SB0123 : Neuronal calcium sensor-1, NCS-1
[ CaMP Format ]
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | neuronal calcium sensor 1 [Rattus norvegicus]; neuronal calcium sensor 1; NCS-1; frequenin-like protein; frequenin-like ubiquitous protein; Neuronal calcium sensor 1; Frequenin homolog; Frequenin-like protein; Frequenin-like ubiquitous protein |
| Gene Names | Ncs1; Flup, Freq; Flup; Freq; neuronal calcium sensor 1 |
| Gene Locus | 3p12; chromosome 3 |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 190 aa
Average Mass: 21.878 kDa
Monoisotopic Mass: 21.865 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| EF-hand domain 1. | 23 | 51 | 33.0 | 0.0 |
| --- cleavage 36 (inside EF-hand domain 23..51) --- | ||||
| Secreted protein acidic and rich in cysteine Ca binding region 1. | 68 | 86 | 93.0 | 0.1 |
| Caleosin related protein 1. | 146 | 179 | 92.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0123.fasta
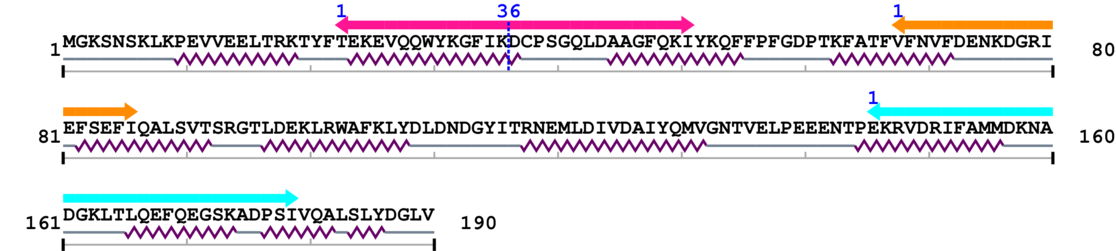
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
1 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 19732951] Blachford C, Celic A, Petri ET, Ehrlich BE, Discrete proteolysis of neuronal calcium sensor-1 (NCS-1) by mu-calpain disrupts calcium binding. Cell Calcium. 2009 Oct;46(4):257-62. doi: 10.1016/j.ceca.2009.08.002. Epub 2009
Cleavage sites (±10aa)
[Site 1] VQQWYKGFIK36-DCPSGQLDAA
Lys36  Asp
Asp
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Val27 | Gln28 | Gln29 | Trp30 | Tyr31 | Lys32 | Gly33 | Phe34 | Ile35 | Lys36 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asp37 | Cys38 | Pro39 | Ser40 | Gly41 | Gln42 | Leu43 | Asp44 | Ala45 | Ala46 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KLKPEVVEELTRKTYFTEKEVQQWYKGFIKDCPSGQLDAAGFQKIYKQFFPFGDPTKFAT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 127.00 | 12 | frequenin homolog |
| 2 | Pongo pygmaeus | 127.00 | 5 | NCS1_PONPY Neuronal calcium sensor 1 (NCS-1) (Fre |
| 3 | Bos taurus | 127.00 | 4 | frequenin |
| 4 | synthetic construct | 127.00 | 2 | frequenin-like |
| 5 | Xenopus tropicalis | 127.00 | 2 | hypothetical protein LOC394570 |
| 6 | synthetic construct | 127.00 | 1 | frequenin homolog (Drosophila) |
| 7 | Homo sapiens | 126.00 | 19 | AF186409_2 frequenin |
| 8 | Xenopus laevis | 125.00 | 5 | frequenin |
| 9 | Danio rerio | 125.00 | 2 | neuronal calcium sensor 1b |
| 10 | Tetraodon nigroviridis | 124.00 | 8 | unnamed protein product |
| 11 | Danio rerio | 121.00 | 7 | neuronal calcium sensor 1 |
| 12 | N/A | 93.20 | 7 | APLC_APLCA Aplycalcin gi |
| 13 | Caenorhabditis elegans | 92.40 | 2 | Neuronal Calcium Sensor family member (ncs-1) |
| 14 | Caenorhabditis briggsae AF16 | 92.40 | 2 | Hypothetical protein CBG08082 |
| 15 | Aspergillus fumigatus Af293 | 90.50 | 1 | calcium sensor (NCS-1) |
| 16 | Apis mellifera | 90.10 | 2 | PREDICTED: similar to CG5907-PA, isoform A |
| 17 | Drosophila melanogaster | 89.40 | 3 | Frequenin 2 CG5907-PA, isoform A |
| 18 | Aspergillus oryzae RIB40 | 87.80 | 1 | hypothetical protein |
| 19 | Neurospora crassa OR74A | 87.40 | 1 | neuronal calcium sensor 1 |
| 20 | Aspergillus nidulans FGSC A4 | 87.40 | 1 | hypothetical protein AN5341.2 |
| 21 | Yarrowia lipolytica | 87.00 | 1 | hypothetical protein |
| 22 | Drosophila melanogaster | 86.70 | 1 | Neurocalcin CG7641-PA |
| 23 | Schizosaccharomyces pombe 972h- | 86.70 | 1 | related to neuronal calcium sensor Ncs1 |
| 24 | Magnaporthe grisea 70-15 | 85.50 | 1 | calcium sensor NCS-1 |
| 25 | Magnaporthe grisea | 85.50 | 1 | NCS homolog |
| 26 | Aedes aegypti | 85.50 | 1 | hippocalcin |
| 27 | Gallus gallus | 84.70 | 3 | neurocalcin |
| 28 | Rattus norvegicus | 84.30 | 5 | visinin-like 1 |
| 29 | Ustilago maydis 521 | 84.30 | 1 | hypothetical protein UM00520.1 |
| 30 | Macaca fascicularis | 84.30 | 1 | hypothetical protein |
| 31 | Gibberella zeae PH-1 | 83.60 | 1 | hypothetical protein FG09591.1 |
| 32 | Homo sapiens | 83.20 | 2 | hippocalcin-like protein 4 |
| 33 | Chaetomium globosum CBS 148.51 | 83.20 | 1 | hypothetical protein CHGG_04888 |
| 34 | Canis familiaris | 82.80 | 5 | PREDICTED: similar to Hippocalcin-like protein 4 |
| 35 | Xenopus tropicalis | 81.60 | 1 | hippocalcin |
| 36 | Candida albicans SC5314 | 80.50 | 1 | hypothetical protein CaO19_4726 |
| 37 | Procambarus clarkii | 79.00 | 1 | frequenin |
| 38 | Cryptococcus neoformans var. neoformans B-3501A | 78.60 | 1 | hypothetical protein CNBG2650 |
| 39 | Ashbya gossypii ATCC 10895 | 76.30 | 1 | ACL163Wp |
| 40 | Panulirus interruptus | 73.60 | 1 | frequenin |
| 41 | cattle, brain, Peptide Partial, 165 aa | 68.90 | 1 | neurocalcin beta=calcium-binding protein |
| 42 | Schistosoma japonicum | 66.60 | 1 | SJCHGC06847 protein |
| 43 | Branchiostoma floridae | 65.90 | 1 | frequenin-like |
| 44 | Ornithorhynchus anatinus | 65.50 | 1 | PREDICTED: similar to visinin-like 1, partial |
| 45 | Canis lupus familiaris | 64.70 | 1 | recoverin |
| 46 | Strongylocentrotus purpuratus | 64.70 | 1 | PREDICTED: similar to ENSANGP00000015900 |
| 47 | Mustela putorius furo | 63.90 | 2 | AF454386_1 Kv channel interacting protein 2a |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 |
| Pongo pygmaeus | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 |
| Bos taurus | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 |
| synthetic construct | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 |
| Xenopus tropicalis | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 |
| synthetic construct | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 |
| Homo sapiens | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 |
| Xenopus laevis | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||||||||||||||||||||||||||||#||||||||| |||||||||||||||||||| Sbjct 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDATGFQKIYKQFFPFGDPTKFAT 66 |
| Danio rerio | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||+ Sbjct 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAS 66 |
| Tetraodon nigroviridis | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||||||||| ||||||||||||||||||#|||||||||||||||||||||||||||||+ Sbjct 7 KLKPEVVEELTSKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAS 66 |
| Danio rerio | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||||||+| |||||||||||||||||||#||||||||++|||||||||||||||||||| Sbjct 7 KLKPEVVEDLCRKTYFTEKEVQQWYKGFIK#DCPSGQLDSSGFQKIYKQFFPFGDPTKFAT 66 |
| N/A | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||||| |||| ++||||| |++||+||| |#||| |+| || |||+||||||||+||| Sbjct 7 KLKPEEVEELKQQTYFTEAEIKQWHKGFRK#DCPDGKLTLEGFTKIYQQFFPFGDPSKFAN 66 |
| Caenorhabditis elegans | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||| + +| +|||||||++||||||++#|||+| | |||||||||||| |||+ ||+ Sbjct 7 KLKSSQIRDLAEQTYFTEKEIKQWYKGFVR#DCPNGMLTEAGFQKIYKQFFPQGDPSDFAS 66 |
| Caenorhabditis briggsae AF16 | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||| + || +|||||||++||||||++#|||+| | |||||||||||| |||+ ||+ Sbjct 7 KLKGSQIRELAEQTYFTEKEIKQWYKGFVR#DCPNGMLTEAGFQKIYKQFFPQGDPSDFAS 66 |
| Aspergillus fumigatus Af293 | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 || | +||| | |+| +||+|||||||+|#||||| | |||||+||||||||+ || Sbjct 7 KLSPSQLEELQRATHFDKKELQQWYKGFLK#DCPSGTLTKEEFQKIYRQFFPFGDPSSFAN 66 |
| Apis mellifera | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||| + ++ || |||||||++||+|||+|#||| | | || |||||||| |||+|||+ Sbjct 7 KLKQDTIDRLTSDTYFTEKEIRQWHKGFLK#DCPDGLLTEQGFIKIYKQFFPHGDPSKFAS 66 |
| Drosophila melanogaster | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 ||| + ++ || |||||||++||+|||+|#|||+| | || |||||||| |||+|||+ Sbjct 7 KLKQDTIDRLTTDTYFTEKEIRQWHKGFLK#DCPNGLLTEQGFIKIYKQFFPDGDPSKFAS 66 |
| Aspergillus oryzae RIB40 | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 || | ++|| + |+| +||+|||||||+|#||||| | |||||+||||||||+ || Sbjct 7 KLSPSQLDELQKATHFDKKELQQWYKGFLK#DCPSGTLTKEEFQKIYRQFFPFGDPSSFAN 66 |
| Neurospora crassa OR74A | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 ||+ + +||| + |+| +||+|||||||+|#||||| | | ||||| ||||||||+ || Sbjct 7 KLERDKLEELEKATHFDKKELQQWYKGFLK#DCPSGMLTKAEFQKIYAQFFPFGDPSSFA 65 |
| Aspergillus nidulans FGSC A4 | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 || | +||| + |+| +||+|||||||+|#||||||| || ||++|||||||+ || Sbjct 6 KLSPTQLEELQKATHFDKKELQQWYKGFLK#DCPSGQLTKEEFQDIYRKFFPFGDPSSFAN 65 |
| Yarrowia lipolytica | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || + + || + ||| ||+|||||||+|#|||||||+ |||||+||||||||+ |+ Sbjct 7 KLSKDELTELKKATYFDRKELQQWYKGFLK#DCPSGQLNKEEFQKIYRQFFPFGDPSMFS 65 |
| Drosophila melanogaster | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 ||||||+|+| + | ||+ |+|+|||||+|#||||| | |+||| |||+|| +||| Sbjct 7 KLKPEVLEDLKQNTEFTDAEIQEWYKGFLK#DCPSGHLSVEEFKKIYGNFFPYGDASKFA 65 |
| Schizosaccharomyces pombe 972h- | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || + +++| | | | +||+||||||| |#||||| |+ + ||||||||||||||+ || Sbjct 7 KLSQDQLQDLVRSTRFDKKELQQWYKGFFK#DCPSGHLNKSEFQKIYKQFFPFGDPSAFA 65 |
| Magnaporthe grisea 70-15 | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || | + || + |+| +||+|||||||+|#||||| | |||||+||||||||+ || Sbjct 7 KLSQEQLAELQKSTHFDKKELQQWYKGFLK#DCPSGTLTKEEFQKIYRQFFPFGDPSSFA 65 |
| Magnaporthe grisea | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || | + || + |+| +||+|||||||+|#||||| | |||||+||||||||+ || Sbjct 7 KLSQEQLAELQKSTHFDKKELQQWYKGFLK#DCPSGTLTKEEFQKIYRQFFPFGDPSSFA 65 |
| Aedes aegypti | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 ||||||+|+| + | |++ |+|+|||||+|#||||| | |+||| |||+|| +||| Sbjct 7 KLKPEVLEDLKQNTEFSDAEIQEWYKGFLK#DCPSGHLSVEEFKKIYGNFFPYGDASKFA 65 |
| Gallus gallus | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 ||+|||+++| | ||| |+|+|||||++#||||| | |+||| |||+|| +||| Sbjct 7 KLRPEVMQDLLESTDFTEHEIQEWYKGFLR#DCPSGHLSMEEFKKIYGNFFPYGDASKFA 65 |
| Rattus norvegicus | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || |||+|+| + | | | |++||||||+|#|||||+|+ ||++| +|||+|| +||| Sbjct 7 KLAPEVMEDLVKSTEFNEHELKQWYKGFLK#DCPSGRLNLEEFQQLYVKFFPYGDASKFA 65 |
| Ustilago maydis 521 | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || || + +| + || |||+|||||||+|#||||| || | +|||||||||||+ || Sbjct 7 KLSPEQLSDLQKNTY-CEKELQQWYKGFLK#DCPSGTLDKQEFCRIYKQFFPFGDPSTFA 64 |
| Macaca fascicularis | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || |||+|+| + | | | |++||||||+|#|||||+|+ ||++| +|||+|| +||| Sbjct 7 KLAPEVMEDLVKSTEFNEHELKQWYKGFLK#DCPSGRLNLEEFQQLYVKFFPYGDASKFA 65 |
| Gibberella zeae PH-1 | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || + + || + |+| +||+|||||||+|#||||| | |||||+||||||||+ || Sbjct 7 KLSQDQLVELQKSTHFDKKELQQWYKGFLK#DCPSGTLTKEEFQKIYRQFFPFGDPSSFA 65 |
| Homo sapiens | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || |||+|+| + | |+|+|++||||||+|#||||| |+ ||++| +|||+|| +||| Sbjct 7 KLAPEVLEDLVQNTEFSEQELKQWYKGFLK#DCPSGILNLEEFQQLYIKFFPYGDASKFA 65 |
| Chaetomium globosum CBS 148.51 | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || + ++|| | | | +||+||||+||+|#||||| | + ||||| ||||||||+ || Sbjct 8 KLPDDQLKELQRSTNFDKKELQQWYRGFLK#DCPSGMLTKSEFQKIYAQFFPFGDPSTFA 66 |
| Canis familiaris | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || |||+|+| + | |+|+|++||||||+|#||||| |+ ||++| +|||+|| +||| Sbjct 63 KLAPEVLEDLVQNTEFSEQELKQWYKGFLK#DCPSGILNLEEFQQLYIKFFPYGDASKFA 121 |
| Xenopus tropicalis | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 ||+||++++| | |++ |+|+|||||+|#||||| |+ |+||| |||+|| +||| Sbjct 7 KLRPEMLQDLRENTEFSDHELQEWYKGFLK#DCPSGILNVEEFKKIYANFFPYGDASKFA 65 |
| Candida albicans SC5314 | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || + + +| + ||| ++|+|||||||++#||||||| | |+||||||||||| + Sbjct 7 KLSKDDLRQLRQATYFDKRELQQWYKGFLR#DCPSGQLSEEEFVKVYKQFFPFGDPTDYC 65 |
| Procambarus clarkii | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYK----GFIK#DCPSGQLDAAGFQKIYKQFFPFGDPT 62 ||| | +++| +|||++||++||+| ||+|#|||+| | || |||||||| |||| Sbjct 7 KLKQETIQKLCEETYFSDKEIKQWHKQWHKGFLK#DCPNGLLTETGFIKIYKQFFPQGDPT 66 Query 63 KFAT 66 | |+ Sbjct 67 KVAS 70 |
| Cryptococcus neoformans var. neoformans B-3501A | Query 7 KLKPEVVEELTRKTYFTEK----------EVQQWYKGFIK#DCPSGQLDAAG---FQKIYK 53 || | + || + ||| +| |+|||||||+|#|||||||+ | |+|||+ Sbjct 7 KLSSEELAELQKNTYFDKKVRDICIDWRTELQQWYKGFLK#DCPSGQLNKEGCTEFKKIYR 66 Query 54 QFFPFGDPTKFA 65 ||||||||++|| Sbjct 67 QFFPFGDPSQFA 78 |
| Ashbya gossypii ATCC 10895 | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 || + ++ | + ||| +|+ ||+|||++#|||+||| | |||||||||| | ||| Sbjct 7 KLSKDDLQSLKQSTYFDRREILQWHKGFLR#DCPNGQLTQEEFVKIYKQFFPFGAPEKFA 65 |
| Panulirus interruptus | Query 23 TEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 |+||++||+|||+|#|||+| | || |||||||| |||||||+ Sbjct 20 TDKEIKQWHKGFLK#DCPNGLLTETGFIKIYKQFFPQGDPTKFAS 63 |
| cattle, brain, Peptide Partial, 165 aa | Query 8 LKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 | |||+|+| + | | |+|++| ||||+|#| |+| |+ ||++| +|||+|| ||| Sbjct 1 LAPEVLEDLVQNTEFYEQELKQLYKGFLK#DGPAGILNLEEFQQLYIKFFPYGDAXKFA 58 |
| Schistosoma japonicum | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 || | +| | | ||+|+++|||+|||+#||||||| | ++| ||| || +| | Sbjct 10 KLPKEDLEFLRTNTNFTKKQIKQWYRGFIR#DCPSGQLSKKKFIEVYSGFFPDGDAEEFCT 69 |
| Branchiostoma floridae | Query 17 TRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 | + | | +|||+|#|||+| |+ ||||||||||||||+|||+ Sbjct 16 TVEAYSIEDRTMHLHKGFLK#DCPNGTLNEEEFQKIYKQFFPFGDPSKFAS 65 |
| Ornithorhynchus anatinus | Query 7 KLKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIY 52 || |||+|+| + | | | |++||||||+|#|||||+|+ ||++| Sbjct 7 KLAPEVMEDLVKSTEFNEHELKQWYKGFLK#DCPSGRLNLEEFQQLY 52 |
| Canis lupus familiaris | Query 8 LKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFA 65 | |++||| | |||+|+ ||+ |+|#+||||++ || || +||| || +| Sbjct 9 LSKEILEELQLNTKFTEEELCSWYQSFLK#ECPSGRITKQEFQSIYSKFFPEADPKAYA 66 |
| Strongylocentrotus purpuratus | Query 8 LKPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKF 64 | + + +| + | |+ +|+|||++ | |#||| | | ||++| +||| |||||| Sbjct 10 LDTQALSDLQKHTSFSVEELQQWFQDFKK#DCPKGSLSQPDFQRVYTKFFPKGDPTKF 66 |
| Mustela putorius furo | Query 9 KPEVVEELTRKTYFTEKEVQQWYKGFIK#DCPSGQLDAAGFQKIYKQFFPFGDPTKFAT 66 +|| +|+| +| || ||+| |+|| #+|||| ++ |++|| |||| || + +|| Sbjct 39 RPEGLEQLQEQTKFTRKELQVLYRGFKN#ECPSGIVNEENFKQIYSQFFPQGDSSTYAT 96 |
* References
[PubMed ID: 22521426] Drumond LE, Mourao FA, Leite HR, Abreu RV, Reis HJ, Moraes MF, Pereira GS, Massensini AR, Differential effects of swimming training on neuronal calcium sensor-1 expression in rat hippocampus/cortex and in object recognition memory tasks. Brain Res Bull. 2012 Jul 1;88(4):385-91. doi: 10.1016/j.brainresbull.2012.04.005.
[PubMed ID: 21035569] ... Chandra K, Sharma Y, Chary KV, Characterization of low-energy excited states in the native state ensemble of non-myristoylated and myristoylated neuronal calcium sensor-1. Biochim Biophys Acta. 2011 Feb;1814(2):334-44. doi: 10.1016/j.bbapap.2010.10.007.
[PubMed ID: 20838877] ... Souza BR, Torres KC, Miranda DM, Motta BS, Caetano FS, Rosa DV, Souza RP, Giovani A Jr, Carneiro DS, Guimaraes MM, Martins-Silva C, Reis HJ, Gomez MV, Jeromin A, Romano-Silva MA, Downregulation of the cAMP/PKA pathway in PC12 cells overexpressing NCS-1. Cell Mol Neurobiol. 2011 Jan;31(1):135-43. doi: 10.1007/s10571-010-9562-4. Epub
[PubMed ID: 20668007] ... McCue HV, Haynes LP, Burgoyne RD, The diversity of calcium sensor proteins in the regulation of neuronal function. Cold Spring Harb Perspect Biol. 2010 Aug;2(8):a004085. doi:
[PubMed ID: 20585375] ... Yip PK, Wong LF, Sears TA, Yanez-Munoz RJ, McMahon SB, Cortical overexpression of neuronal calcium sensor-1 induces functional plasticity in spinal cord following unilateral pyramidal tract injury in rat. PLoS Biol. 2010 Jun 22;8(6):e1000399. doi: 10.1371/journal.pbio.1000399.
[PubMed ID: 12006624] ... Scalettar BA, Rosa P, Taverna E, Francolini M, Tsuboi T, Terakawa S, Koizumi S, Roder J, Jeromin A, Neuronal calcium sensor-1 binds to regulated secretory organelles and functions in basal and stimulated exocytosis in PC12 cells. J Cell Sci. 2002 Jun 1;115(Pt 11):2399-412.
[PubMed ID: 11825672] ... Chen C, Yu L, Zhang P, Jiang J, Zhang Y, Chen X, Wu Q, Wu Q, Zhao S, Human neuronal calcium sensor-1 shows the highest expression level in cerebral cortex. Neurosci Lett. 2002 Feb 15;319(2):67-70.
[PubMed ID: 10514519] ... McFerran BW, Weiss JL, Burgoyne RD, Neuronal Ca(2+) sensor 1. Characterization of the myristoylated protein, its cellular effects in permeabilized adrenal chromaffin cells, Ca(2+)-independent membrane association, and interaction with binding proteins, suggesting a role in rapid Ca(2+) signal transduction. J Biol Chem. 1999 Oct 15;274(42):30258-65.
[PubMed ID: 9712909] ... McFerran BW, Graham ME, Burgoyne RD, Neuronal Ca2+ sensor 1, the mammalian homologue of frequenin, is expressed in chromaffin and PC12 cells and regulates neurosecretion from dense-core granules. J Biol Chem. 1998 Aug 28;273(35):22768-72.
[PubMed ID: 7488079] ... De Castro E, Nef S, Fiumelli H, Lenz SE, Kawamura S, Nef P, Regulation of rhodopsin phosphorylation by a family of neuronal calcium sensors. Biochem Biophys Res Commun. 1995 Nov 2;216(1):133-40.