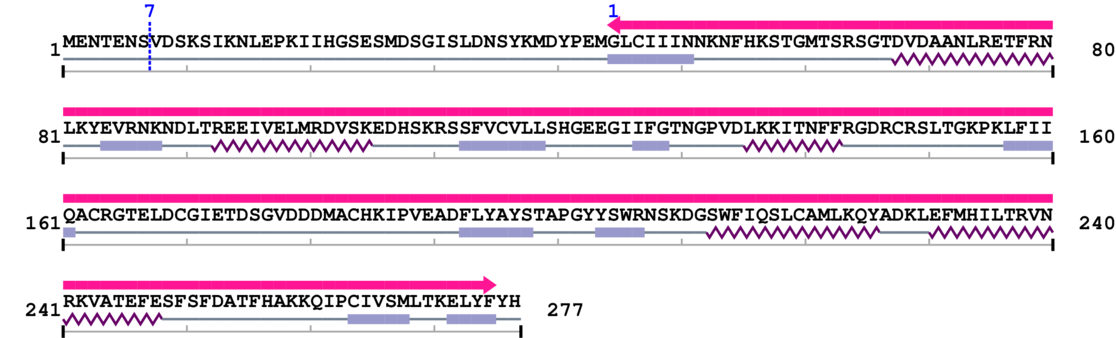
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1CP3 (X-ray; 230 A; A/B=1-277), 1GFW (X-ray; 280 A; A=29-175, B=181-277), 1I3O (X-ray; 270 A; A/C=1-175, B/D=176-277), 1NME (X-ray; 160 A; A=29-174, B=186-277), 1NMQ (X-ray; 240 A; A/B=29-277), 1NMS (X-ray; 170 A; A/B=29-277), 1PAU (X-ray; 250 A; A=29-175, B=176-277), 1QX3 (X-ray; 190 A; A=29-277), 1RE1 (X-ray; 250 A; A=29-175, B=176-277), 1RHJ (X-ray; 220 A; A/C=29-175, B/D=176-277), 1RHK (X-ray; 250 A; A=29-175, B=176-277), 1RHM (X-ray; 250 A; A/C=29-175, B/D=176-277), 1RHQ (X-ray; 300 A; A/D=29-175, B/E=176-277), 1RHR (X-ray; 300 A; A=29-175, B=176-277), 1RHU (X-ray; 251 A; A=29-175, B=176-277), 2C1E (X-ray; 177 A; A=29-175, B=176-277), 2C2K (X-ray; 187 A; A=29-175, B=176-277), 2C2M (X-ray; 194 A; A=29-175, B=176-277), 2C2O (X-ray; 245 A; A=29-175, B=176-277), 2CDR (X-ray; 170 A; A=29-175, B=176-277), 2CJX (X-ray; 170 A; A=29-175, B=176-277), 2CJY (X-ray; 167 A; A=29-175, B=176-277), 2CNK (X-ray; 175 A; A=29-175, B=176-277), 2CNL (X-ray; 167 A; A=29-175, B=176-277), 2CNN (X-ray; 170 A; A=29-175, B=176-277), 2CNO (X-ray; 195 A; A=29-175, B=176-277), 2DKO (X-ray; 106 A; A=29-174, B=175-277), 2H5I (X-ray; 169 A; A=29-174, B=184-277), 2H5J (X-ray; 200 A; A/C=29-174, B/D=184-277), 2H65 (X-ray; 230 A; A/C=29-174, B/D=184-277), 2J30 (X-ray; 140 A; A=29-277), 2J31 (X-ray; 150 A; A=29-277), 2J32 (X-ray; 130 A; A=29-277), 2J33 (X-ray; 200 A; A=29-277), 2XYG (X-ray; 154 A; A=29-174, B=185-277), 2XYH (X-ray; 189 A; A=29-174, B=185-277), 2XYP (X-ray; 186 A; A=29-174, B=185-277), 2XZD (X-ray; 210 A; A/C=27-175, B/D=176-277), 2XZT (X-ray; 270 A; A/C=29-175, B/D=176-277), 2Y0B (X-ray; 210 A; A/C=29-175, B/D=176-277), 3DEH (X-ray; 250 A; A/B/C/D=29-277), 3DEI (X-ray; 280 A; A/B/C/D=29-277), 3DEJ (X-ray; 260 A; A/B/C/D=29-277), 3DEK (X-ray; 240 A; A/B/C/D=29-277), 3EDQ (X-ray; 161 A; A/C=29-175, B/D=176-277), 3GJQ (X-ray; 260 A; A/C=29-175, B/D=176-277), 3GJR (X-ray; 220 A; A/C=29-175, B/D=176-277), 3GJS (X-ray; 190 A; A/C=29-175, B/D=176-277), 3GJT (X-ray; 220 A; A/C=29-175, B/D=176-277), 3H0E (X-ray; 200 A; A/B=29-277), 3ITN (X-ray; 163 A; A=29-277), 3KJF (X-ray; 200 A; A=29-175, B=176-277), 3PCX (X-ray; 150 A; A=29-277), 3PD0 (X-ray; 200 A; A=29-277), 3PD1 (X-ray; 162 A; A=29-277), 4DCJ (X-ray; 170 A; A/D=29-175, B/E=176-277), 4DCO (X-ray; 170 A; A/D=29-175, B/E=176-277), 4DCP (X-ray; 170 A; A/D=29-175, B/E=176-277), 4EHA (X-ray; 170 A; A/C=1-277), 4EHD (X-ray; 158 A; A=1-277), 4EHF (X-ray; 166 A; A=1-277), 4EHH (X-ray; 178 A; A=1-277), 4EHK (X-ray; 167 A; A/C=1-277), 4EHL (X-ray; 180 A; A/C=1-277), 4EHN (X-ray; 169 A; A=1-277), 4JJE (X-ray; 148 A; A=29-277), 4JQY (X-ray; 250 A; A/B=34-277), 4JQZ (X-ray; 289 A; A/B=34-277), 4JR0 (X-ray; 180 A; A/B=34-277), 4PRY (X-ray; 170 A; A=1-277), 4PS0 (X-ray; 163 A; A/B=1-277), 4QTX (X-ray; 197 A; A=1-277), 4QTY (X-ray; 160 A; A=29-277), 4QU0 (X-ray; 195 A; A=1-277), 4QU5 (X-ray; 191 A; A=1-277), 4QU8 (X-ray; 172 A; A=1-277), 4QU9 (X-ray; 156 A; A=1-277), 4QUA (X-ray; 189 A; A=1-277), 4QUB (X-ray; 169 A; A=1-277), 4QUD (X-ray; 200 A; A/B=1-277), 4QUE (X-ray; 184 A; A/C=1-277), 4QUG (X-ray; 192 A; A/C=1-277), 4QUH (X-ray; 176 A; A/C=1-277), 4QUI (X-ray; 176 A; A/B=1-277), 4QUJ (X-ray; 150 A; A=1-277), 4QUL (X-ray; 190 A; A/C=1-277)
* Cleavage Information
1 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 10477693] Wolf BB, Goldstein JC, Stennicke HR, Beere H, Amarante-Mendes GP, Salvesen GS, Green DR, Calpain functions in a caspase-independent manner to promote apoptosis-like events during platelet activation. Blood. 1999 Sep 1;94(5):1683-92.
Cleavage sites (±10aa)
[Site 1] MENTENS7-VDSKSIKNLE
Ser7  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | - | Met1 | Glu2 | Asn3 | Thr4 | Glu5 | Asn6 | Ser7 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val8 | Asp9 | Ser10 | Lys11 | Ser12 | Ile13 | Lys14 | Asn15 | Leu16 | Glu17 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24553140] Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR, van Lookeren Campagne M, A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014 Feb 27;506(7489):456-62. doi: 10.1038/nature13044. Epub 2014 Feb 19.