SB0120 : Caspase-9
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | caspase-9 isoform alpha precursor [Homo sapiens]; caspase-9 isoform alpha precursor; apoptotic protease MCH-6; apoptotic protease activating factor 3; ICE-like apoptotic protease 6; protein phosphatase 1, regulatory subunit 56; Caspase-9; CASP-9; 3.4.22.62; Apoptotic protease Mch-6; Apoptotic protease-activating factor 3; APAF-3; ICE-LAP6; Caspase-9 subunit p35; Caspase-9 subunit p10 |
| Gene Names | CASP9; MCH6; caspase 9, apoptosis-related cysteine peptidase |
| Gene Locus | 1p36.21; chromosome 1 |
| GO Function | Not available |
* Information From OMIM
Function: Li et al. (1997) determined that caspase-9 and APAF1 (OMIM:602233) bind to each other via their respective NH2-terminal CED-3 homologous domains in the presence of cytochrome c (OMIM:123970) and dATP, an event that leads to caspase-9 activation. Activated caspase-9 in turn cleaves and activates caspase-3. Depletion of caspase-9 from S-100 extracts diminished caspase-3 activation. Mutation of the active site of caspase-9 (cys287 to ala) attenuated the activation of caspase-3 and cellular apoptotic response in vivo, indicating that caspase-9 is the most upstream member of the apoptotic protease cascade that is triggered by cytochrome c and dATP.
* Structure Information
1. Primary Information
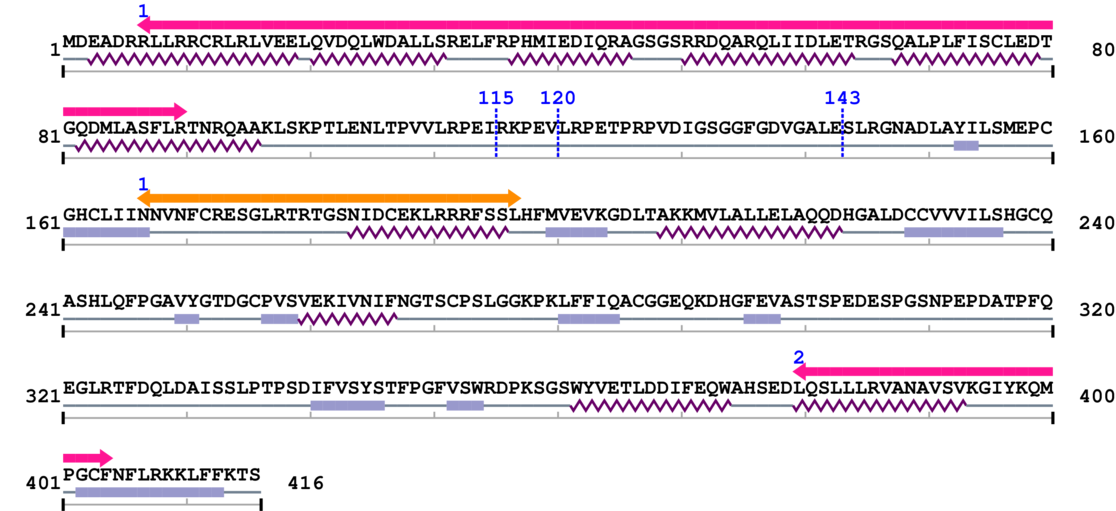
Length: 416 aa
Average Mass: 46.280 kDa
Monoisotopic Mass: 46.251 kDa
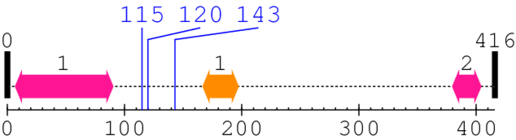
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Caspase recruitment domain 1. | 7 | 90 | 2.0 | 0.4 |
| --- cleavage 115 --- | ||||
| --- cleavage 120 --- | ||||
| --- cleavage 143 --- | ||||
| Anti-Sigma Factor A 1. | 167 | 197 | 31.0 | 0.0 |
| Caspase recruitment domain 2. | 380 | 404 | 9.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0120.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
3 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 10477693] Wolf BB, Goldstein JC, Stennicke HR, Beere H, Amarante-Mendes GP, Salvesen GS, Green DR, Calpain functions in a caspase-independent manner to promote apoptosis-like events during platelet activation. Blood. 1999 Sep 1;94(5):1683-92.
Cleavage sites (±10aa)
[Site 1] LTPVVLRPEI115-RKPEVLRPET
Ile115  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu106 | Thr107 | Pro108 | Val109 | Val110 | Leu111 | Arg112 | Pro113 | Glu114 | Ile115 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg116 | Lys117 | Pro118 | Glu119 | Val120 | Leu121 | Arg122 | Pro123 | Glu124 | Thr125 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
[Site 2] LRPEIRKPEV120-LRPETPRPVD
Val120  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu111 | Arg112 | Pro113 | Glu114 | Ile115 | Arg116 | Lys117 | Pro118 | Glu119 | Val120 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu121 | Arg122 | Pro123 | Glu124 | Thr125 | Pro126 | Arg127 | Pro128 | Val129 | Asp130 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
[Site 3] GGFGDVGALE143-SLRGNADLAY
Glu143  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly134 | Gly135 | Phe136 | Gly137 | Asp138 | Val139 | Gly140 | Ala141 | Leu142 | Glu143 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser144 | Leu145 | Arg146 | Gly147 | Asn148 | Ala149 | Asp150 | Leu151 | Ala152 | Tyr153 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24424093] Sharoar MG, Islam MI, Shahnawaz M, Shin SY, Park IS, Amyloid beta binds procaspase-9 to inhibit assembly of Apaf-1 apoptosome and intrinsic apoptosis pathway. Biochim Biophys Acta. 2014 Apr;1843(4):685-93. doi: 10.1016/j.bbamcr.2014.01.008.