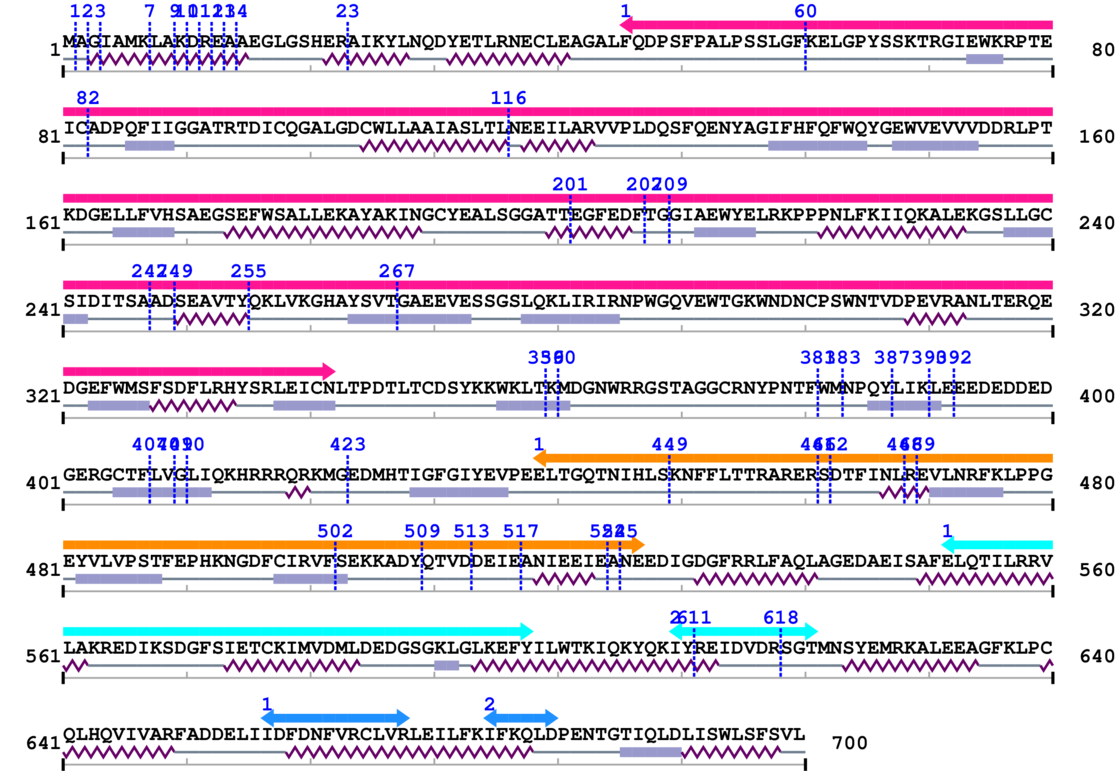
SB0129 : CAPN2, m-calpain catalytic subunit, mCL
[ CaMP Format ]
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | calpain-2 catalytic subunit precursor [Rattus norvegicus]; calpain-2 catalytic subunit precursor; calpain-2 catalytic subunit; M-calpain; calpain M-type; millimolar-calpain; calpain-2 large subunit; calcium-activated neutral proteinase 2; CANP 2; Calpain-2 catalytic subunit; 3.4.22.53; Calcium-activated neutral proteinase 2; Calpain M-type; Calpain-2 large subunit; Millimolar-calpain |
| Gene Names | Capn2; calpain 2, (m/II) large subunit |
| Gene Locus | 13q26; chromosome 13 |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 700 aa
Average Mass: 79.918 kDa
Monoisotopic Mass: 79.869 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 1 --- | ||||
| --- cleavage 2 --- | ||||
| --- cleavage 3 --- | ||||
| --- cleavage 7 --- | ||||
| --- cleavage 9 --- | ||||
| --- cleavage 10 --- | ||||
| --- cleavage 11 --- | ||||
| --- cleavage 12 --- | ||||
| --- cleavage 13 --- | ||||
| --- cleavage 14 --- | ||||
| --- cleavage 23 --- | ||||
| Calpain family cysteine protease 1. | 46 | 342 | 1.0 | 0.0 |
| --- cleavage 60 (inside Calpain family cysteine protease 46..342) --- | ||||
| --- cleavage 82 (inside Calpain family cysteine protease 46..342) --- | ||||
| --- cleavage 116 (inside Calpain family cysteine protease 46..342) --- | ||||
| --- cleavage 201 (inside Calpain family cysteine protease 46..342) --- | ||||
| --- cleavage 207 (inside Calpain family cysteine protease 46..342) --- | ||||
| --- cleavage 209 (inside Calpain family cysteine protease 46..342) --- | ||||
| --- cleavage 247 (inside Calpain family cysteine protease 46..342) --- | ||||
| --- cleavage 249 (inside Calpain family cysteine protease 46..342) --- | ||||
| --- cleavage 255 (inside Calpain family cysteine protease 46..342) --- | ||||
| --- cleavage 267 (inside Calpain family cysteine protease 46..342) --- | ||||
| --- cleavage 359 --- | ||||
| --- cleavage 360 --- | ||||
| --- cleavage 381 --- | ||||
| --- cleavage 383 --- | ||||
| --- cleavage 387 --- | ||||
| --- cleavage 390 --- | ||||
| --- cleavage 392 --- | ||||
| --- cleavage 407 --- | ||||
| --- cleavage 409 --- | ||||
| --- cleavage 410 --- | ||||
| --- cleavage 423 --- | ||||
| CorA-like Mg2+ transporter protein 1. | 439 | 527 | 67.0 | 0.1 |
| --- cleavage 449 (inside CorA-like Mg2+ transporter protein 439..527) --- | ||||
| --- cleavage 461 (inside CorA-like Mg2+ transporter protein 439..527) --- | ||||
| --- cleavage 462 (inside CorA-like Mg2+ transporter protein 439..527) --- | ||||
| --- cleavage 468 (inside CorA-like Mg2+ transporter protein 439..527) --- | ||||
| --- cleavage 469 (inside CorA-like Mg2+ transporter protein 439..527) --- | ||||
| --- cleavage 502 (inside CorA-like Mg2+ transporter protein 439..527) --- | ||||
| --- cleavage 509 (inside CorA-like Mg2+ transporter protein 439..527) --- | ||||
| --- cleavage 513 (inside CorA-like Mg2+ transporter protein 439..527) --- | ||||
| --- cleavage 517 (inside CorA-like Mg2+ transporter protein 439..527) --- | ||||
| --- cleavage 524 (inside CorA-like Mg2+ transporter protein 439..527) --- | ||||
| --- cleavage 525 (inside CorA-like Mg2+ transporter protein 439..527) --- | ||||
| EF-hand domain pair 1. | 552 | 598 | 20.0 | 0.1 |
| EF-hand domain pair 2. | 610 | 621 | 31.0 | 0.0 |
| --- cleavage 611 (inside EF-hand domain pair 610..621) --- | ||||
| --- cleavage 618 (inside EF-hand domain pair 610..621) --- | ||||
| EF hand 1. | 657 | 668 | 17.0 | 0.0 |
| EF hand 2. | 675 | 680 | 4.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0129.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
45 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 21549862] Chou JS, Impens F, Gevaert K, Davies PL, m-Calpain activation in vitro does not require autolysis or subunit dissociation. Biochim Biophys Acta. 2011 Jul;1814(7):864-72. doi: 10.1016/j.bbapap.2011.04.007.
Cleavage sites (±10aa)
[Site 1] M1-AGIAMKLAKD
Met1  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | Met1 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala2 | Gly3 | Ile4 | Ala5 | Met6 | Lys7 | Leu8 | Ala9 | Lys10 | Asp11 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQD |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 65.90 | 9 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 65.90 | 4 | calpain 2 |
| 3 | Mus musculus | 64.30 | 14 | m-calpain large subunit |
| 4 | Homo sapiens | 63.50 | 4 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 63.50 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 62.40 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus laevis | 45.80 | 3 | calpain 8 |
| 8 | Coturnix coturnix | 42.40 | 1 | quail calpain |
| 9 | Gallus gallus | 42.40 | 1 | calpain 1, (mu/I) large subunit |
| 10 | Canis familiaris | 42.00 | 3 | PREDICTED: similar to calpain 8 |
| 11 | Xenopus tropicalis | 41.20 | 4 | calpain 2, (m/II) large subunit |
| 12 | Stizostedion vitreum vitreum | 39.30 | 1 | putative calpain-like protein |
| 13 | Oncorhynchus mykiss | 39.30 | 1 | m-calpain |
| 14 | Xenopus laevis | 38.50 | 1 | mu/m-calpain large subunit |
| 15 | Tetraodon nigroviridis | 38.10 | 4 | unnamed protein product |
| 16 | Danio rerio | 38.10 | 3 | calpain 2, (m/II) large subunit a |
| 17 | Rattus norvegicus | 36.60 | 1 | calpain 1, large subunit |
| 18 | Bos taurus | 35.40 | 2 | micromolar calcium activated neutral protease 1 |
| 19 | synthetic construct | 35.40 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 35.00 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Sus scrofa | 33.50 | 1 | calpain 1, large subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#|||||||||||||||||||||||||||||| Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |
| Rattus norvegicus | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#|||||||||||||||||||||||||||||| Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |
| Mus musculus | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#||||+||||||||||||||||||||||||| Sbjct 1 M#AGIAIKLAKDREAAEGLGSHERAIKYLNQD 31 |
| Homo sapiens | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#|||| ||||||||||||||||||||||||| Sbjct 1 M#AGIAAKLAKDREAAEGLGSHERAIKYLNQD 31 |
| Homo sapiens | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#|||| ||||||||||||||||||||||||| Sbjct 1 M#AGIAAKLAKDREAAEGLGSHERAIKYLNQD 31 |
| Macaca mulatta | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#|||| || |||||||||||||||||||||| Sbjct 1 M#AGIAAKLVKDREAAEGLGSHERAIKYLNQD 31 |
| Xenopus laevis | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#+||| |+|||| |+|||++++ +|||||| Sbjct 1 M#SGIASKVAKDRVLAQGLGTNDKPLKYLNQD 31 |
| Coturnix coturnix | Query 2 AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 ||| +| +|| |||+| | |+|||||| Sbjct 5 GGIAARLQRDRLRAEGVGEHNNAVKYLNQD 34 |
| Gallus gallus | Query 2 AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 ||| +| +|| |||+| | |+|||||| Sbjct 5 GGIAARLQRDRLRAEGVGEHNNAVKYLNQD 34 |
| Canis familiaris | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#+ +| ++|| + || |||||++|+||| || Sbjct 94 M#SALAARIAKQQAAAGGLGSHQKAVKYLGQD 124 |
| Xenopus tropicalis | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#+|+| +|||+| | |||++|+|+ ||+|| Sbjct 1 M#SGVADRLAKERALASGLGTNEKALPYLSQD 31 |
| Stizostedion vitreum vitreum | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#+||| | + || +|+|| +|+|||||| Sbjct 1 M#SGIAATLQRRREKEQGIGSCSQAVKYLNQD 31 |
| Oncorhynchus mykiss | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#+|+| ||| | | | |++ |+|||||| Sbjct 1 M#SGMASNLAKKRALAAGFGTNANAVKYLNQD 31 |
| Xenopus laevis | Query 2 AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 ||| || || | |+|||+ |+|| ||| Sbjct 5 GGIASKLKIDRLKASGVGSHDCAVKYQNQD 34 |
| Tetraodon nigroviridis | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#+||| || |+ |+| |+ +| |||||| Sbjct 1 M#SGIAAKLQHHRDRAQGFGTASQAQKYLNQD 31 |
| Danio rerio | Query 1 M#AGIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |#+|+| ||| | | | |++ |+|||||+ Sbjct 1 M#SGVASTLAKKRALAAGFGTNSNAVKYLNQN 31 |
| Rattus norvegicus | Query 3 GIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |++ ++ | |+ ||| || ||||| || Sbjct 13 GVSAQVQKQRDKELGLGRHENAIKYLGQD 41 |
| Bos taurus | Query 3 GIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |++ ++ | | ||| || ||||| || Sbjct 13 GVSAQVQKQRAKELGLGRHENAIKYLGQD 41 |
| synthetic construct | Query 3 GIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |++ ++ | | ||| || ||||| || Sbjct 13 GVSAQVQKQRARELGLGRHENAIKYLGQD 41 |
| Pongo pygmaeus | Query 3 GIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |++ ++ | | ||| || ||||| || Sbjct 13 GVSAQVQKQRARELGLGRHENAIKYLGQD 41 |
| Sus scrofa | Query 3 GIAMKLAKDREAAEGLGSHERAIKYLNQD 31 |++ ++ | | ||| || ||||| || Sbjct 13 GVSAQVQKLRAKELGLGRHENAIKYLGQD 41 |
[Site 2] MA2-GIAMKLAKDR
Ala2  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | Met1 | Ala2 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly3 | Ile4 | Ala5 | Met6 | Lys7 | Leu8 | Ala9 | Lys10 | Asp11 | Arg12 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDY |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 68.90 | 10 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 68.90 | 4 | calpain 2 |
| 3 | Mus musculus | 67.40 | 14 | m-calpain |
| 4 | Homo sapiens | 66.20 | 4 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 66.20 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 65.10 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus laevis | 48.50 | 3 | calpain 8 |
| 8 | Coturnix coturnix | 45.40 | 1 | quail calpain |
| 9 | Gallus gallus | 45.40 | 1 | calpain 1, (mu/I) large subunit |
| 10 | Xenopus tropicalis | 43.90 | 4 | calpain 2, (m/II) large subunit |
| 11 | Canis familiaris | 43.50 | 3 | PREDICTED: similar to calpain 8 |
| 12 | Stizostedion vitreum vitreum | 42.40 | 1 | putative calpain-like protein |
| 13 | Xenopus laevis | 41.20 | 1 | mu/m-calpain large subunit |
| 14 | Oncorhynchus mykiss | 40.40 | 1 | m-calpain |
| 15 | Danio rerio | 39.70 | 5 | PREDICTED: similar to Calpain 1, (mu/I) large sub |
| 16 | Tetraodon nigroviridis | 39.30 | 5 | unnamed protein product |
| 17 | Rattus norvegicus | 39.30 | 1 | calpain 1, large subunit |
| 18 | Bos taurus | 38.10 | 2 | micromolar calcium activated neutral protease 1 |
| 19 | synthetic construct | 38.10 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 37.70 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Sus scrofa | 36.20 | 1 | calpain 1, large subunit |
| 22 | Gallus gallus | 32.30 | 1 | mu-calpain large subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 ||#|||||||||||||||||||||||||||||| Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |
| Rattus norvegicus | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 ||#|||||||||||||||||||||||||||||| Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |
| Mus musculus | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 ||#|||+|||||||||||||||||||||||||| Sbjct 1 MA#GIAIKLAKDREAAEGLGSHERAIKYLNQDY 32 |
| Homo sapiens | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 ||#||| |||||||||||||||||||||||||| Sbjct 1 MA#GIAAKLAKDREAAEGLGSHERAIKYLNQDY 32 |
| Homo sapiens | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 ||#||| |||||||||||||||||||||||||| Sbjct 1 MA#GIAAKLAKDREAAEGLGSHERAIKYLNQDY 32 |
| Macaca mulatta | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 ||#||| || ||||||||||||||||||||||| Sbjct 1 MA#GIAAKLVKDREAAEGLGSHERAIKYLNQDY 32 |
| Xenopus laevis | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |+#||| |+|||| |+|||++++ +||||||| Sbjct 1 MS#GIASKVAKDRVLAQGLGTNDKPLKYLNQDY 32 |
| Coturnix coturnix | Query 2 A#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 #||| +| +|| |||+| | |+||||||| Sbjct 5 G#GIAARLQRDRLRAEGVGEHNNAVKYLNQDY 35 |
| Gallus gallus | Query 2 A#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 #||| +| +|| |||+| | |+||||||| Sbjct 5 G#GIAARLQRDRLRAEGVGEHNNAVKYLNQDY 35 |
| Xenopus tropicalis | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |+#|+| +|||+| | |||++|+|+ ||+||| Sbjct 1 MS#GVADRLAKERALASGLGTNEKALPYLSQDY 32 |
| Canis familiaris | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |+# +| ++|| + || |||||++|+||| ||+ Sbjct 94 MS#ALAARIAKQQAAAGGLGSHQKAVKYLGQDF 125 |
| Stizostedion vitreum vitreum | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |+#||| | + || +|+|| +|+||||||| Sbjct 1 MS#GIAATLQRRREKEQGIGSCSQAVKYLNQDY 32 |
| Xenopus laevis | Query 2 A#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 #||| || || | |+|||+ |+|| |||| Sbjct 5 G#GIASKLKIDRLKASGVGSHDCAVKYQNQDY 35 |
| Oncorhynchus mykiss | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |+#|+| ||| | | | |++ |+||||||+ Sbjct 1 MS#GMASNLAKKRALAAGFGTNANAVKYLNQDF 32 |
| Danio rerio | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 + #|++ ++ |+| |||+|+++ |||+||||| Sbjct 4 IG#GVSTRIYKERLQAEGMGANDLAIKFLNQDY 35 |
| Tetraodon nigroviridis | Query 1 MA#GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |+#||| || |+ |+| |+ +| ||||||+ Sbjct 1 MS#GIAAKLQHHRDRAQGFGTASQAQKYLNQDF 32 |
| Rattus norvegicus | Query 3 GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |++ ++ | |+ ||| || ||||| ||| Sbjct 13 GVSAQVQKQRDKELGLGRHENAIKYLGQDY 42 |
| Bos taurus | Query 3 GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |++ ++ | | ||| || ||||| ||| Sbjct 13 GVSAQVQKQRAKELGLGRHENAIKYLGQDY 42 |
| synthetic construct | Query 3 GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |++ ++ | | ||| || ||||| ||| Sbjct 13 GVSAQVQKQRARELGLGRHENAIKYLGQDY 42 |
| Pongo pygmaeus | Query 3 GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |++ ++ | | ||| || ||||| ||| Sbjct 13 GVSAQVQKQRARELGLGRHENAIKYLGQDY 42 |
| Sus scrofa | Query 3 GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |++ ++ | | ||| || ||||| ||| Sbjct 13 GVSAQVQKLRAKELGLGRHENAIKYLGQDY 42 |
| Gallus gallus | Query 3 GIAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |++ ++ + | | ||| |+ |+++ ||| Sbjct 13 GVSAQVQRQRAKALGLGQHQNAVRFRGQDY 42 |
[Site 3] MAG3-IAMKLAKDRE
Gly3  Ile
Ile
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | Met1 | Ala2 | Gly3 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ile4 | Ala5 | Met6 | Lys7 | Leu8 | Ala9 | Lys10 | Asp11 | Arg12 | Glu13 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDYE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 70.10 | 11 | A Chain A, Crystal Structure Of A Mu-Like Calpain |
| 2 | Rattus norvegicus | 69.70 | 4 | calpain 2 |
| 3 | Mus musculus | 68.20 | 14 | m-calpain |
| 4 | Homo sapiens | 67.40 | 4 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 67.40 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 66.20 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus laevis | 50.40 | 3 | calpain 8 |
| 8 | Coturnix coturnix | 47.00 | 1 | quail calpain |
| 9 | Gallus gallus | 47.00 | 1 | calpain 1, (mu/I) large subunit |
| 10 | Xenopus tropicalis | 45.80 | 4 | calpain 2, (m/II) large subunit |
| 11 | Canis familiaris | 45.40 | 3 | PREDICTED: similar to calpain 8 |
| 12 | Stizostedion vitreum vitreum | 43.90 | 1 | putative calpain-like protein |
| 13 | Xenopus laevis | 42.70 | 1 | mu/m-calpain large subunit |
| 14 | Oncorhynchus mykiss | 42.40 | 1 | m-calpain |
| 15 | Danio rerio | 41.20 | 5 | calpain 2, (m/II) large subunit a |
| 16 | Tetraodon nigroviridis | 40.80 | 5 | unnamed protein product |
| 17 | Rattus norvegicus | 40.80 | 1 | calpain 1, large subunit |
| 18 | Bos taurus | 40.00 | 2 | calpain 1, (mu/I) large subunit |
| 19 | synthetic construct | 39.70 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 39.70 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Sus scrofa | 38.10 | 1 | calpain 1, large subunit |
| 22 | Gallus gallus | 32.00 | 1 | mu-calpain large subunit |
| 23 | Canis familiaris | 31.20 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 24 | Danio rerio | 30.40 | 1 | calpain 1, large subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |||#|||||||||||||||||||||||||||||| Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |
| Rattus norvegicus | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |||#|||||||||||||||||||||||||||||| Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |
| Mus musculus | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |||#||+||||||||||||||||||||||||||| Sbjct 1 MAG#IAIKLAKDREAAEGLGSHERAIKYLNQDYE 33 |
| Homo sapiens | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |||#|| ||||||||||||||||||||||||||| Sbjct 1 MAG#IAAKLAKDREAAEGLGSHERAIKYLNQDYE 33 |
| Homo sapiens | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |||#|| ||||||||||||||||||||||||||| Sbjct 1 MAG#IAAKLAKDREAAEGLGSHERAIKYLNQDYE 33 |
| Macaca mulatta | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |||#|| || |||||||||||||||||||||||| Sbjct 1 MAG#IAAKLVKDREAAEGLGSHERAIKYLNQDYE 33 |
| Xenopus laevis | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |+|#|| |+|||| |+|||++++ +|||||||| Sbjct 1 MSG#IASKVAKDRVLAQGLGTNDKPLKYLNQDYE 33 |
| Coturnix coturnix | Query 2 AG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |#|| +| +|| |||+| | |+|||||||| Sbjct 5 GG#IAARLQRDRLRAEGVGEHNNAVKYLNQDYE 36 |
| Gallus gallus | Query 2 AG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |#|| +| +|| |||+| | |+|||||||| Sbjct 5 GG#IAARLQRDRLRAEGVGEHNNAVKYLNQDYE 36 |
| Xenopus tropicalis | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |+|#+| +|||+| | |||++|+|+ ||+|||| Sbjct 1 MSG#VADRLAKERALASGLGTNEKALPYLSQDYE 33 |
| Canis familiaris | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |+ #+| ++|| + || |||||++|+||| ||+| Sbjct 94 MSA#LAARIAKQQAAAGGLGSHQKAVKYLGQDFE 126 |
| Stizostedion vitreum vitreum | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |+|#|| | + || +|+|| +|+|||||||| Sbjct 1 MSG#IAATLQRRREKEQGIGSCSQAVKYLNQDYE 33 |
| Xenopus laevis | Query 2 AG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |#|| || || | |+|||+ |+|| ||||| Sbjct 5 GG#IASKLKIDRLKASGVGSHDCAVKYQNQDYE 36 |
| Oncorhynchus mykiss | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |+|#+| ||| | | | |++ |+||||||+| Sbjct 1 MSG#MASNLAKKRALAAGFGTNANAVKYLNQDFE 33 |
| Danio rerio | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |+|#+| ||| | | | |++ |+|||||++| Sbjct 1 MSG#VASTLAKKRALAAGFGTNSNAVKYLNQNFE 33 |
| Tetraodon nigroviridis | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |+|#|| || |+ |+| |+ +| ||||||+| Sbjct 1 MSG#IAAKLQHHRDRAQGFGTASQAQKYLNQDFE 33 |
| Rattus norvegicus | Query 3 G#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |#++ ++ | |+ ||| || ||||| |||| Sbjct 13 G#VSAQVQKQRDKELGLGRHENAIKYLGQDYE 43 |
| Bos taurus | Query 3 G#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |#++ ++ | | ||| || ||||| |||| Sbjct 13 G#VSAQVQKQRAKELGLGRHENAIKYLGQDYE 43 |
| synthetic construct | Query 3 G#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |#++ ++ | | ||| || ||||| |||| Sbjct 13 G#VSAQVQKQRARELGLGRHENAIKYLGQDYE 43 |
| Pongo pygmaeus | Query 3 G#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |#++ ++ | | ||| || ||||| |||| Sbjct 13 G#VSAQVQKQRARELGLGRHENAIKYLGQDYE 43 |
| Sus scrofa | Query 3 G#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |#++ ++ | | ||| || ||||| |||| Sbjct 13 G#VSAQVQKLRAKELGLGRHENAIKYLGQDYE 43 |
| Gallus gallus | Query 3 G#IAMKLAKDREAAEGLGSHERAIKYLNQDY 32 |#++ ++ + | | ||| |+ |+++ ||| Sbjct 13 G#VSAQVQRQRAKALGLGQHQNAVRFRGQDY 42 |
| Canis familiaris | Query 1 MAG#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 + |#+| ++ | |+||| | | | ||++| Sbjct 83 LEG#MAAHISHSRLKAKGLGQHHNAQNYNNQNFE 115 |
| Danio rerio | Query 3 G#IAMKLAKDREAAEGLGSHERAIKYLNQDYE 33 |#+| +| + ||| + |+|+| |||| Sbjct 8 G#MAARLRSQWDRDAGLGQNHNAVKFLGQDYE 38 |
[Site 4] MAGIAMK7-LAKDREAAEG
Lys7  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | - | Met1 | Ala2 | Gly3 | Ile4 | Ala5 | Met6 | Lys7 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu8 | Ala9 | Lys10 | Asp11 | Arg12 | Glu13 | Ala14 | Ala15 | Glu16 | Gly17 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDYETLRN |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 78.20 | 11 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 78.20 | 4 | calpain 2 |
| 3 | Mus musculus | 76.60 | 14 | m-calpain |
| 4 | Homo sapiens | 74.30 | 4 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 74.30 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 72.80 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus laevis | 52.80 | 4 | calpain 8 |
| 8 | Xenopus tropicalis | 52.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Canis familiaris | 49.30 | 3 | PREDICTED: similar to calpain 8 |
| 10 | Coturnix coturnix | 49.30 | 1 | quail calpain |
| 11 | Gallus gallus | 49.30 | 1 | calpain 1, (mu/I) large subunit |
| 12 | Stizostedion vitreum vitreum | 47.80 | 1 | putative calpain-like protein |
| 13 | Danio rerio | 47.40 | 5 | calpain 2, (m/II) large subunit a |
| 14 | Oncorhynchus mykiss | 46.20 | 1 | m-calpain |
| 15 | Xenopus laevis | 46.20 | 1 | mu/m-calpain large subunit |
| 16 | Tetraodon nigroviridis | 45.40 | 5 | unnamed protein product |
| 17 | Rattus norvegicus | 44.70 | 1 | calpain 1, large subunit |
| 18 | Bos taurus | 43.50 | 2 | micromolar calcium activated neutral protease 1 |
| 19 | synthetic construct | 43.10 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 43.10 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Sus scrofa | 41.60 | 1 | calpain 1, large subunit |
| 22 | Gallus gallus | 36.20 | 1 | mu-calpain large subunit |
| 23 | Danio rerio | 35.80 | 1 | calpain 1, large subunit |
| 24 | Canis familiaris | 33.10 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 |||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 |
| Rattus norvegicus | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 |||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 |
| Mus musculus | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 |||||+|#|||||||||||||||||||||||||||||| Sbjct 1 MAGIAIK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 |
| Homo sapiens | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 ||||| |#|||||||||||||||||||||||||| ||| Sbjct 1 MAGIAAK#LAKDREAAEGLGSHERAIKYLNQDYEALRN 37 |
| Homo sapiens | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 ||||| |#|||||||||||||||||||||||||| ||| Sbjct 1 MAGIAAK#LAKDREAAEGLGSHERAIKYLNQDYEALRN 37 |
| Macaca mulatta | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 ||||| |#| |||||||||||||||||||||||| ||| Sbjct 1 MAGIAAK#LVKDREAAEGLGSHERAIKYLNQDYEALRN 37 |
| Xenopus laevis | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 |+||| |#+|||| |+|||++++ +|||||||| |+ Sbjct 1 MSGIASK#VAKDRVLAQGLGTNDKPLKYLNQDYEQLK 36 |
| Xenopus tropicalis | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 |+|+| +#|||+| | |||++|+|+ ||+|||| ||| Sbjct 1 MSGVADR#LAKERALASGLGTNEKALPYLSQDYEALRN 37 |
| Canis familiaris | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 |+ +| +#+|| + || |||||++|+||| ||+| || Sbjct 94 MSALAAR#IAKQQAAAGGLGSHQKAVKYLGQDFEKLR 129 |
| Coturnix coturnix | Query 2 AGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 ||| +#| +|| |||+| | |+|||||||| |+ Sbjct 5 GGIAAR#LQRDRLRAEGVGEHNNAVKYLNQDYEALK 39 |
| Gallus gallus | Query 2 AGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 ||| +#| +|| |||+| | |+|||||||| |+ Sbjct 5 GGIAAR#LQRDRLRAEGVGEHNNAVKYLNQDYEALK 39 |
| Stizostedion vitreum vitreum | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 |+||| #| + || +|+|| +|+|||||||| || Sbjct 1 MSGIAAT#LQRRREKEQGIGSCSQAVKYLNQDYEALR 36 |
| Danio rerio | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 |+|+| #||| | | | |++ |+|||||++||||+ Sbjct 1 MSGVAST#LAKKRALAAGFGTNSNAVKYLNQNFETLRS 37 |
| Oncorhynchus mykiss | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 |+|+| #||| | | | |++ |+||||||+| || Sbjct 1 MSGMASN#LAKKRALAAGFGTNANAVKYLNQDFEALR 36 |
| Xenopus laevis | Query 2 AGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 ||| |#| || | |+|||+ |+|| |||||+|+ Sbjct 5 GGIASK#LKIDRLKASGVGSHDCAVKYQNQDYESLK 39 |
| Tetraodon nigroviridis | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 |+||| |#| |+ |+| |+ +| ||||||+|+|| Sbjct 1 MSGIAAK#LQHHRDRAQGFGTASQAQKYLNQDFESLR 36 |
| Rattus norvegicus | Query 3 GIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 |++ +#+ | |+ ||| || ||||| |||| || Sbjct 13 GVSAQ#VQKQRDKELGLGRHENAIKYLGQDYENLR 46 |
| Bos taurus | Query 3 GIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 |++ +#+ | | ||| || ||||| |||| || Sbjct 13 GVSAQ#VQKQRAKELGLGRHENAIKYLGQDYEQLR 46 |
| synthetic construct | Query 3 GIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 |++ +#+ | | ||| || ||||| |||| || Sbjct 13 GVSAQ#VQKQRARELGLGRHENAIKYLGQDYEQLR 46 |
| Pongo pygmaeus | Query 3 GIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 |++ +#+ | | ||| || ||||| |||| || Sbjct 13 GVSAQ#VQKQRARELGLGRHENAIKYLGQDYEQLR 46 |
| Sus scrofa | Query 3 GIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 |++ +#+ | | ||| || ||||| |||| || Sbjct 13 GVSAQ#VQKLRAKELGLGRHENAIKYLGQDYEQLR 46 |
| Gallus gallus | Query 3 GIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLRN 37 |++ +#+ + | | ||| |+ |+++ ||| ||+ Sbjct 13 GVSAQ#VQRQRAKALGLGQHQNAVRFRGQDYAALRD 47 |
| Danio rerio | Query 3 GIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 |+| +#| + ||| + |+|+| ||||||| Sbjct 8 GMAAR#LRSQWDRDAGLGQNHNAVKFLGQDYETLR 41 |
| Canis familiaris | Query 1 MAGIAMK#LAKDREAAEGLGSHERAIKYLNQDYETLR 36 + |+| #++ | |+||| | | | ||++| |+ Sbjct 83 LEGMAAH#ISHSRLKAKGLGQHHNAQNYNNQNFEDLQ 118 |
[Site 5] MAGIAMKLA9-KDREAAEGLG
Ala9  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | Met1 | Ala2 | Gly3 | Ile4 | Ala5 | Met6 | Lys7 | Leu8 | Ala9 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys10 | Asp11 | Arg12 | Glu13 | Ala14 | Ala15 | Glu16 | Gly17 | Leu18 | Gly19 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDYETLRNEC |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 84.00 | 11 | A Chain A, Crystal Structure Of A Mu-Like Calpain |
| 2 | Rattus norvegicus | 83.20 | 4 | calpain 2 |
| 3 | Mus musculus | 81.60 | 14 | unnamed protein product |
| 4 | Homo sapiens | 79.30 | 4 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 79.30 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 77.80 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus tropicalis | 57.40 | 4 | calpain 2, (m/II) large subunit |
| 8 | Gallus gallus | 55.10 | 1 | calpain 1, (mu/I) large subunit |
| 9 | Xenopus laevis | 54.70 | 4 | calpain 8 |
| 10 | Canis familiaris | 53.90 | 3 | PREDICTED: similar to calpain 8 |
| 11 | Danio rerio | 52.80 | 6 | calpain 2, (m/II) large subunit a |
| 12 | Coturnix coturnix | 52.80 | 1 | quail calpain |
| 13 | Stizostedion vitreum vitreum | 50.80 | 1 | putative calpain-like protein |
| 14 | Xenopus laevis | 50.40 | 2 | mu/m-calpain large subunit |
| 15 | Oncorhynchus mykiss | 50.10 | 1 | m-calpain |
| 16 | Tetraodon nigroviridis | 48.10 | 6 | unnamed protein product |
| 17 | Rattus norvegicus | 47.80 | 1 | calpain 1, large subunit |
| 18 | Bos taurus | 46.20 | 2 | micromolar calcium activated neutral protease 1 |
| 19 | synthetic construct | 45.80 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 45.40 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Sus scrofa | 44.70 | 1 | calpain 1, large subunit |
| 22 | Gallus gallus | 40.40 | 1 | mu-calpain large subunit |
| 23 | Danio rerio | 36.20 | 1 | calpain 1, large subunit |
| 24 | Canis familiaris | 35.80 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |
| Rattus norvegicus | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |
| Mus musculus | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |||||+|||#|||||||||||||||||||||||||||||| Sbjct 1 MAGIAIKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |
| Homo sapiens | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 ||||| |||#|||||||||||||||||||||||| ||||| Sbjct 1 MAGIAAKLA#KDREAAEGLGSHERAIKYLNQDYEALRNEC 39 |
| Homo sapiens | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 ||||| |||#|||||||||||||||||||||||| ||||| Sbjct 1 MAGIAAKLA#KDREAAEGLGSHERAIKYLNQDYEALRNEC 39 |
| Macaca mulatta | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 ||||| || #|||||||||||||||||||||||| ||||| Sbjct 1 MAGIAAKLV#KDREAAEGLGSHERAIKYLNQDYEALRNEC 39 |
| Xenopus tropicalis | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+|+| +||#|+| | |||++|+|+ ||+|||| ||||| Sbjct 1 MSGVADRLA#KERALASGLGTNEKALPYLSQDYEALRNEC 39 |
| Gallus gallus | Query 2 AGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 ||| +| #+|| |||+| | |+|||||||| |+ || Sbjct 5 GGIAARLQ#RDRLRAEGVGEHNNAVKYLNQDYEALKQEC 42 |
| Xenopus laevis | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+||| |+|#||| |+|||++++ +|||||||| |+ | Sbjct 1 MSGIASKVA#KDRVLAQGLGTNDKPLKYLNQDYEQLKARC 39 |
| Canis familiaris | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+ +| ++|#| + || |||||++|+||| ||+| || +| Sbjct 94 MSALAARIA#KQQAAAGGLGSHQKAVKYLGQDFEKLRQQC 132 |
| Danio rerio | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+|+| ||#| | | | |++ |+|||||++||||+|| Sbjct 1 MSGVASTLA#KKRALAAGFGTNSNAVKYLNQNFETLRSEC 39 |
| Coturnix coturnix | Query 2 AGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 ||| +| #+|| |||+| | |+|||||||| |+ | Sbjct 5 GGIAARLQ#RDRLRAEGVGEHNNAVKYLNQDYEALKQAC 42 |
| Stizostedion vitreum vitreum | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+||| | #+ || +|+|| +|+|||||||| || | Sbjct 1 MSGIAATLQ#RRREKEQGIGSCSQAVKYLNQDYEALRRRC 39 |
| Xenopus laevis | Query 2 AGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 ||| || # || | |+|||+ |+|| |||||+|+ +| Sbjct 5 GGIASKLK#IDRLKASGVGSHDCAVKYQNQDYESLKQQC 42 |
| Oncorhynchus mykiss | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+|+| ||#| | | | |++ |+||||||+| || +| Sbjct 1 MSGMASNLA#KKRALAAGFGTNANAVKYLNQDFEALRAQC 39 |
| Tetraodon nigroviridis | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |+||| || # |+ |+| |+ +| ||||||+|+|| | Sbjct 1 MSGIAAKLQ#HHRDRAQGFGTASQAQKYLNQDFESLRESC 39 |
| Rattus norvegicus | Query 3 GIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |++ ++ #| |+ ||| || ||||| |||| || | Sbjct 13 GVSAQVQ#KQRDKELGLGRHENAIKYLGQDYENLRARC 49 |
| Bos taurus | Query 3 GIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |++ ++ #| | ||| || ||||| |||| || | Sbjct 13 GVSAQVQ#KQRAKELGLGRHENAIKYLGQDYEQLRVHC 49 |
| synthetic construct | Query 3 GIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |++ ++ #| | ||| || ||||| |||| || | Sbjct 13 GVSAQVQ#KQRARELGLGRHENAIKYLGQDYEQLRVRC 49 |
| Pongo pygmaeus | Query 3 GIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |++ ++ #| | ||| || ||||| |||| || | Sbjct 13 GVSAQVQ#KQRARELGLGRHENAIKYLGQDYEQLRVRC 49 |
| Sus scrofa | Query 3 GIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |++ ++ #| | ||| || ||||| |||| || | Sbjct 13 GVSAQVQ#KLRAKELGLGRHENAIKYLGQDYEQLRAHC 49 |
| Gallus gallus | Query 3 GIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 |++ ++ #+ | | ||| |+ |+++ ||| ||++| Sbjct 13 GVSAQVQ#RQRAKALGLGQHQNAVRFRGQDYAALRDDC 49 |
| Danio rerio | Query 3 GIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNE 38 |+| +| # + ||| + |+|+| ||||||| + Sbjct 8 GMAARLR#SQWDRDAGLGQNHNAVKFLGQDYETLRAQ 43 |
| Canis familiaris | Query 1 MAGIAMKLA#KDREAAEGLGSHERAIKYLNQDYETLRNEC 39 + |+| ++# | |+||| | | | ||++| |+ | Sbjct 83 LEGMAAHIS#HSRLKAKGLGQHHNAQNYNNQNFEDLQAAC 121 |
[Site 6] MAGIAMKLAK10-DREAAEGLGS
Lys10  Asp
Asp
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Met1 | Ala2 | Gly3 | Ile4 | Ala5 | Met6 | Lys7 | Leu8 | Ala9 | Lys10 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asp11 | Arg12 | Glu13 | Ala14 | Ala15 | Glu16 | Gly17 | Leu18 | Gly19 | Ser20 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDYETLRNECL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 85.90 | 12 | A Chain A, Crystal Structure Of A Mu-Like Calpain |
| 2 | Rattus norvegicus | 85.10 | 5 | calpain 2 |
| 3 | Mus musculus | 83.60 | 14 | m-calpain |
| 4 | Homo sapiens | 81.30 | 6 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 81.30 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 79.70 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus tropicalis | 59.30 | 4 | calpain 2, (m/II) large subunit |
| 8 | Xenopus laevis | 56.60 | 4 | calpain 8 |
| 9 | Gallus gallus | 55.80 | 1 | calpain 1, (mu/I) large subunit |
| 10 | Canis familiaris | 55.50 | 3 | PREDICTED: similar to calpain 8 |
| 11 | Danio rerio | 54.70 | 6 | calpain 2, (m/II) large subunit a |
| 12 | Coturnix coturnix | 53.50 | 1 | quail calpain |
| 13 | Stizostedion vitreum vitreum | 52.80 | 1 | putative calpain-like protein |
| 14 | Oncorhynchus mykiss | 52.00 | 1 | m-calpain |
| 15 | Xenopus laevis | 50.80 | 2 | mu/m-calpain large subunit |
| 16 | Tetraodon nigroviridis | 50.10 | 7 | unnamed protein product |
| 17 | Rattus norvegicus | 49.30 | 1 | calpain 1, large subunit |
| 18 | Bos taurus | 47.80 | 2 | calpain 1, (mu/I) large subunit |
| 19 | synthetic construct | 47.40 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 47.40 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Sus scrofa | 46.20 | 1 | calpain 1, large subunit |
| 22 | Gallus gallus | 42.00 | 1 | mu-calpain large subunit |
| 23 | Canis familiaris | 37.40 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 24 | Danio rerio | 36.20 | 1 | calpain 1, large subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |
| Rattus norvegicus | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 ||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |
| Mus musculus | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |||||+||||#|||||||||||||||||||||||||||||| Sbjct 1 MAGIAIKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |
| Homo sapiens | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 ||||| ||||#||||||||||||||||||||||| |||||| Sbjct 1 MAGIAAKLAK#DREAAEGLGSHERAIKYLNQDYEALRNECL 40 |
| Homo sapiens | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 ||||| ||||#||||||||||||||||||||||| |||||| Sbjct 1 MAGIAAKLAK#DREAAEGLGSHERAIKYLNQDYEALRNECL 40 |
| Macaca mulatta | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 ||||| || |#||||||||||||||||||||||| |||||| Sbjct 1 MAGIAAKLVK#DREAAEGLGSHERAIKYLNQDYEALRNECL 40 |
| Xenopus tropicalis | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |+|+| +|||#+| | |||++|+|+ ||+|||| |||||| Sbjct 1 MSGVADRLAK#ERALASGLGTNEKALPYLSQDYEALRNECL 40 |
| Xenopus laevis | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |+||| |+||#|| |+|||++++ +|||||||| |+ || Sbjct 1 MSGIASKVAK#DRVLAQGLGTNDKPLKYLNQDYEQLKARCL 40 |
| Gallus gallus | Query 2 AGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 ||| +| +#|| |||+| | |+|||||||| |+ ||+ Sbjct 5 GGIAARLQR#DRLRAEGVGEHNNAVKYLNQDYEALKQECI 43 |
| Canis familiaris | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |+ +| ++||# + || |||||++|+||| ||+| || +|| Sbjct 94 MSALAARIAK#QQAAAGGLGSHQKAVKYLGQDFEKLRQQCL 133 |
| Danio rerio | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |+|+| |||# | | | |++ |+|||||++||||+||| Sbjct 1 MSGVASTLAK#KRALAAGFGTNSNAVKYLNQNFETLRSECL 40 |
| Coturnix coturnix | Query 2 AGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 ||| +| +#|| |||+| | |+|||||||| |+ |+ Sbjct 5 GGIAARLQR#DRLRAEGVGEHNNAVKYLNQDYEALKQACI 43 |
| Stizostedion vitreum vitreum | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |+||| | +# || +|+|| +|+|||||||| || || Sbjct 1 MSGIAATLQR#RREKEQGIGSCSQAVKYLNQDYEALRRRCL 40 |
| Oncorhynchus mykiss | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |+|+| |||# | | | |++ |+||||||+| || +|| Sbjct 1 MSGMASNLAK#KRALAAGFGTNANAVKYLNQDFEALRAQCL 40 |
| Xenopus laevis | Query 2 AGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 ||| || #|| | |+|||+ |+|| |||||+|+ +|+ Sbjct 5 GGIASKLKI#DRLKASGVGSHDCAVKYQNQDYESLKQQCV 43 |
| Tetraodon nigroviridis | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |+||| || # |+ |+| |+ +| ||||||+|+|| || Sbjct 1 MSGIAAKLQH#HRDRAQGFGTASQAQKYLNQDFESLRESCL 40 |
| Rattus norvegicus | Query 3 GIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |++ ++ |# |+ ||| || ||||| |||| || || Sbjct 13 GVSAQVQK#QRDKELGLGRHENAIKYLGQDYENLRARCL 50 |
| Bos taurus | Query 3 GIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |++ ++ |# | ||| || ||||| |||| || || Sbjct 13 GVSAQVQK#QRAKELGLGRHENAIKYLGQDYEQLRVHCL 50 |
| synthetic construct | Query 3 GIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |++ ++ |# | ||| || ||||| |||| || || Sbjct 13 GVSAQVQK#QRARELGLGRHENAIKYLGQDYEQLRVRCL 50 |
| Pongo pygmaeus | Query 3 GIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |++ ++ |# | ||| || ||||| |||| || || Sbjct 13 GVSAQVQK#QRARELGLGRHENAIKYLGQDYEQLRVRCL 50 |
| Sus scrofa | Query 3 GIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |++ ++ |# | ||| || ||||| |||| || || Sbjct 13 GVSAQVQK#LRAKELGLGRHENAIKYLGQDYEQLRAHCL 50 |
| Gallus gallus | Query 3 GIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 |++ ++ +# | | ||| |+ |+++ ||| ||++|| Sbjct 13 GVSAQVQR#QRAKALGLGQHQNAVRFRGQDYAALRDDCL 50 |
| Canis familiaris | Query 1 MAGIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNECL 40 + |+| ++ # | |+||| | | | ||++| |+ || Sbjct 83 LEGMAAHISH#SRLKAKGLGQHHNAQNYNNQNFEDLQAACL 122 |
| Danio rerio | Query 3 GIAMKLAK#DREAAEGLGSHERAIKYLNQDYETLRNE 38 |+| +| # + ||| + |+|+| ||||||| + Sbjct 8 GMAARLRS#QWDRDAGLGQNHNAVKFLGQDYETLRAQ 43 |
[Site 7] AGIAMKLAKD11-REAAEGLGSH
Asp11  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala2 | Gly3 | Ile4 | Ala5 | Met6 | Lys7 | Leu8 | Ala9 | Lys10 | Asp11 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg12 | Glu13 | Ala14 | Ala15 | Glu16 | Gly17 | Leu18 | Gly19 | Ser20 | His21 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDYETLRNECLE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 87.00 | 12 | A Chain A, Crystal Structure Of A Mu-Like Calpain |
| 2 | Rattus norvegicus | 86.30 | 5 | calpain 2 |
| 3 | Mus musculus | 84.30 | 14 | m-calpain |
| 4 | Homo sapiens | 82.40 | 6 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 82.40 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 80.90 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus tropicalis | 59.70 | 4 | calpain 2, (m/II) large subunit |
| 8 | Xenopus laevis | 58.20 | 4 | calpain 8 |
| 9 | Gallus gallus | 57.40 | 2 | calpain 1, (mu/I) large subunit |
| 10 | Canis familiaris | 55.80 | 3 | PREDICTED: similar to calpain 8 |
| 11 | Coturnix coturnix | 55.10 | 1 | quail calpain |
| 12 | Danio rerio | 54.30 | 6 | calpain 2, (m/II) large subunit a |
| 13 | Stizostedion vitreum vitreum | 54.30 | 1 | putative calpain-like protein |
| 14 | Xenopus laevis | 52.40 | 1 | mu/m-calpain large subunit |
| 15 | Tetraodon nigroviridis | 51.60 | 7 | unnamed protein product |
| 16 | Oncorhynchus mykiss | 51.60 | 1 | m-calpain |
| 17 | Rattus norvegicus | 49.70 | 1 | calpain 1, large subunit |
| 18 | Bos taurus | 48.10 | 2 | micromolar calcium activated neutral protease 1 |
| 19 | synthetic construct | 47.80 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 47.80 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Sus scrofa | 47.00 | 1 | calpain 1, large subunit |
| 22 | Gallus gallus | 41.60 | 1 | mu-calpain large subunit |
| 23 | Canis familiaris | 37.40 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 24 | Danio rerio | 36.20 | 1 | calpain 1, large subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |
| Rattus norvegicus | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |
| Mus musculus | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |||||+|||||#|||||||||||||||||||||||||||||| Sbjct 1 MAGIAIKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |
| Homo sapiens | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 ||||| |||||#|||||||||||||||||||||| ||||||| Sbjct 1 MAGIAAKLAKD#REAAEGLGSHERAIKYLNQDYEALRNECLE 41 |
| Homo sapiens | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 ||||| |||||#|||||||||||||||||||||| ||||||| Sbjct 1 MAGIAAKLAKD#REAAEGLGSHERAIKYLNQDYEALRNECLE 41 |
| Macaca mulatta | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 ||||| || ||#|||||||||||||||||||||| ||||||| Sbjct 1 MAGIAAKLVKD#REAAEGLGSHERAIKYLNQDYEALRNECLE 41 |
| Xenopus tropicalis | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |+|+| +|||+#| | |||++|+|+ ||+|||| ||||||+ Sbjct 1 MSGVADRLAKE#RALASGLGTNEKALPYLSQDYEALRNECLQ 41 |
| Xenopus laevis | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |+||| |+|||#| |+|||++++ +|||||||| |+ ||| Sbjct 1 MSGIASKVAKD#RVLAQGLGTNDKPLKYLNQDYEQLKARCLE 41 |
| Gallus gallus | Query 2 AGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 ||| +| +|#| |||+| | |+|||||||| |+ ||+| Sbjct 5 GGIAARLQRD#RLRAEGVGEHNNAVKYLNQDYEALKQECIE 44 |
| Canis familiaris | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |+ +| ++|| #+ || |||||++|+||| ||+| || +||+ Sbjct 94 MSALAARIAKQ#QAAAGGLGSHQKAVKYLGQDFEKLRQQCLD 134 |
| Coturnix coturnix | Query 2 AGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 ||| +| +|#| |||+| | |+|||||||| |+ |+| Sbjct 5 GGIAARLQRD#RLRAEGVGEHNNAVKYLNQDYEALKQACIE 44 |
| Danio rerio | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECL 40 |+|+| ||| #| | | |++ |+|||||++||||+||| Sbjct 1 MSGVASTLAKK#RALAAGFGTNSNAVKYLNQNFETLRSECL 40 |
| Stizostedion vitreum vitreum | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |+||| | + #|| +|+|| +|+|||||||| || ||| Sbjct 1 MSGIAATLQRR#REKEQGIGSCSQAVKYLNQDYEALRRRCLE 41 |
| Xenopus laevis | Query 2 AGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 ||| || |#| | |+|||+ |+|| |||||+|+ +|+| Sbjct 5 GGIASKLKID#RLKASGVGSHDCAVKYQNQDYESLKQQCVE 44 |
| Tetraodon nigroviridis | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |+||| || #|+ |+| |+ +| ||||||+|+|| ||| Sbjct 1 MSGIAAKLQHH#RDRAQGFGTASQAQKYLNQDFESLRESCLE 41 |
| Oncorhynchus mykiss | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECL 40 |+|+| ||| #| | | |++ |+||||||+| || +|| Sbjct 1 MSGMASNLAKK#RALAAGFGTNANAVKYLNQDFEALRAQCL 40 |
| Rattus norvegicus | Query 3 GIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |++ ++ | #|+ ||| || ||||| |||| || ||+ Sbjct 13 GVSAQVQKQ#RDKELGLGRHENAIKYLGQDYENLRARCLQ 51 |
| Bos taurus | Query 3 GIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |++ ++ | #| ||| || ||||| |||| || ||+ Sbjct 13 GVSAQVQKQ#RAKELGLGRHENAIKYLGQDYEQLRVHCLQ 51 |
| synthetic construct | Query 3 GIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |++ ++ | #| ||| || ||||| |||| || ||+ Sbjct 13 GVSAQVQKQ#RARELGLGRHENAIKYLGQDYEQLRVRCLQ 51 |
| Pongo pygmaeus | Query 3 GIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |++ ++ | #| ||| || ||||| |||| || ||+ Sbjct 13 GVSAQVQKQ#RARELGLGRHENAIKYLGQDYEQLRVRCLQ 51 |
| Sus scrofa | Query 3 GIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECLE 41 |++ ++ | #| ||| || ||||| |||| || ||+ Sbjct 13 GVSAQVQKL#RAKELGLGRHENAIKYLGQDYEQLRAHCLQ 51 |
| Gallus gallus | Query 3 GIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECL 40 |++ ++ + #| | ||| |+ |+++ ||| ||++|| Sbjct 13 GVSAQVQRQ#RAKALGLGQHQNAVRFRGQDYAALRDDCL 50 |
| Canis familiaris | Query 1 MAGIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNECL 40 + |+| ++ #| |+||| | | | ||++| |+ || Sbjct 83 LEGMAAHISHS#RLKAKGLGQHHNAQNYNNQNFEDLQAACL 122 |
| Danio rerio | Query 3 GIAMKLAKD#REAAEGLGSHERAIKYLNQDYETLRNE 38 |+| +| # + ||| + |+|+| ||||||| + Sbjct 8 GMAARLRSQ#WDRDAGLGQNHNAVKFLGQDYETLRAQ 43 |
[Site 8] GIAMKLAKDR12-EAAEGLGSHE
Arg12  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly3 | Ile4 | Ala5 | Met6 | Lys7 | Leu8 | Ala9 | Lys10 | Asp11 | Arg12 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu13 | Ala14 | Ala15 | Glu16 | Gly17 | Leu18 | Gly19 | Ser20 | His21 | Glu22 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDYETLRNECLEA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 89.00 | 12 | A Chain A, Crystal Structure Of A Mu-Like Calpain |
| 2 | Rattus norvegicus | 87.80 | 5 | calpain 2 |
| 3 | Mus musculus | 86.30 | 14 | calpain 2 |
| 4 | Homo sapiens | 84.30 | 6 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 84.30 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 82.80 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus tropicalis | 60.10 | 4 | calpain 2, (m/II) large subunit |
| 8 | Xenopus laevis | 58.90 | 4 | calpain 8 |
| 9 | Gallus gallus | 57.80 | 2 | calpain 1, (mu/I) large subunit |
| 10 | Canis familiaris | 57.40 | 3 | PREDICTED: similar to calpain 8 |
| 11 | Coturnix coturnix | 55.50 | 1 | quail calpain |
| 12 | Stizostedion vitreum vitreum | 54.70 | 1 | putative calpain-like protein |
| 13 | Danio rerio | 54.30 | 6 | calpain 2, (m/II) large subunit a |
| 14 | Xenopus laevis | 53.10 | 1 | mu/m-calpain large subunit |
| 15 | Tetraodon nigroviridis | 51.60 | 7 | unnamed protein product |
| 16 | Oncorhynchus mykiss | 51.60 | 1 | m-calpain |
| 17 | Rattus norvegicus | 49.70 | 1 | calpain 1, large subunit |
| 18 | Bos taurus | 48.50 | 2 | micromolar calcium activated neutral protease 1 |
| 19 | synthetic construct | 48.50 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 48.10 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Sus scrofa | 47.40 | 1 | calpain 1, large subunit |
| 22 | Gallus gallus | 42.40 | 1 | mu-calpain large subunit |
| 23 | Canis familiaris | 37.40 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 24 | Danio rerio | 36.20 | 1 | calpain 1, large subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 ||||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |
| Rattus norvegicus | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 ||||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |
| Mus musculus | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |||||+||||||#|||||||||||||||||||||||||||||| Sbjct 1 MAGIAIKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |
| Homo sapiens | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 ||||| ||||||#||||||||||||||||||||| |||||||| Sbjct 1 MAGIAAKLAKDR#EAAEGLGSHERAIKYLNQDYEALRNECLEA 42 |
| Homo sapiens | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 ||||| ||||||#||||||||||||||||||||| |||||||| Sbjct 1 MAGIAAKLAKDR#EAAEGLGSHERAIKYLNQDYEALRNECLEA 42 |
| Macaca mulatta | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 ||||| || |||#||||||||||||||||||||| |||||||| Sbjct 1 MAGIAAKLVKDR#EAAEGLGSHERAIKYLNQDYEALRNECLEA 42 |
| Xenopus tropicalis | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |+|+| +|||+|# | |||++|+|+ ||+|||| ||||||++ Sbjct 1 MSGVADRLAKER#ALASGLGTNEKALPYLSQDYEALRNECLQS 42 |
| Xenopus laevis | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |+||| |+||||# |+|||++++ +|||||||| |+ |||+ Sbjct 1 MSGIASKVAKDR#VLAQGLGTNDKPLKYLNQDYEQLKARCLES 42 |
| Gallus gallus | Query 2 AGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 ||| +| +||# |||+| | |+|||||||| |+ ||+|+ Sbjct 5 GGIAARLQRDR#LRAEGVGEHNNAVKYLNQDYEALKQECIES 45 |
| Canis familiaris | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |+ +| ++|| +# || |||||++|+||| ||+| || +||+| Sbjct 94 MSALAARIAKQQ#AAAGGLGSHQKAVKYLGQDFEKLRQQCLDA 135 |
| Coturnix coturnix | Query 2 AGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 ||| +| +||# |||+| | |+|||||||| |+ |+|+ Sbjct 5 GGIAARLQRDR#LRAEGVGEHNNAVKYLNQDYEALKQACIES 45 |
| Stizostedion vitreum vitreum | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |+||| | + |#| +|+|| +|+|||||||| || |||+ Sbjct 1 MSGIAATLQRRR#EKEQGIGSCSQAVKYLNQDYEALRRRCLES 42 |
| Danio rerio | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECL 40 |+|+| ||| |# | | |++ |+|||||++||||+||| Sbjct 1 MSGVASTLAKKR#ALAAGFGTNSNAVKYLNQNFETLRSECL 40 |
| Xenopus laevis | Query 2 AGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 ||| || ||# | |+|||+ |+|| |||||+|+ +|+|+ Sbjct 5 GGIASKLKIDR#LKASGVGSHDCAVKYQNQDYESLKQQCVES 45 |
| Tetraodon nigroviridis | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLE 41 |+||| || |#+ |+| |+ +| ||||||+|+|| ||| Sbjct 1 MSGIAAKLQHHR#DRAQGFGTASQAQKYLNQDFESLRESCLE 41 |
| Oncorhynchus mykiss | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECL 40 |+|+| ||| |# | | |++ |+||||||+| || +|| Sbjct 1 MSGMASNLAKKR#ALAAGFGTNANAVKYLNQDFEALRAQCL 40 |
| Rattus norvegicus | Query 3 GIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLE 41 |++ ++ | |#+ ||| || ||||| |||| || ||+ Sbjct 13 GVSAQVQKQR#DKELGLGRHENAIKYLGQDYENLRARCLQ 51 |
| Bos taurus | Query 3 GIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLE 41 |++ ++ | |# ||| || ||||| |||| || ||+ Sbjct 13 GVSAQVQKQR#AKELGLGRHENAIKYLGQDYEQLRVHCLQ 51 |
| synthetic construct | Query 3 GIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |++ ++ | |# ||| || ||||| |||| || ||++ Sbjct 13 GVSAQVQKQR#ARELGLGRHENAIKYLGQDYEQLRVRCLQS 52 |
| Pongo pygmaeus | Query 3 GIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |++ ++ | |# ||| || ||||| |||| || ||++ Sbjct 13 GVSAQVQKQR#ARELGLGRHENAIKYLGQDYEQLRVRCLQS 52 |
| Sus scrofa | Query 3 GIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |++ ++ | |# ||| || ||||| |||| || ||++ Sbjct 13 GVSAQVQKLR#AKELGLGRHENAIKYLGQDYEQLRAHCLQS 52 |
| Gallus gallus | Query 3 GIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECLEA 42 |++ ++ + |# | ||| |+ |+++ ||| ||++|| + Sbjct 13 GVSAQVQRQR#AKALGLGQHQNAVRFRGQDYAALRDDCLRS 52 |
| Canis familiaris | Query 1 MAGIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNECL 40 + |+| ++ |# |+||| | | | ||++| |+ || Sbjct 83 LEGMAAHISHSR#LKAKGLGQHHNAQNYNNQNFEDLQAACL 122 |
| Danio rerio | Query 3 GIAMKLAKDR#EAAEGLGSHERAIKYLNQDYETLRNE 38 |+| +| #+ ||| + |+|+| ||||||| + Sbjct 8 GMAARLRSQW#DRDAGLGQNHNAVKFLGQDYETLRAQ 43 |
[Site 9] IAMKLAKDRE13-AAEGLGSHER
Glu13  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile4 | Ala5 | Met6 | Lys7 | Leu8 | Ala9 | Lys10 | Asp11 | Arg12 | Glu13 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala14 | Ala15 | Glu16 | Gly17 | Leu18 | Gly19 | Ser20 | His21 | Glu22 | Arg23 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDYETLRNECLEAG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 91.70 | 12 | A Chain A, Crystal Structure Of A Mu-Like Calpain |
| 2 | Rattus norvegicus | 90.10 | 5 | calpain 2 |
| 3 | Mus musculus | 88.60 | 14 | calpain 2 |
| 4 | Homo sapiens | 86.70 | 6 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 86.70 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 85.10 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus tropicalis | 62.40 | 4 | calpain 2, (m/II) large subunit |
| 8 | Gallus gallus | 60.10 | 2 | calpain 1, (mu/I) large subunit |
| 9 | Xenopus laevis | 59.30 | 4 | calpain 8 |
| 10 | Canis familiaris | 59.30 | 3 | PREDICTED: similar to calpain 8 |
| 11 | Coturnix coturnix | 57.80 | 1 | quail calpain |
| 12 | Danio rerio | 55.80 | 6 | calpain 2, (m/II) large subunit a |
| 13 | Xenopus laevis | 55.50 | 1 | mu/m-calpain large subunit |
| 14 | Stizostedion vitreum vitreum | 54.70 | 1 | putative calpain-like protein |
| 15 | Tetraodon nigroviridis | 53.50 | 7 | unnamed protein product |
| 16 | Oncorhynchus mykiss | 53.10 | 1 | m-calpain |
| 17 | Rattus norvegicus | 51.60 | 1 | calpain 1, large subunit |
| 18 | synthetic construct | 50.80 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 19 | Pongo pygmaeus | 50.80 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | Bos taurus | 50.10 | 2 | micromolar calcium activated neutral protease 1 |
| 21 | Sus scrofa | 49.70 | 1 | calpain 1, large subunit |
| 22 | Gallus gallus | 44.70 | 1 | mu-calpain large subunit |
| 23 | Canis familiaris | 38.90 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 24 | Danio rerio | 36.20 | 1 | calpain 1, large subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |||||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |
| Rattus norvegicus | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |||||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |
| Mus musculus | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |||||+|||||||#|||||||||||||||||||||||||||||| Sbjct 1 MAGIAIKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |
| Homo sapiens | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 ||||| |||||||#|||||||||||||||||||| ||||||||| Sbjct 1 MAGIAAKLAKDRE#AAEGLGSHERAIKYLNQDYEALRNECLEAG 43 |
| Homo sapiens | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 ||||| |||||||#|||||||||||||||||||| ||||||||| Sbjct 1 MAGIAAKLAKDRE#AAEGLGSHERAIKYLNQDYEALRNECLEAG 43 |
| Macaca mulatta | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 ||||| || ||||#|||||||||||||||||||| ||||||||| Sbjct 1 MAGIAAKLVKDRE#AAEGLGSHERAIKYLNQDYEALRNECLEAG 43 |
| Xenopus tropicalis | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |+|+| +|||+| # | |||++|+|+ ||+|||| ||||||++| Sbjct 1 MSGVADRLAKERA#LASGLGTNEKALPYLSQDYEALRNECLQSG 43 |
| Gallus gallus | Query 2 AGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 ||| +| +|| # |||+| | |+|||||||| |+ ||+|+| Sbjct 5 GGIAARLQRDRL#RAEGVGEHNNAVKYLNQDYEALKQECIESG 46 |
| Xenopus laevis | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEA 42 |+||| |+|||| # |+|||++++ +|||||||| |+ |||+ Sbjct 1 MSGIASKVAKDRV#LAQGLGTNDKPLKYLNQDYEQLKARCLES 42 |
| Canis familiaris | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |+ +| ++|| + #|| |||||++|+||| ||+| || +||+|| Sbjct 94 MSALAARIAKQQA#AAGGLGSHQKAVKYLGQDFEKLRQQCLDAG 136 |
| Coturnix coturnix | Query 2 AGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 ||| +| +|| # |||+| | |+|||||||| |+ |+|+| Sbjct 5 GGIAARLQRDRL#RAEGVGEHNNAVKYLNQDYEALKQACIESG 46 |
| Danio rerio | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |+|+| ||| | # | | |++ |+|||||++||||+||| | Sbjct 1 MSGVASTLAKKRA#LAAGFGTNSNAVKYLNQNFETLRSECLSRG 43 |
| Xenopus laevis | Query 2 AGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 ||| || || # | |+|||+ |+|| |||||+|+ +|+|+| Sbjct 5 GGIASKLKIDRL#KASGVGSHDCAVKYQNQDYESLKQQCVESG 46 |
| Stizostedion vitreum vitreum | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEA 42 |+||| | + ||# +|+|| +|+|||||||| || |||+ Sbjct 1 MSGIAATLQRRRE#KEQGIGSCSQAVKYLNQDYEALRRRCLES 42 |
| Tetraodon nigroviridis | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |+||| || |+# |+| |+ +| ||||||+|+|| ||| | Sbjct 1 MSGIAAKLQHHRD#RAQGFGTASQAQKYLNQDFESLRESCLERG 43 |
| Oncorhynchus mykiss | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |+|+| ||| | # | | |++ |+||||||+| || +|| | Sbjct 1 MSGMASNLAKKRA#LAAGFGTNANAVKYLNQDFEALRAQCLNRG 43 |
| Rattus norvegicus | Query 3 GIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |++ ++ | |+# ||| || ||||| |||| || ||+ | Sbjct 13 GVSAQVQKQRD#KELGLGRHENAIKYLGQDYENLRARCLQNG 53 |
| synthetic construct | Query 3 GIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |++ ++ | | # ||| || ||||| |||| || ||++| Sbjct 13 GVSAQVQKQRA#RELGLGRHENAIKYLGQDYEQLRVRCLQSG 53 |
| Pongo pygmaeus | Query 3 GIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |++ ++ | | # ||| || ||||| |||| || ||++| Sbjct 13 GVSAQVQKQRA#RELGLGRHENAIKYLGQDYEQLRVRCLQSG 53 |
| Bos taurus | Query 3 GIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |++ ++ | | # ||| || ||||| |||| || ||+ | Sbjct 13 GVSAQVQKQRA#KELGLGRHENAIKYLGQDYEQLRVHCLQRG 53 |
| Sus scrofa | Query 3 GIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |++ ++ | | # ||| || ||||| |||| || ||++| Sbjct 13 GVSAQVQKLRA#KELGLGRHENAIKYLGQDYEQLRAHCLQSG 53 |
| Gallus gallus | Query 3 GIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 |++ ++ + | # | ||| |+ |+++ ||| ||++|| +| Sbjct 13 GVSAQVQRQRA#KALGLGQHQNAVRFRGQDYAALRDDCLRSG 53 |
| Canis familiaris | Query 1 MAGIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNECLEAG 43 + |+| ++ | # |+||| | | | ||++| |+ || | Sbjct 83 LEGMAAHISHSRL#KAKGLGQHHNAQNYNNQNFEDLQAACLRKG 125 |
| Danio rerio | Query 3 GIAMKLAKDRE#AAEGLGSHERAIKYLNQDYETLRNE 38 |+| +| +# ||| + |+|+| ||||||| + Sbjct 8 GMAARLRSQWD#RDAGLGQNHNAVKFLGQDYETLRAQ 43 |
[Site 10] AMKLAKDREA14-AEGLGSHERA
Ala14  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala5 | Met6 | Lys7 | Leu8 | Ala9 | Lys10 | Asp11 | Arg12 | Glu13 | Ala14 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala15 | Glu16 | Gly17 | Leu18 | Gly19 | Ser20 | His21 | Glu22 | Arg23 | Ala24 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDYETLRNECLEAGA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 93.60 | 12 | A Chain A, Crystal Structure Of A Mu-Like Calpain |
| 2 | Rattus norvegicus | 92.00 | 5 | calpain 2 |
| 3 | Mus musculus | 90.50 | 14 | calpain 2 |
| 4 | Homo sapiens | 87.00 | 2 | calpain 2, large subunit |
| 5 | Homo sapiens | 86.70 | 6 | AF261089_1 calpain large polypeptide L2 |
| 6 | Macaca mulatta | 85.50 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus tropicalis | 62.80 | 4 | calpain 2, (m/II) large subunit |
| 8 | Gallus gallus | 60.10 | 2 | calpain 1, (mu/I) large subunit |
| 9 | Xenopus laevis | 59.30 | 4 | calpain 8 |
| 10 | Canis familiaris | 59.30 | 3 | PREDICTED: similar to calpain 8 |
| 11 | Coturnix coturnix | 59.30 | 1 | quail calpain |
| 12 | Danio rerio | 55.80 | 6 | calpain 2, (m/II) large subunit a |
| 13 | Xenopus laevis | 55.50 | 2 | mu/m-calpain large subunit |
| 14 | Stizostedion vitreum vitreum | 54.70 | 1 | putative calpain-like protein |
| 15 | Tetraodon nigroviridis | 53.50 | 7 | unnamed protein product |
| 16 | Oncorhynchus mykiss | 53.10 | 1 | m-calpain |
| 17 | Bos taurus | 52.00 | 2 | micromolar calcium activated neutral protease 1 |
| 18 | Rattus norvegicus | 52.00 | 1 | calpain 1, large subunit |
| 19 | synthetic construct | 51.20 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 50.80 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Sus scrofa | 50.10 | 1 | calpain 1, large subunit |
| 22 | Gallus gallus | 45.10 | 1 | mu-calpain large subunit |
| 23 | Canis familiaris | 38.90 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 24 | Danio rerio | 36.20 | 1 | calpain 1, large subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAGA 44 ||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAGA 44 |
| Rattus norvegicus | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAGA 44 ||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAGA 44 |
| Mus musculus | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAGA 44 |||||+||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MAGIAIKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAGA 44 |
| Homo sapiens | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 ||||| ||||||||#||||||||||||||||||| ||||||||| Sbjct 1 MAGIAAKLAKDREA#AEGLGSHERAIKYLNQDYEALRNECLEAG 43 |
| Homo sapiens | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 ||||| ||||||||#||||||||||||||||||| ||||||||| Sbjct 1 MAGIAAKLAKDREA#AEGLGSHERAIKYLNQDYEALRNECLEAG 43 |
| Macaca mulatta | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 ||||| || |||||#||||||||||||||||||| ||||||||| Sbjct 1 MAGIAAKLVKDREA#AEGLGSHERAIKYLNQDYEALRNECLEAG 43 |
| Xenopus tropicalis | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 |+|+| +|||+| #| |||++|+|+ ||+|||| ||||||++| Sbjct 1 MSGVADRLAKERAL#ASGLGTNEKALPYLSQDYEALRNECLQSG 43 |
| Gallus gallus | Query 2 AGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 ||| +| +|| #|||+| | |+|||||||| |+ ||+|+| Sbjct 5 GGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQECIESG 46 |
| Xenopus laevis | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEA 42 |+||| |+|||| #|+|||++++ +|||||||| |+ |||+ Sbjct 1 MSGIASKVAKDRVL#AQGLGTNDKPLKYLNQDYEQLKARCLES 42 |
| Canis familiaris | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 |+ +| ++|| + |#| |||||++|+||| ||+| || +||+|| Sbjct 94 MSALAARIAKQQAA#AGGLGSHQKAVKYLGQDFEKLRQQCLDAG 136 |
| Coturnix coturnix | Query 2 AGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAGA 44 ||| +| +|| #|||+| | |+|||||||| |+ |+|+|| Sbjct 5 GGIAARLQRDRLR#AEGVGEHNNAVKYLNQDYEALKQACIESGA 47 |
| Danio rerio | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 |+|+| ||| | #| | |++ |+|||||++||||+||| | Sbjct 1 MSGVASTLAKKRAL#AAGFGTNSNAVKYLNQNFETLRSECLSRG 43 |
| Xenopus laevis | Query 2 AGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 ||| || || #| |+|||+ |+|| |||||+|+ +|+|+| Sbjct 5 GGIASKLKIDRLK#ASGVGSHDCAVKYQNQDYESLKQQCVESG 46 |
| Stizostedion vitreum vitreum | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEA 42 |+||| | + || # +|+|| +|+|||||||| || |||+ Sbjct 1 MSGIAATLQRRREK#EQGIGSCSQAVKYLNQDYEALRRRCLES 42 |
| Tetraodon nigroviridis | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 |+||| || |+ #|+| |+ +| ||||||+|+|| ||| | Sbjct 1 MSGIAAKLQHHRDR#AQGFGTASQAQKYLNQDFESLRESCLERG 43 |
| Oncorhynchus mykiss | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAGA 44 |+|+| ||| | #| | |++ |+||||||+| || +|| | Sbjct 1 MSGMASNLAKKRAL#AAGFGTNANAVKYLNQDFEALRAQCLNRGG 44 |
| Bos taurus | Query 3 GIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAGA 44 |++ ++ | | # ||| || ||||| |||| || ||+ || Sbjct 13 GVSAQVQKQRAK#ELGLGRHENAIKYLGQDYEQLRVHCLQRGA 54 |
| Rattus norvegicus | Query 3 GIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 |++ ++ | |+ # ||| || ||||| |||| || ||+ | Sbjct 13 GVSAQVQKQRDK#ELGLGRHENAIKYLGQDYENLRARCLQNG 53 |
| synthetic construct | Query 3 GIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 |++ ++ | | # ||| || ||||| |||| || ||++| Sbjct 13 GVSAQVQKQRAR#ELGLGRHENAIKYLGQDYEQLRVRCLQSG 53 |
| Pongo pygmaeus | Query 3 GIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 |++ ++ | | # ||| || ||||| |||| || ||++| Sbjct 13 GVSAQVQKQRAR#ELGLGRHENAIKYLGQDYEQLRVRCLQSG 53 |
| Sus scrofa | Query 3 GIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAGA 44 |++ ++ | | # ||| || ||||| |||| || ||++|+ Sbjct 13 GVSAQVQKLRAK#ELGLGRHENAIKYLGQDYEQLRAHCLQSGS 54 |
| Gallus gallus | Query 3 GIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAGA 44 |++ ++ + | #| ||| |+ |+++ ||| ||++|| +|+ Sbjct 13 GVSAQVQRQRAK#ALGLGQHQNAVRFRGQDYAALRDDCLRSGS 54 |
| Canis familiaris | Query 1 MAGIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNECLEAG 43 + |+| ++ | #|+||| | | | ||++| |+ || | Sbjct 83 LEGMAAHISHSRLK#AKGLGQHHNAQNYNNQNFEDLQAACLRKG 125 |
| Danio rerio | Query 3 GIAMKLAKDREA#AEGLGSHERAIKYLNQDYETLRNE 38 |+| +| + # ||| + |+|+| ||||||| + Sbjct 8 GMAARLRSQWDR#DAGLGQNHNAVKFLGQDYETLRAQ 43 |
[Site 11] AAEGLGSHER23-AIKYLNQDYE
Arg23  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala14 | Ala15 | Glu16 | Gly17 | Leu18 | Gly19 | Ser20 | His21 | Glu22 | Arg23 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala24 | Ile25 | Lys26 | Tyr27 | Leu28 | Asn29 | Gln30 | Asp31 | Tyr32 | Glu33 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MAGIAMKLAKDREAAEGLGSHERAIKYLNQDYETLRNECLEAGALFQDPSFPA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 112.00 | 12 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 111.00 | 10 | calpain 2 |
| 3 | Mus musculus | 110.00 | 19 | m-calpain large subunit |
| 4 | Homo sapiens | 105.00 | 11 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 105.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 104.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus tropicalis | 77.40 | 5 | calpain 2, (m/II) large subunit |
| 8 | Gallus gallus | 77.40 | 2 | calpain 1, (mu/I) large subunit |
| 9 | Coturnix coturnix | 76.30 | 1 | quail calpain |
| 10 | Canis familiaris | 75.10 | 5 | PREDICTED: similar to calpain 8 |
| 11 | Xenopus laevis | 73.90 | 5 | calpain 8 |
| 12 | Xenopus laevis | 72.40 | 2 | mu/m-calpain large subunit |
| 13 | Danio rerio | 71.60 | 6 | calpain 2, (m/II) large subunit a |
| 14 | Oncorhynchus mykiss | 70.50 | 1 | m-calpain |
| 15 | Stizostedion vitreum vitreum | 68.90 | 1 | putative calpain-like protein |
| 16 | Rattus norvegicus | 65.50 | 1 | calpain 1, large subunit |
| 17 | Tetraodon nigroviridis | 64.30 | 12 | unnamed protein product |
| 18 | Bos taurus | 63.90 | 6 | calpain 1, (mu/I) large subunit |
| 19 | synthetic construct | 63.20 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 62.80 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Sus scrofa | 62.40 | 2 | calpain 1, large subunit |
| 22 | Gallus gallus | 57.40 | 1 | mu-calpain large subunit |
| 23 | Canis familiaris | 51.20 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 24 | Danio rerio | 48.10 | 1 | calpain 1, large subunit |
| 25 | Drosophila melanogaster | 42.00 | 4 | CAA55297.1 |
| 26 | Drosophila melanogaster | 42.00 | 2 | Calpain-A CG7563-PB, isoform B |
| 27 | Oryctolagus cuniculus | 41.20 | 1 | calpain 3, (p94) |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |
| Rattus norvegicus | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |
| Mus musculus | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |||||+|||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 MAGIAIKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |
| Homo sapiens | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 ||||| |||||||||||||||||#|||||||||| ||||||||| ||||||||| Sbjct 1 MAGIAAKLAKDREAAEGLGSHER#AIKYLNQDYEALRNECLEAGTLFQDPSFPA 53 |
| Homo sapiens | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 ||||| |||||||||||||||||#|||||||||| ||||||||| ||||||||| Sbjct 1 MAGIAAKLAKDREAAEGLGSHER#AIKYLNQDYEALRNECLEAGTLFQDPSFPA 53 |
| Macaca mulatta | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 ||||| || ||||||||||||||#|||||||||| ||||||||| ||||||||| Sbjct 1 MAGIAAKLVKDREAAEGLGSHER#AIKYLNQDYEALRNECLEAGTLFQDPSFPA 53 |
| Xenopus tropicalis | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |+|+| +|||+| | |||++|+#|+ ||+|||| ||||||++| ||+| |||| Sbjct 1 MSGVADRLAKERALASGLGTNEK#ALPYLSQDYEALRNECLQSGTLFKDLSFPA 53 |
| Gallus gallus | Query 2 AGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 ||| +| +|| |||+| | #|+|||||||| |+ ||+|+| ||+|| ||| Sbjct 5 GGIAARLQRDRLRAEGVGEHNN#AVKYLNQDYEALKQECIESGTLFRDPQFPA 56 |
| Coturnix coturnix | Query 2 AGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 ||| +| +|| |||+| | #|+|||||||| |+ |+|+||||+|| ||| Sbjct 5 GGIAARLQRDRLRAEGVGEHNN#AVKYLNQDYEALKQACIESGALFRDPQFPA 56 |
| Canis familiaris | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |+ +| ++|| + || |||||++#|+||| ||+| || +||+|| ||+|| ||| Sbjct 94 MSALAARIAKQQAAAGGLGSHQK#AVKYLGQDFEKLRQQCLDAGVLFKDPEFPA 146 |
| Xenopus laevis | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |+||| |+|||| |+|||++++# +|||||||| |+ |||+ ||+| +||| Sbjct 1 MSGIASKVAKDRVLAQGLGTNDK#PLKYLNQDYEQLKARCLESNTLFEDETFPA 53 |
| Xenopus laevis | Query 2 AGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 ||| || || | |+|||+ #|+|| |||||+|+ +|+|+| ||+||+||| Sbjct 5 GGIASKLKIDRLKASGVGSHDC#AVKYQNQDYESLKQQCVESGILFEDPNFPA 56 |
| Danio rerio | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |+|+| ||| | | | |++ #|+|||||++||||+||| | || ||+||| Sbjct 1 MSGVASTLAKKRALAAGFGTNSN#AVKYLNQNFETLRSECLSRGELFCDPAFPA 53 |
| Oncorhynchus mykiss | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |+|+| ||| | | | |++ #|+||||||+| || +|| | || ||+||| Sbjct 1 MSGMASNLAKKRALAAGFGTNAN#AVKYLNQDFEALRAQCLNRGGLFSDPTFPA 53 |
| Stizostedion vitreum vitreum | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |+||| | + || +|+|| +#|+|||||||| || |||+ +||+| +||| Sbjct 1 MSGIAATLQRRREKEQGIGSCSQ#AVKYLNQDYEALRRRCLESESLFKDDAFPA 53 |
| Rattus norvegicus | Query 3 GIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFP 52 |++ ++ | |+ ||| || #||||| |||| || ||+ | |||| +|| Sbjct 13 GVSAQVQKQRDKELGLGRHEN#AIKYLGQDYENLRARCLQNGVLFQDDAFP 62 |
| Tetraodon nigroviridis | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |+||| || |+ |+| |+ +#| ||||||+|+|| ||| | |||| | | Sbjct 1 MSGIAAKLQHHRDRAQGFGTASQ#AQKYLNQDFESLRESCLERGELFQDEYFEA 53 |
| Bos taurus | Query 3 GIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFP 52 |++ ++ | | ||| || #||||| |||| || ||+ ||||+| +|| Sbjct 13 GVSAQVQKQRAKELGLGRHEN#AIKYLGQDYEQLRVHCLQRGALFRDEAFP 62 |
| synthetic construct | Query 3 GIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFP 52 |++ ++ | | ||| || #||||| |||| || ||++| ||+| +|| Sbjct 13 GVSAQVQKQRARELGLGRHEN#AIKYLGQDYEQLRVRCLQSGTLFRDEAFP 62 |
| Pongo pygmaeus | Query 3 GIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFP 52 |++ ++ | | ||| || #||||| |||| || ||++| ||+| +|| Sbjct 13 GVSAQVQKQRARELGLGRHEN#AIKYLGQDYEQLRVRCLQSGTLFRDEAFP 62 |
| Sus scrofa | Query 3 GIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFP 52 |++ ++ | | ||| || #||||| |||| || ||++|+||+| +|| Sbjct 13 GVSAQVQKLRAKELGLGRHEN#AIKYLGQDYEQLRAHCLQSGSLFRDEAFP 62 |
| Gallus gallus | Query 3 GIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFP 52 |++ ++ + | | ||| |+ #|+++ ||| ||++|| +|+||+| +|| Sbjct 13 GVSAQVQRQRAKALGLGQHQN#AVRFRGQDYAALRDDCLRSGSLFRDETFP 62 |
| Canis familiaris | Query 1 MAGIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 + |+| ++ | |+||| | #| | ||++| |+ || | ||+| ||| Sbjct 83 LEGMAAHISHSRLKAKGLGQHHN#AQNYNNQNFEDLQAACLRKGELFEDSFFPA 135 |
| Danio rerio | Query 3 GIAMKLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 |+| +| + ||| + #|+|+| ||||||| + ++ ||+|| | | Sbjct 8 GMAARLRSQWDRDAGLGQNHN#AVKFLGQDYETLRAQSQQSRRLFEDPMFTA 58 |
| Drosophila melanogaster | Query 30 QDYETLRNECLEAGALFQDPSFPA 53 |||||+ | || +|+||+|| ||| Sbjct 50 QDYETILNSCLASGSLFEDPLFPA 73 |
| Drosophila melanogaster | Query 30 QDYETLRNECLEAGALFQDPSFPA 53 |||||+ | || +|+||+|| ||| Sbjct 73 QDYETILNSCLASGSLFEDPLFPA 96 |
| Oryctolagus cuniculus | Query 7 KLAKDREAAEGLGSHER#AIKYLNQDYETLRNECLEAGALFQDPSFPA 53 ++ ++|+ | |+ +# ||| ||+ || || ||+| ||| Sbjct 12 RVIRERDRRNGEGTVTQ#PIKYEGQDFVVLRQRCLAQKCLFEDRVFPA 58 |
[Site 12] FPALPSSLGF60-KELGPYSSKT
Phe60  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe51 | Pro52 | Ala53 | Leu54 | Pro55 | Ser56 | Ser57 | Leu58 | Gly59 | Phe60 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys61 | Glu62 | Leu63 | Gly64 | Pro65 | Tyr66 | Ser67 | Ser68 | Lys69 | Thr70 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DYETLRNECLEAGALFQDPSFPALPSSLGFKELGPYSSKTRGIEWKRPTEICADPQFIIG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 129.00 | 13 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 128.00 | 11 | calpain 2 |
| 3 | Mus musculus | 127.00 | 21 | m-calpain |
| 4 | Homo sapiens | 123.00 | 7 | AF261089_1 calpain large polypeptide L2 |
| 5 | Macaca mulatta | 123.00 | 2 | PREDICTED: calpain 2, large subunit |
| 6 | Homo sapiens | 123.00 | 2 | calpain 2, large subunit |
| 7 | Gallus gallus | 107.00 | 3 | calpain 1, (mu/I) large subunit |
| 8 | Xenopus laevis | 105.00 | 5 | hypothetical protein LOC398772 |
| 9 | Coturnix coturnix | 104.00 | 1 | quail calpain |
| 10 | Xenopus laevis | 102.00 | 1 | mu/m-calpain large subunit |
| 11 | Danio rerio | 99.40 | 5 | PREDICTED: similar to Calpain 1, (mu/I) large sub |
| 12 | Tetraodon nigroviridis | 99.00 | 13 | unnamed protein product |
| 13 | Xenopus tropicalis | 95.50 | 5 | CAPN2 protein |
| 14 | Oncorhynchus mykiss | 93.20 | 1 | m-calpain |
| 15 | Bos taurus | 92.80 | 5 | micromolar calcium activated neutral protease 1 |
| 16 | Canis familiaris | 91.30 | 7 | PREDICTED: similar to calpain 8 |
| 17 | Sus scrofa | 89.40 | 4 | calpain 1, large subunit |
| 18 | Gallus gallus | 89.00 | 1 | mu-calpain large subunit |
| 19 | Rattus norvegicus | 88.60 | 1 | calpain 1, large subunit |
| 20 | Pongo pygmaeus | 87.00 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | synthetic construct | 87.00 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 22 | Stizostedion vitreum vitreum | 84.30 | 1 | putative calpain-like protein |
| 23 | Danio rerio | 82.00 | 1 | calpain 1, large subunit |
| 24 | Canis familiaris | 67.80 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 25 | Oryctolagus cuniculus | 62.40 | 1 | calpain 3, (p94) |
| 26 | Oncorhynchus mykiss | 61.20 | 1 | gill-specific calpain |
| 27 | Drosophila melanogaster | 57.80 | 5 | CAA55297.1 |
| 28 | Drosophila melanogaster | 57.80 | 3 | Calpain-A CG7563-PB, isoform B |
| 29 | Drosophila pseudoobscura | 52.80 | 1 | GA20829-PA |
| 30 | Caenorhabditis elegans | 52.40 | 2 | CaLPain family member (clp-4) |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |
| Rattus norvegicus | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |
| Mus musculus | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |||||||||||||||||||||||||||||+#|||||||||||||||||||||||||||||| Sbjct 31 DYETLRNECLEAGALFQDPSFPALPSSLGY#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |
| Homo sapiens | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 ||| ||||||||| |||||||||+||+|||#|||||||||||||||||||||||||||||| Sbjct 31 DYEALRNECLEAGTLFQDPSFPAIPSALGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |
| Macaca mulatta | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 ||| ||||||||| |||||||||+||+|||#|||||||||||||||||||||||||||||| Sbjct 31 DYEALRNECLEAGTLFQDPSFPAIPSALGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |
| Homo sapiens | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 ||| ||||||||| |||||||||+||+|||#|||||||||||||||||||||||||||||| Sbjct 31 DYEALRNECLEAGTLFQDPSFPAIPSALGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |
| Gallus gallus | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 ||| |+ ||+|+| ||+|| ||| |++|||#||||||||||||+|||||+|+ |||||+| Sbjct 34 DYEALKQECIESGTLFRDPQFPAGPTALGF#KELGPYSSKTRGVEWKRPSELVDDPQFIVG 93 |
| Xenopus laevis | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |||+|+ +|||+| ||+|| ||| ||||||#||||| ||||||++||||+|| |||||+| Sbjct 34 DYESLKQQCLESGILFEDPHFPATPSSLGF#KELGPGSSKTRGVQWKRPSEIVDDPQFIVG 93 |
| Coturnix coturnix | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 ||| |+ |+|+||||+|| ||| |++|||#|||||+||||||+|||||+|+ |||||+| Sbjct 34 DYEALKQACIESGALFRDPQFPAGPTALGF#KELGPHSSKTRGVEWKRPSELVDDPQFIVG 93 |
| Xenopus laevis | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |||+|+ +|+|+| ||+||+|||+|| |||#||||| ||||||++||||++| ||||||| Sbjct 34 DYESLKQQCVESGILFEDPNFPAIPSILGF#KELGPGSSKTRGVQWKRPSDIVDDPQFIIG 93 |
| Danio rerio | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 ||| || ||+|+| ||+|| |||+| ||||#||| | ||||||++| ||||+ +||||+| Sbjct 34 DYEELRRECVESGRLFEDPCFPAVPQSLGF#KELAPNSSKTRGVKWIRPTELSENPQFIVG 93 |
| Tetraodon nigroviridis | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 ||| |+ +|+|+| ||+|| | | |+||||#||| |+||||||+||+||||+ |||||+| Sbjct 29 DYEALKQQCVESGCLFEDPLFLAQPASLGF#KELAPHSSKTRGVEWQRPTELTGDPQFIVG 88 |
| Xenopus tropicalis | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |||+|+ +| |+|+||+||+|| + |||||#||||| +|||+|++|||| +| |||||+| Sbjct 34 DYESLKQQCRESGSLFEDPNFPPIGSSLGF#KELGPGTSKTKGVQWKRPHDIADDPQFIVG 93 |
| Oncorhynchus mykiss | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |+| || +|| | || ||+||| | ||||# |||| | ||| ++|||| |+|+ |+||+| Sbjct 31 DFEALRAQCLNRGGLFSDPTFPAAPESLGF#NELGPGSYKTRAVQWKRPGELCSSPEFIVG 90 |
| Bos taurus | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 ||| || ||+ ||||+| +|| +| ||||#||||| |||| ||+||||||+ ++||||+ Sbjct 41 DYEQLRVHCLQRGALFRDEAFPPVPQSLGF#KELGPNSSKTYGIKWKRPTELFSNPQFIV 99 |
| Canis familiaris | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |+| || +||+|| ||+|| ||| ||+|| #|+||| | +|+|| |||| |+||+||||+| Sbjct 124 DFEKLRQQCLDAGVLFKDPEFPACPSALGH#KDLGPQSPQTQGIVWKRPPELCANPQFIVG 183 |
| Sus scrofa | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 ||| || ||++|+||+| +|| +| ||||#||||| |||| |++||||||+ ++||||+ Sbjct 41 DYEQLRAHCLQSGSLFRDEAFPPVPQSLGF#KELGPNSSKTYGVKWKRPTELFSNPQFIV 99 |
| Gallus gallus | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 || ||++|| +|+||+| +|| |||||#+|||| ||||||+ ||||||+| ||||+ Sbjct 41 DYAALRDDCLRSGSLFRDETFPPSASSLGF#RELGPGSSKTRGVTWKRPTELCRHPQFIV 99 |
| Rattus norvegicus | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 ||| || ||+ | |||| +|| + ||||#||||| |||| ||+||||||+ ++||||+ Sbjct 41 DYENLRARCLQNGVLFQDDAFPPVSHSLGF#KELGPNSSKTYGIKWKRPTELLSNPQFIV 99 |
| Pongo pygmaeus | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 ||| || ||++| ||+| +|| +| |||+#|+||| |||| ||+||||||+ ++||||+ Sbjct 41 DYEQLRVRCLQSGTLFRDEAFPPVPQSLGY#KDLGPNSSKTYGIKWKRPTELLSNPQFIV 99 |
| synthetic construct | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 ||| || ||++| ||+| +|| +| |||+#|+||| |||| ||+||||||+ ++||||+ Sbjct 41 DYEQLRVRCLQSGTLFRDEAFPPVPQSLGY#KDLGPNSSKTYGIKWKRPTELLSNPQFIV 99 |
| Stizostedion vitreum vitreum | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 ||| || |||+ +||+| +||| |||||#|||| | | ||+ |+|||| +|||||+ Sbjct 31 DYEALRRRCLESESLFKDDAFPAETSSLGF#KELGVGSYKIRGVSWERPTEFTSDPQFIV 89 |
| Danio rerio | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 |||||| + ++ ||+|| | | |||||# |||| ||||+|+ | || |+|| ||||+ Sbjct 36 DYETLRAQSQQSRRLFEDPMFTASSSSLGF#NELGPRSSKTQGVRWMRPKEMCARPQFIV 94 |
| Canis familiaris | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 ++| |+ || | ||+| ||| ||||||#|+||| | + | |+|| +| + | ||+ Sbjct 113 NFEDLQAACLRKGELFEDSFFPAEPSSLGF#KDLGPNSRNVQNICWQRPGQITSSPHFIV 171 |
| Oryctolagus cuniculus | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 |+ || || ||+| ||| ||| # || +| + | |||| ||| +|+|||| Sbjct 36 DFVVLRQRCLAQKCLFEDRVFPAGVQSLGA#HELSQ-KTKMKAITWKRPQEICENPRFIIG 94 |
| Oncorhynchus mykiss | Query 32 YETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFIIG 90 +| || |||+ ||+|| ||| ||| +# + | + ||||| |+| +|+||+| Sbjct 15 FEQLRQECLQKKKLFEDPDFPACDSSLYY#SQSVPIN-----FEWKRPGELCENPEFIVG 68 |
| Drosophila melanogaster | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 ||||+ | || +|+||+|| ||| || |# | ||| || || +||| + Sbjct 51 DYETILNSCLASGSLFEDPLFPASNESLQF#SR-----RPDRHIEWLRPHEIAENPQFFV 104 |
| Drosophila melanogaster | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 ||||+ | || +|+||+|| ||| || |# | ||| || || +||| + Sbjct 74 DYETILNSCLASGSLFEDPLFPASNESLQF#SR-----RPDRHIEWLRPHEIAENPQFFV 127 |
| Drosophila pseudoobscura | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFII 89 |+ +||+ |+ | +|+|| ||| +|| |# + | ||| || +| |||| + Sbjct 251 DFASLRDSCVINGTMFEDPDFPANNASLMF#SK-----RPDRYIEWLRPGDIVGDPQFFV 304 |
| Caenorhabditis elegans | Query 31 DYETLRNECLEAGALFQDPSFPALPSSLGF#KELGPYSSKTRGIEWKRPTEICADPQFI 88 |+|| | +|| ||+|| ||| +|| +#+ + | |||| || |+|| | Sbjct 226 DFETEREKCLRNKTLFEDPEFPATAASLYY#R-----TPPRDRIIWKRPGEIIANPQLI 278 |
[Site 13] IEWKRPTEIC82-ADPQFIIGGA
Cys82  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile73 | Glu74 | Trp75 | Lys76 | Arg77 | Pro78 | Thr79 | Glu80 | Ile81 | Cys82 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala83 | Asp84 | Pro85 | Gln86 | Phe87 | Ile88 | Ile89 | Gly90 | Gly91 | Ala92 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ALPSSLGFKELGPYSSKTRGIEWKRPTEICADPQFIIGGATRTDICQGALGDCWLLAAIA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Rattus norvegicus | 127.00 | 13 | calpain 2 |
| 2 | Mus musculus | 126.00 | 21 | m-calpain large subunit |
| 3 | Homo sapiens | 125.00 | 9 | AF261089_1 calpain large polypeptide L2 |
| 4 | Homo sapiens | 125.00 | 2 | calpain 2, large subunit |
| 5 | Macaca mulatta | 125.00 | 2 | PREDICTED: calpain 2, large subunit |
| 6 | N/A | 124.00 | 12 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 7 | Tetraodon nigroviridis | 119.00 | 9 | unnamed protein product |
| 8 | Gallus gallus | 119.00 | 3 | calpain 1, (mu/I) large subunit |
| 9 | Xenopus laevis | 117.00 | 4 | hypothetical protein LOC398772 |
| 10 | Coturnix coturnix | 116.00 | 1 | quail calpain |
| 11 | Danio rerio | 115.00 | 5 | calpain 1, (mu/I) large subunit a |
| 12 | Xenopus laevis | 115.00 | 1 | mu/m-calpain large subunit |
| 13 | Bos taurus | 111.00 | 5 | calpain 1, (mu/I) large subunit |
| 14 | Gallus gallus | 110.00 | 1 | mu-calpain large subunit |
| 15 | Sus scrofa | 109.00 | 4 | calpain 1, large subunit |
| 16 | Canis familiaris | 108.00 | 23 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 17 | Oncorhynchus mykiss | 108.00 | 1 | m-calpain |
| 18 | Rattus norvegicus | 108.00 | 1 | calpain 1, large subunit |
| 19 | Pongo pygmaeus | 108.00 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | synthetic construct | 108.00 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 21 | Xenopus tropicalis | 107.00 | 4 | CAPN2 protein |
| 22 | Danio rerio | 107.00 | 1 | calpain 1, large subunit |
| 23 | Stizostedion vitreum vitreum | 100.00 | 1 | putative calpain-like protein |
| 24 | Oryctolagus cuniculus | 85.10 | 1 | calpain 3, (p94) |
| 25 | Canis familiaris | 84.00 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 26 | Oncorhynchus mykiss | 82.00 | 1 | gill-specific calpain |
| 27 | Ovis aries | 77.80 | 1 | skeletal muscle-specific calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| Rattus norvegicus | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |
| Mus musculus | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |||||||+||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 53 ALPSSLGYKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |
| Homo sapiens | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |+||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 53 AIPSALGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |
| Homo sapiens | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |+||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 53 AIPSALGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |
| Macaca mulatta | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |+||+|||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 53 AIPSALGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |
| N/A | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 ||||||||||||||||||||||||||||||#|||||||||||||||||||||| ||||||| Sbjct 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDSWLLAAIA 112 |
| Tetraodon nigroviridis | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 | |+||||||| |+||||||+||+||||+ # |||||+||||||||||||||||||||||| Sbjct 51 AQPASLGFKELAPHSSKTRGVEWQRPTELT#GDPQFIVGGATRTDICQGALGDCWLLAAIA 110 |
| Gallus gallus | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 | |++|||||||||||||||+|||||+|+ # |||||+|||||||||||||||||||||| Sbjct 56 AGPTALGFKELGPYSSKTRGVEWKRPSELV#DDPQFIVGGATRTDICQGALGDCWLLAAIG 115 |
| Xenopus laevis | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 | ||||||||||| ||||||++||||+|| # |||||+|||||||||||||||||||||| Sbjct 56 ATPSSLGFKELGPGSSKTRGVQWKRPSEIV#DDPQFIVGGATRTDICQGALGDCWLLAAIG 115 |
| Coturnix coturnix | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 | |++||||||||+||||||+|||||+|+ # |||||+|||||||||||||||||||||| Sbjct 56 AGPTALGFKELGPHSSKTRGVEWKRPSELV#DDPQFIVGGATRTDICQGALGDCWLLAAIG 115 |
| Danio rerio | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 | | ||||||| |+||||| +|| ||||+|# |||||+|||||||||||||||||||||| Sbjct 56 AEPPSLGFKELAPHSSKTRDVEWMRPTELC#DDPQFIVGGATRTDICQGALGDCWLLAAIG 115 |
| Xenopus laevis | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |+|| |||||||| ||||||++||||++| # |||||||||||||||||||||||||||| Sbjct 56 AIPSILGFKELGPGSSKTRGVQWKRPSDIV#DDPQFIIGGATRTDICQGALGDCWLLAAIG 115 |
| Bos taurus | Query 54 LPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 +| ||||||||| |||| ||+||||||+ #++||||+ |||||||||||||||||||||| Sbjct 64 VPQSLGFKELGPNSSKTYGIKWKRPTELF#SNPQFIVDGATRTDICQGALGDCWLLAAIA 122 |
| Gallus gallus | Query 56 SSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |||||+|||| ||||||+ ||||||+|# ||||+ |||||||||||||||||||||| Sbjct 66 SSLGFRELGPGSSKTRGVTWKRPTELC#RHPQFIVDGATRTDICQGALGDCWLLAAIA 122 |
| Sus scrofa | Query 54 LPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 +| ||||||||| |||| |++||||||+ #++||||+ |||||||||||||||||||||| Sbjct 64 VPQSLGFKELGPNSSKTYGVKWKRPTELF#SNPQFIVDGATRTDICQGALGDCWLLAAIA 122 |
| Canis familiaris | Query 54 LPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 +| |||+|+||| |||| ||+||||||+ #++||||+ |||||||||||||||||||||| Sbjct 64 VPQSLGYKDLGPNSSKTYGIKWKRPTELL#SNPQFIVDGATRTDICQGALGDCWLLAAIA 122 |
| Oncorhynchus mykiss | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 | | |||| |||| | ||| ++|||| |+|#+ |+||+||||||||||||||||||||||| Sbjct 53 AAPESLGFNELGPGSYKTRAVQWKRPGELC#SSPEFIVGGATRTDICQGALGDCWLLAAIA 112 |
| Rattus norvegicus | Query 57 SLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 ||||||||| |||| ||+||||||+ #++||||+ |||||||||||||||||||||| Sbjct 67 SLGFKELGPNSSKTYGIKWKRPTELL#SNPQFIVDGATRTDICQGALGDCWLLAAIA 122 |
| Pongo pygmaeus | Query 54 LPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 +| |||+|+||| |||| ||+||||||+ #++||||+ |||||||||||||||||||||| Sbjct 64 VPQSLGYKDLGPNSSKTYGIKWKRPTELL#SNPQFIVDGATRTDICQGALGDCWLLAAIA 122 |
| synthetic construct | Query 54 LPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 +| |||+|+||| |||| ||+||||||+ #++||||+ |||||||||||||||||||||| Sbjct 64 VPQSLGYKDLGPNSSKTYGIKWKRPTELL#SNPQFIVDGATRTDICQGALGDCWLLAAIA 122 |
| Xenopus tropicalis | Query 56 SSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |||||||||| +|||+|++|||| +| # |||||+|||||||||||||||||||||| Sbjct 59 SSLGFKELGPGTSKTKGVQWKRPHDIA#DDPQFIVGGATRTDICQGALGDCWLLAAIG 115 |
| Danio rerio | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 | ||||| |||| ||||+|+ | || |+|#| ||||+ |||||||||||||||||||||| Sbjct 58 ASSSSLGFNELGPRSSKTQGVRWMRPKEMC#ARPQFIVDGATRTDICQGALGDCWLLAAIA 117 |
| Stizostedion vitreum vitreum | Query 56 SSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 ||||||||| | | ||+ |+|||| #+|||||+ ||+||||||||||||||||||| Sbjct 56 SSLGFKELGVGSYKIRGVSWERPTEFT#SDPQFIVSGASRTDICQGALGDCWLLAAIA 112 |
| Oryctolagus cuniculus | Query 57 SLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 ||| || +| + | |||| |||# +|+|||||| ||||||| ||||| ||||| Sbjct 62 SLGAHELSQ-KTKMKAITWKRPQEIC#ENPRFIIGGANRTDICQGDLGDCWFLAAIA 116 |
| Canis familiaris | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 | |||||||+||| | + | |+|| +| #+ | ||+ | + |||||| |||||||||| Sbjct 135 AEPSSLGFKDLGPNSRNVQNICWQRPGQIT#SSPHFIVNGFSPTDICQGVLGDCWLLAAIG 194 |
| Oncorhynchus mykiss | Query 53 ALPSSLGFKELGPYSSKTRGIEWKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 | ||| + + | + ||||| |+|# +|+||+||| ||||||||||||||||||| Sbjct 36 ACDSSLYYSQSVPIN-----FEWKRPGELC#ENPEFIVGGADRTDICQGALGDCWLLAAIA 90 |
| Ovis aries | Query 75 WKRPTEIC#ADPQFIIGGATRTDICQGALGDCWLLAAIA 112 |||| |||# +|+|||||| ||||||| ||||| ||||| Sbjct 99 WKRPPEIC#ENPRFIIGGANRTDICQGDLGDCWFLAAIA 136 |
[Site 14] LLAAIASLTL116-NEEILARVVP
Leu116  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu107 | Leu108 | Ala109 | Ala110 | Ile111 | Ala112 | Ser113 | Leu114 | Thr115 | Leu116 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn117 | Glu118 | Glu119 | Ile120 | Leu121 | Ala122 | Arg123 | Val124 | Val125 | Pro126 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| FIIGGATRTDICQGALGDCWLLAAIASLTLNEEILARVVPLDQSFQENYAGIFHFQFWQY |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Rattus norvegicus | 133.00 | 12 | calpain 2 |
| 2 | Homo sapiens | 132.00 | 11 | CAPN2 protein |
| 3 | N/A | 131.00 | 14 | neutral protease large subunit |
| 4 | Macaca mulatta | 131.00 | 2 | PREDICTED: calpain 2, large subunit |
| 5 | Homo sapiens | 131.00 | 2 | calpain 2, large subunit |
| 6 | Mus musculus | 130.00 | 20 | unnamed protein product |
| 7 | Xenopus tropicalis | 127.00 | 4 | calpain 2, (m/II) large subunit |
| 8 | Tetraodon nigroviridis | 119.00 | 8 | unnamed protein product |
| 9 | Danio rerio | 119.00 | 5 | calpain 2, (m/II) large subunit a |
| 10 | Gallus gallus | 119.00 | 3 | calpain 2, (m/II) large subunit |
| 11 | Oncorhynchus mykiss | 119.00 | 1 | m-calpain |
| 12 | Xenopus laevis | 116.00 | 1 | mu/m-calpain large subunit |
| 13 | Coturnix coturnix | 116.00 | 1 | quail calpain |
| 14 | Canis familiaris | 114.00 | 23 | PREDICTED: similar to Calpain-3 (Calpain L3) (Cal |
| 15 | Xenopus laevis | 114.00 | 4 | hypothetical protein LOC398772 |
| 16 | Ovis aries | 114.00 | 1 | skeletal muscle-specific calpain |
| 17 | Rattus norvegicus | 114.00 | 1 | calpain 1, large subunit |
| 18 | Sus scrofa | 113.00 | 3 | calpain 3 |
| 19 | Bos taurus | 112.00 | 4 | calpain 3 |
| 20 | Oryctolagus cuniculus | 112.00 | 1 | calpain 3, (p94) |
| 21 | Oncorhynchus mykiss | 112.00 | 1 | gill-specific calpain |
| 22 | Gallus gallus | 110.00 | 1 | mu-calpain large subunit |
| 23 | Macaca fascicularis | 109.00 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 24 | Stizostedion vitreum vitreum | 109.00 | 1 | putative calpain-like protein |
| 25 | Pongo pygmaeus | 109.00 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 26 | synthetic construct | 109.00 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 27 | Danio rerio | 107.00 | 1 | calpain 1, large subunit |
| 28 | Canis familiaris | 98.20 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
Top-ranked sequences
| organism | matching |
|---|---|
| Rattus norvegicus | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 |
| Homo sapiens | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||||||||||||||||||||||||||||||#|||||||||||+|||||||||||||||||| Sbjct 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLNQSFQENYAGIFHFQFWQY 146 |
| N/A | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||||||||||||||||||||||||||||||#|||||||||||+|||||||||||||||||| Sbjct 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLNQSFQENYAGIFHFQFWQY 146 |
| Macaca mulatta | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||||||||||||||||||||||||||||||#|||||||||||+|||||||||||||||||| Sbjct 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLNQSFQENYAGIFHFQFWQY 146 |
| Homo sapiens | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||||||||||||||||||||||||||||||#|||||||||||+|||||||||||||||||| Sbjct 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLNQSFQENYAGIFHFQFWQY 146 |
| Mus musculus | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||||||||||||||||||||||||||||||#|||||||||| ||||||||||||||||||| Sbjct 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPPDQSFQENYAGIFHFQFWQY 146 |
| Xenopus tropicalis | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 || |||||||||||||||||||||||||||#||+||+|||| ||||||||||||||||||| Sbjct 87 FIAGGATRTDICQGALGDCWLLAAIASLTL#NEQILSRVVPKDQSFQENYAGIFHFQFWQY 146 |
| Tetraodon nigroviridis | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+|||||||||||||||||||||||||||#|| +|||||| || | |+|||||||||||+ Sbjct 87 FIVGGATRTDICQGALGDCWLLAAIASLTL#NEFVLARVVPGDQGFGEDYAGIFHFQFWQF 146 |
| Danio rerio | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+|||||||||||||||||||||||||||#||++|||||| || | ++|||||||||||+ Sbjct 87 FIVGGATRTDICQGALGDCWLLAAIASLTL#NEDVLARVVPSDQGFGQDYAGIFHFQFWQF 146 |
| Gallus gallus | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 || |||||||||||||||||||||||||||#|||||||||| |||||+ |||||||||||| Sbjct 87 FIAGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPRDQSFQDEYAGIFHFQFWQY 146 |
| Oncorhynchus mykiss | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+|||||||||||||||||||||||||||#||++++||||+ ||| |+|||||||||||+ Sbjct 87 FIVGGATRTDICQGALGDCWLLAAIASLTL#NEDVMSRVVPVGQSFGESYAGIFHFQFWQF 146 |
| Xenopus laevis | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||||||||||||||||||||||||| ||||#||++| |||| |||||+|||||||| ||+ Sbjct 90 FIIGGATRTDICQGALGDCWLLAAIGSLTL#NEDLLHRVVPHGQSFQEDYAGIFHFQIWQF 149 |
| Coturnix coturnix | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+|||||||||||||||||||||| ||||#|||+| |||| |||||+|||||||| ||+ Sbjct 90 FIVGGATRTDICQGALGDCWLLAAIGSLTL#NEELLHRVVPHGQSFQEDYAGIFHFQIWQF 149 |
| Canis familiaris | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 |||||| ||||||| ||||| ||||| |||#|| +| ||+| |||| ||||||||||||+| Sbjct 111 FIIGGANRTDICQGDLGDCWFLAAIACLTL#NERLLFRVIPHDQSFTENYAGIFHFQFWRY 170 |
| Xenopus laevis | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+|||||||||||||||||||||| ||||#||++| |||| ||||| |||||||| ||+ Sbjct 90 FIVGGATRTDICQGALGDCWLLAAIGSLTL#NEDLLHRVVPHGQSFQEEYAGIFHFQIWQF 149 |
| Ovis aries | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 |||||| ||||||| ||||| ||||| |||#|| +| ||+| |||| ||||||||||||+| Sbjct 111 FIIGGANRTDICQGDLGDCWFLAAIACLTL#NERLLFRVIPHDQSFTENYAGIFHFQFWRY 170 |
| Rattus norvegicus | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+ ||||||||||||||||||||||||||#|| || |||| ||||| |||||||| ||+ Sbjct 97 FIVDGATRTDICQGALGDCWLLAAIASLTL#NETILHRVVPYGQSFQEGYAGIFHFQLWQF 156 |
| Sus scrofa | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 |||||| ||||||| ||||| ||||| |||#|+ +| ||+| |||| ||||||||||||+| Sbjct 110 FIIGGANRTDICQGDLGDCWFLAAIACLTL#NKRLLFRVIPHDQSFTENYAGIFHFQFWRY 169 |
| Bos taurus | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+||| ||||||| ||||| ||||| |||#|+ +| ||+| |||| ||||||||||||+| Sbjct 111 FIVGGANRTDICQGDLGDCWFLAAIACLTL#NKRLLFRVIPHDQSFTENYAGIFHFQFWRY 170 |
| Oryctolagus cuniculus | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 |||||| ||||||| ||||| ||||| |||#|| +| ||+| |||| ||||||||||||+| Sbjct 91 FIIGGANRTDICQGDLGDCWFLAAIACLTL#NERLLFRVIPHDQSFTENYAGIFHFQFWRY 150 |
| Oncorhynchus mykiss | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+||| |||||||||||||||||||||||#++| |||||| || | +|||||||||||+ Sbjct 65 FIVGGADRTDICQGALGDCWLLAAIASLTL#HKETLARVVPNDQGFDRSYAGIFHFQFWQH 124 |
| Gallus gallus | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+ ||||||||||||||||||||||||||#|| || |||| |||| |||||||| ||+ Sbjct 97 FIVDGATRTDICQGALGDCWLLAAIASLTL#NETILHRVVPHGQSFQNGYAGIFHFQIWQF 156 |
| Macaca fascicularis | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||| || ||||||| ||||| ||||| |||#|+ +| ||+| |||| ||||||||||||+| Sbjct 111 FIIDGANRTDICQGDLGDCWFLAAIACLTL#NQRLLFRVIPHDQSFIENYAGIFHFQFWRY 170 |
| Stizostedion vitreum vitreum | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQE-NYAGIFHFQFWQ 145 ||+ ||+|||||||||||||||||||||||#|+|+|||||| ||| + ||||||||||| Sbjct 87 FIVSGASRTDICQGALGDCWLLAAIASLTL#NQEVLARVVPHGQSFSKGEYAGIFHFQFWQ 146 Query 146 Y 146 + Sbjct 147 F 147 |
| Pongo pygmaeus | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+ ||||||||||||||||||||||||||#|+ +| |||| |||| |||||||| ||+ Sbjct 97 FIVDGATRTDICQGALGDCWLLAAIASLTL#NDTLLHRVVPHGQSFQNGYAGIFHFQLWQF 156 |
| synthetic construct | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+ ||||||||||||||||||||||||||#|+ +| |||| |||| |||||||| ||+ Sbjct 97 FIVDGATRTDICQGALGDCWLLAAIASLTL#NDTLLHRVVPHGQSFQNGYAGIFHFQLWQF 156 |
| Danio rerio | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+ ||||||||||||||||||||||||||#|+ +| |||| | | |||||||||||+ Sbjct 92 FIVDGATRTDICQGALGDCWLLAAIASLTL#NDNLLHRVVPHGQDFDSRYAGIFHFQFWQF 151 |
| Canis familiaris | Query 87 FIIGGATRTDICQGALGDCWLLAAIASLTL#NEEILARVVPLDQSFQENYAGIFHFQFWQY 146 ||+ | + |||||| |||||||||| ||| # ++| |||| |||++||||||||||||+ Sbjct 169 FIVNGFSPTDICQGVLGDCWLLAAIGSLTT#CPKLLHRVVPKGQSFRKNYAGIFHFQFWQF 228 |
[Site 15] YEALSGGATT201-EGFEDFTGGI
Thr201  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Tyr192 | Glu193 | Ala194 | Leu195 | Ser196 | Gly197 | Gly198 | Ala199 | Thr200 | Thr201 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu202 | Gly203 | Phe204 | Glu205 | Asp206 | Phe207 | Thr208 | Gly209 | Gly210 | Ile211 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EGSEFWSALLEKAYAKINGCYEALSGGATTEGFEDFTGGIAEWYELRKPPPNLFKIIQKA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 126.00 | 23 | calpain 2 |
| 2 | N/A | 126.00 | 14 | A Chain A, Crystal Structure Of M-Calpain |
| 3 | Rattus norvegicus | 126.00 | 13 | calpain 2 |
| 4 | Homo sapiens | 124.00 | 11 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 124.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 124.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus tropicalis | 115.00 | 4 | calpain 2, (m/II) large subunit |
| 8 | Oncorhynchus mykiss | 115.00 | 1 | m-calpain |
| 9 | Gallus gallus | 114.00 | 2 | calpain 2, (m/II) large subunit |
| 10 | Tetraodon nigroviridis | 112.00 | 7 | unnamed protein product |
| 11 | Gallus gallus | 112.00 | 1 | mu-calpain large subunit |
| 12 | Danio rerio | 108.00 | 5 | calpain 2, (m/II) large subunit a |
| 13 | Stizostedion vitreum vitreum | 107.00 | 1 | putative calpain-like protein |
| 14 | synthetic construct | 106.00 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 15 | Pongo pygmaeus | 106.00 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 16 | Canis familiaris | 105.00 | 12 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 17 | Xenopus laevis | 105.00 | 4 | calpain |
| 18 | Bos taurus | 104.00 | 4 | micromolar calcium activated neutral protease 1 |
| 19 | Sus scrofa | 103.00 | 3 | calpain 1, large subunit |
| 20 | Danio rerio | 102.00 | 1 | calpain 1, large subunit |
| 21 | Rattus norvegicus | 99.80 | 1 | calpain 1, large subunit |
| 22 | Xenopus laevis | 98.60 | 1 | mu/m-calpain large subunit |
| 23 | Coturnix coturnix | 98.60 | 1 | quail calpain |
| 24 | Drosophila melanogaster | 92.00 | 3 | Calpain-A CG7563-PA, isoform A |
| 25 | Drosophila pseudoobscura | 91.70 | 1 | GA20829-PA |
| 26 | Drosophila melanogaster | 90.50 | 5 | CAA55297.1 |
| 27 | Canis familiaris | 90.50 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 28 | Macaca fascicularis | 85.10 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 29 | Oryctolagus cuniculus | 85.10 | 1 | calpain 3, (p94) |
| 30 | Oncorhynchus mykiss | 84.00 | 1 | gill-specific calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 |
| N/A | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 |
| Rattus norvegicus | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 |
| Homo sapiens | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 ||||||||||||||||||||||||||||||#||||||||||||||||+||||||||||||| Sbjct 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELKKPPPNLFKIIQKA 231 |
| Homo sapiens | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 ||||||||||||||||||||||||||||||#||||||||||||||||+||||||||||||| Sbjct 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELKKPPPNLFKIIQKA 231 |
| Macaca mulatta | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 ||||||||||||||||||||||||||||||#||||||||||||||||+||||||||||||| Sbjct 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELKKPPPNLFKIIQKA 231 |
| Xenopus tropicalis | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 |||||||||+||||||+|| ||||||| ||#|||||||||+|||||||| ||||||||||| Sbjct 172 EGSEFWSALMEKAYAKLNGSYEALSGGTTT#EGFEDFTGGLAEWYELRKAPPNLFKIIQKA 231 |
| Oncorhynchus mykiss | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 ||||||||||||||||+||||||||||+||#||||||||| || |+|||||||+|+||+|| Sbjct 172 EGSEFWSALLEKAYAKVNGCYEALSGGSTT#EGFEDFTGGNAENYDLRKPPPNMFQIIKKA 231 |
| Gallus gallus | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 ||||||||||||||||+|| ||||||| ||#||||||||||||||||+| ||||||||||| Sbjct 172 EGSEFWSALLEKAYAKLNGSYEALSGGTTT#EGFEDFTGGIAEWYELQKAPPNLFKIIQKA 231 |
| Tetraodon nigroviridis | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 || |||||||||||||+||||||||||+||#|||||||||||| |+||||| |||+|++|| Sbjct 172 EGREFWSALLEKAYAKVNGCYEALSGGSTT#EGFEDFTGGIAENYDLRKPPSNLFQILKKA 231 |
| Gallus gallus | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 ||+|||||||||||||+||||||||||+|+#|||||||||+ |||+||||| +|++|| || Sbjct 182 EGTEFWSALLEKAYAKVNGCYEALSGGSTS#EGFEDFTGGVTEWYDLRKPPADLYQIILKA 241 |
| Danio rerio | Query 173 GSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 |||||||||||||||+||||||||||+||#|||||||||||| |||+ | |||+||+|| Sbjct 173 GSEFWSALLEKAYAKVNGCYEALSGGSTT#EGFEDFTGGIAEMYELKSAPTNLFQIIKKA 231 |
| Stizostedion vitreum vitreum | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 || |||||||||||||+||||||||||+||#|||||||||||| |||++| || ||||| Sbjct 173 EGREFWSALLEKAYAKLNGCYEALSGGSTT#EGFEDFTGGIAEVYELKQPDARLFHIIQKA 232 |
| synthetic construct | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 ||+|||||||||||||+|| |||||||+|+#|||||||||+ ||||||| | +|++|| || Sbjct 182 EGNEFWSALLEKAYAKVNGSYEALSGGSTS#EGFEDFTGGVTEWYELRKAPSDLYQIILKA 241 |
| Pongo pygmaeus | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 ||+|||||||||||||+|| |||||||+|+#|||||||||+ ||||||| | +|++|| || Sbjct 182 EGNEFWSALLEKAYAKVNGSYEALSGGSTS#EGFEDFTGGVTEWYELRKAPSDLYQIILKA 241 |
| Canis familiaris | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 +|+|||||||||||||+|| |||||||+|+#|||||||||+ ||||||| | +|++|| || Sbjct 182 QGNEFWSALLEKAYAKVNGSYEALSGGSTS#EGFEDFTGGVTEWYELRKAPSDLYQIILKA 241 |
| Xenopus laevis | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 || |||||||||||||+|| ||||+||+| #|||||||||||| |||+| |||||+||||| Sbjct 172 EGDEFWSALLEKAYAKLNGSYEALTGGSTI#EGFEDFTGGIAEVYELKKAPPNLFQIIQKA 231 |
| Bos taurus | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 +|+|||||||||||||+|| |||||||+|+#|||||||||+ ||||||| | +|+ || || Sbjct 182 QGNEFWSALLEKAYAKVNGSYEALSGGSTS#EGFEDFTGGVTEWYELRKAPSDLYNIILKA 241 |
| Sus scrofa | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 +|+|||||||||||||+|| |||||||+|+#|||||||||+ ||||||| | +|+ || || Sbjct 182 QGNEFWSALLEKAYAKVNGSYEALSGGSTS#EGFEDFTGGVTEWYELRKAPSDLYSIILKA 241 |
| Danio rerio | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 || |||||||||||||+||||||||||+| #|||||||||+ | |||+| | +|| || +| Sbjct 177 EGGEFWSALLEKAYAKLNGCYEALSGGSTC#EGFEDFTGGVTEMYELKKAPADLFSIIGRA 236 |
| Rattus norvegicus | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 +|+|||||||||||||+|| ||||||| |+#| |||||||+ |||+|+| | +|++|| || Sbjct 182 QGNEFWSALLEKAYAKVNGSYEALSGGCTS#EAFEDFTGGVTEWYDLQKAPSDLYQIILKA 241 |
| Xenopus laevis | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 | +|||||||||||||+|| ||+||||+||#|||||||||+|| ||||| | +| |||++| Sbjct 175 ECTEFWSALLEKAYAKLNGSYESLSGGSTT#EGFEDFTGGVAEMYELRKAPRDLDKIIKRA 234 |
| Coturnix coturnix | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 | +|||||||||||||+|||||+||||+||#|||||||||+|| |+|++ | |+ ||+|| Sbjct 175 ECTEFWSALLEKAYAKLNGCYESLSGGSTT#EGFEDFTGGVAEMYDLKRAPRNMGHIIRKA 234 |
| Drosophila melanogaster | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 | +|||||||||||||++| |||| ||+| #| ||||||++|||+|++ | ||| |+||| Sbjct 210 EKNEFWSALLEKAYAKLHGSYEALKGGSTC#EAMEDFTGGVSEWYDLKEAPGNLFTILQKA 269 |
| Drosophila pseudoobscura | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 | +|||||||||||||++| |||| || | #| ||||||+ |||++++ ||||| |+ || Sbjct 387 ERNEFWSALLEKAYAKLHGSYEALKGGTTC#EAMEDFTGGVTEWYDIKEAPPNLFSIMMKA 446 |
| Drosophila melanogaster | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 | +|||||||||||||++| |||| ||+| #| ||||||++|||+|++ | ||| |+||| Sbjct 187 EKNEFWSALLEKAYAKLHGSYEALKGGSTC#EAMEDFTGGVSEWYDLKEAPGNLFTILQKA 246 |
| Canis familiaris | Query 172 EGSEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 | +|||||||||||||+|| |||||||+| #|||||||||+ +|+||| || ++++|| Sbjct 254 ERTEFWSALLEKAYAKLNGSYEALSGGSTV#EGFEDFTGGVTYTIQLQKPPRNLLRMLRKA 313 |
| Macaca fascicularis | Query 174 SEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 +|||||||||||||++| |||| || ||#| ||||||+ |++|+| | ++ ||++|| Sbjct 198 NEFWSALLEKAYAKLHGSYEALKGGNTT#EAMEDFTGGVTEFFEIRDAPSDMHKIMKKA 255 |
| Oryctolagus cuniculus | Query 174 SEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 +|||||||||||||++| |||| || ||#| ||||||+ |++|++ | +++||++|| Sbjct 178 NEFWSALLEKAYAKLHGSYEALKGGNTT#EAMEDFTGGVTEFFEIKDAPRDMYKIMRKA 235 |
| Oncorhynchus mykiss | Query 174 SEFWSALLEKAYAKINGCYEALSGGATT#EGFEDFTGGIAEWYELRKPPPNLFKIIQKA 231 +|||||||||||||+|| ||+| ||+| #| ||||||+ | || + | +|| |++|| Sbjct 152 NEFWSALLEKAYAKLNGSYESLKGGSTL#EAMEDFTGGVGETYETKSAPTDLFNIMKKA 209 |
[Site 16] GATTEGFEDF207-TGGIAEWYEL
Phe207  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly198 | Ala199 | Thr200 | Thr201 | Glu202 | Gly203 | Phe204 | Glu205 | Asp206 | Phe207 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr208 | Gly209 | Gly210 | Ile211 | Ala212 | Glu213 | Trp214 | Tyr215 | Glu216 | Leu217 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SALLEKAYAKINGCYEALSGGATTEGFEDFTGGIAEWYELRKPPPNLFKIIQKALEKGSL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 124.00 | 22 | unnamed protein product |
| 2 | N/A | 124.00 | 14 | A Chain A, Crystal Structure Of M-Calpain |
| 3 | Rattus norvegicus | 124.00 | 11 | calpain 2 |
| 4 | Homo sapiens | 121.00 | 11 | AF261089_1 calpain large polypeptide L2 |
| 5 | Macaca mulatta | 121.00 | 2 | PREDICTED: calpain 2, large subunit |
| 6 | Homo sapiens | 121.00 | 2 | calpain 2, large subunit |
| 7 | Gallus gallus | 111.00 | 2 | calpain 2, (m/II) large subunit |
| 8 | Gallus gallus | 110.00 | 1 | mu-calpain large subunit |
| 9 | Xenopus tropicalis | 109.00 | 4 | calpain 2, (m/II) large subunit |
| 10 | Oncorhynchus mykiss | 109.00 | 1 | m-calpain |
| 11 | Tetraodon nigroviridis | 108.00 | 6 | unnamed protein product |
| 12 | Xenopus laevis | 105.00 | 3 | calpain 1, (mu/I) large subunit |
| 13 | synthetic construct | 105.00 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 14 | Canis familiaris | 104.00 | 21 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 15 | Danio rerio | 104.00 | 5 | calpain 2, (m/II) large subunit a |
| 16 | Pongo pygmaeus | 104.00 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 17 | Bos taurus | 103.00 | 4 | micromolar calcium activated neutral protease 1 |
| 18 | Sus scrofa | 103.00 | 3 | calpain 1, large subunit |
| 19 | Stizostedion vitreum vitreum | 102.00 | 1 | putative calpain-like protein |
| 20 | Coturnix coturnix | 100.00 | 1 | quail calpain |
| 21 | Danio rerio | 100.00 | 1 | calpain 1, large subunit |
| 22 | Rattus norvegicus | 99.40 | 1 | calpain 1, large subunit |
| 23 | Xenopus laevis | 99.40 | 1 | mu/m-calpain large subunit |
| 24 | Drosophila pseudoobscura | 90.10 | 1 | GA20829-PA |
| 25 | Drosophila melanogaster | 89.70 | 3 | Calpain-B CG8107-PA |
| 26 | Drosophila melanogaster | 88.60 | 5 | CAA55297.1 |
| 27 | Canis familiaris | 87.40 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 28 | Macaca fascicularis | 87.00 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 29 | Oryctolagus cuniculus | 84.30 | 1 | calpain 3, (p94) |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 |
| N/A | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 |
| Rattus norvegicus | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 |
| Homo sapiens | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||||||||||||||||||||||#||||||||||+||||||||||||||+|||| Sbjct 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELKKPPPNLFKIIQKALQKGSL 237 |
| Macaca mulatta | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||||||||||||||||||||||#||||||||||+||||||||||||||+|||| Sbjct 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELKKPPPNLFKIIQKALQKGSL 237 |
| Homo sapiens | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||||||||||||||||||||||#||||||||||+||||||||||||||+|||| Sbjct 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELKKPPPNLFKIIQKALQKGSL 237 |
| Gallus gallus | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+|| ||||||| ||||||||#||||||||||+| ||||||||||||+|||| Sbjct 178 SALLEKAYAKLNGSYEALSGGTTTEGFEDF#TGGIAEWYELQKAPPNLFKIIQKALQKGSL 237 |
| Gallus gallus | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+||||||||||+|+||||||#|||+ |||+||||| +|++|| ||||+||| Sbjct 188 SALLEKAYAKVNGCYEALSGGSTSEGFEDF#TGGVTEWYDLRKPPADLYQIILKALERGSL 247 |
| Xenopus tropicalis | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 |||+||||||+|| ||||||| ||||||||#|||+|||||||| |||||||||||| ||| Sbjct 178 SALMEKAYAKLNGSYEALSGGTTTEGFEDF#TGGLAEWYELRKAPPNLFKIIQKALRTGSL 237 |
| Oncorhynchus mykiss | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+||||||||||+||||||||#||| || |+|||||||+|+||+||+| |+| Sbjct 178 SALLEKAYAKVNGCYEALSGGSTTEGFEDF#TGGNAENYDLRKPPPNMFQIIKKAIESGAL 237 |
| Tetraodon nigroviridis | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+||||||||||+||||||||#|||||| |+||||| |||+|++|||| |+| Sbjct 178 SALLEKAYAKVNGCYEALSGGSTTEGFEDF#TGGIAENYDLRKPPSNLFQILKKALEAGAL 237 |
| Xenopus laevis | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 |||||||||| ||||||||||+|+||||||#|||+ ||||++| | +|| || ||||+||| Sbjct 188 SALLEKAYAKANGCYEALSGGSTSEGFEDF#TGGVTEWYEMKKAPKDLFNIILKALERGSL 247 |
| synthetic construct | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+|| |||||||+|+||||||#|||+ ||||||| | +|++|| ||||+||| Sbjct 188 SALLEKAYAKVNGSYEALSGGSTSEGFEDF#TGGVTEWYELRKAPSDLYQIILKALERGSL 247 |
| Canis familiaris | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+|| |||||||+|+||||||#|||+ ||||||| | +|++|| ||||+||| Sbjct 188 SALLEKAYAKVNGSYEALSGGSTSEGFEDF#TGGVTEWYELRKAPSDLYQIILKALERGSL 247 |
| Danio rerio | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+||||||||||+||||||||#|||||| |||+ | |||+||+|||+ |+| Sbjct 178 SALLEKAYAKVNGCYEALSGGSTTEGFEDF#TGGIAEMYELKSAPTNLFQIIKKALDSGAL 237 |
| Pongo pygmaeus | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+|| |||||||+|+||||||#|||+ ||||||| | +|++|| ||||+||| Sbjct 188 SALLEKAYAKVNGSYEALSGGSTSEGFEDF#TGGVTEWYELRKAPSDLYQIILKALERGSL 247 |
| Bos taurus | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+|| |||||||+|+||||||#|||+ ||||||| | +|+ || ||||+||| Sbjct 188 SALLEKAYAKVNGSYEALSGGSTSEGFEDF#TGGVTEWYELRKAPSDLYNIILKALERGSL 247 |
| Sus scrofa | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+|| |||||||+|+||||||#|||+ ||||||| | +|+ || ||||+||| Sbjct 188 SALLEKAYAKVNGSYEALSGGSTSEGFEDF#TGGVTEWYELRKAPSDLYSIILKALERGSL 247 |
| Stizostedion vitreum vitreum | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+||||||||||+||||||||#|||||| |||++| || ||||| +||| Sbjct 179 SALLEKAYAKLNGCYEALSGGSTTEGFEDF#TGGIAEVYELKQPDARLFHIIQKAQSRGSL 238 |
| Coturnix coturnix | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+|||||+||||+||||||||#|||+|| |+|++ | |+ ||+||||+||| Sbjct 181 SALLEKAYAKLNGCYESLSGGSTTEGFEDF#TGGVAEMYDLKRAPRNMGHIIRKALERGSL 240 |
| Danio rerio | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+||||||||||+| ||||||#|||+ | |||+| | +|| || +|+|+||| Sbjct 183 SALLEKAYAKLNGCYEALSGGSTCEGFEDF#TGGVTEMYELKKAPADLFSIIGRAIERGSL 242 |
| Rattus norvegicus | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+|| ||||||| |+| ||||#|||+ |||+|+| | +|++|| ||||+||| Sbjct 188 SALLEKAYAKVNGSYEALSGGCTSEAFEDF#TGGVTEWYDLQKAPSDLYQIILKALERGSL 247 |
| Xenopus laevis | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+|| ||+||||+||||||||#|||+|| ||||| | +| |||++|||+||| Sbjct 181 SALLEKAYAKLNGSYESLSGGSTTEGFEDF#TGGVAEMYELRKAPRDLDKIIKRALERGSL 240 |
| Drosophila pseudoobscura | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||++| |||| || | | |||#|||+ |||++++ ||||| |+ || |+||+ Sbjct 393 SALLEKAYAKLHGSYEALKGGTTCEAMEDF#TGGVTEWYDIKEAPPNLFSIMMKAAERGSM 452 |
| Drosophila melanogaster | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||++| |||| || | | |||#|||+ |||++++ ||||| |+ || |+||+ Sbjct 387 SALLEKAYAKLHGSYEALKGGTTCEAMEDF#TGGVTEWYDIKEAPPNLFSIMMKAAERGSM 446 |
| Drosophila melanogaster | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||++| |||| ||+| | |||#|||++|||+|++ | ||| |+||| |+ |+ Sbjct 193 SALLEKAYAKLHGSYEALKGGSTCEAMEDF#TGGVSEWYDLKEAPGNLFTILQKAAERNSM 252 |
| Canis familiaris | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||+|| |||||||+| ||||||#|||+ +|+||| || ++++||+|+ || Sbjct 260 SALLEKAYAKLNGSYEALSGGSTVEGFEDF#TGGVTYTIQLQKPPRNLLRMLRKAVERSSL 319 |
| Macaca fascicularis | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||++| |||| || ||| |||#|||+ |++|+| | ++ ||++||+|+||| Sbjct 202 SALLEKAYAKLHGSYEALKGGNTTEAMEDF#TGGVTEFFEIRDAPSDMHKIMKKAIERGSL 261 |
| Oryctolagus cuniculus | Query 178 SALLEKAYAKINGCYEALSGGATTEGFEDF#TGGIAEWYELRKPPPNLFKIIQKALEKGSL 237 ||||||||||++| |||| || ||| |||#|||+ |++|++ | +++||++||+|+| | Sbjct 182 SALLEKAYAKLHGSYEALKGGNTTEAMEDF#TGGVTEFFEIKDAPRDMYKIMRKAIERGFL 241 |
[Site 17] TTEGFEDFTG209-GIAEWYELRK
Gly209  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Thr200 | Thr201 | Glu202 | Gly203 | Phe204 | Glu205 | Asp206 | Phe207 | Thr208 | Gly209 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly210 | Ile211 | Ala212 | Glu213 | Trp214 | Tyr215 | Glu216 | Leu217 | Arg218 | Lys219 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LLEKAYAKINGCYEALSGGATTEGFEDFTGGIAEWYELRKPPPNLFKIIQKALEKGSLLG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 124.00 | 22 | unnamed protein product |
| 2 | N/A | 124.00 | 14 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 3 | Rattus norvegicus | 124.00 | 11 | calpain 2 |
| 4 | Homo sapiens | 121.00 | 11 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 121.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 121.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Gallus gallus | 111.00 | 2 | calpain 2, (m/II) large subunit |
| 8 | Gallus gallus | 111.00 | 1 | mu-calpain large subunit |
| 9 | Tetraodon nigroviridis | 110.00 | 5 | unnamed protein product |
| 10 | Xenopus tropicalis | 110.00 | 4 | calpain 2, (m/II) large subunit |
| 11 | Oncorhynchus mykiss | 110.00 | 1 | m-calpain |
| 12 | Canis familiaris | 105.00 | 21 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 13 | Danio rerio | 105.00 | 5 | calpain 2, (m/II) large subunit a |
| 14 | Xenopus laevis | 105.00 | 3 | calpain 1, (mu/I) large subunit |
| 15 | Pongo pygmaeus | 105.00 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 16 | synthetic construct | 105.00 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 17 | Bos taurus | 104.00 | 4 | calpain 1, (mu/I) large subunit |
| 18 | Sus scrofa | 103.00 | 3 | calpain 1, large subunit |
| 19 | Stizostedion vitreum vitreum | 102.00 | 1 | putative calpain-like protein |
| 20 | Rattus norvegicus | 100.00 | 1 | calpain 1, large subunit |
| 21 | Coturnix coturnix | 100.00 | 1 | quail calpain |
| 22 | Danio rerio | 100.00 | 1 | calpain 1, large subunit |
| 23 | Xenopus laevis | 100.00 | 1 | mu/m-calpain large subunit |
| 24 | Drosophila pseudoobscura | 90.90 | 1 | GA20829-PA |
| 25 | Drosophila melanogaster | 90.10 | 3 | Calpain-B CG8107-PA |
| 26 | Drosophila melanogaster | 88.60 | 5 | CAA55297.1 |
| 27 | Canis familiaris | 87.40 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 28 | Macaca fascicularis | 87.00 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 |
| N/A | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 |
| Rattus norvegicus | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 |
| Homo sapiens | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||||||||||||||||||||||||#||||||||+||||||||||||||+|||||| Sbjct 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELKKPPPNLFKIIQKALQKGSLLG 239 |
| Homo sapiens | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||||||||||||||||||||||||#||||||||+||||||||||||||+|||||| Sbjct 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELKKPPPNLFKIIQKALQKGSLLG 239 |
| Macaca mulatta | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||||||||||||||||||||||||#||||||||+||||||||||||||+|||||| Sbjct 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELKKPPPNLFKIIQKALQKGSLLG 239 |
| Gallus gallus | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+|| ||||||| ||||||||||#||||||||+| ||||||||||||+|||||| Sbjct 180 LLEKAYAKLNGSYEALSGGTTTEGFEDFTG#GIAEWYELQKAPPNLFKIIQKALQKGSLLG 239 |
| Gallus gallus | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+||||||||||+|+||||||||#|+ |||+||||| +|++|| ||||+||||| Sbjct 190 LLEKAYAKVNGCYEALSGGSTSEGFEDFTG#GVTEWYDLRKPPADLYQIILKALERGSLLG 249 |
| Tetraodon nigroviridis | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+||||||||||+||||||||||#|||| |+||||| |||+|++|||| |+||| Sbjct 180 LLEKAYAKVNGCYEALSGGSTTEGFEDFTG#GIAENYDLRKPPSNLFQILKKALEAGALLG 239 |
| Xenopus tropicalis | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 |+||||||+|| ||||||| ||||||||||#|+|||||||| |||||||||||| ||||| Sbjct 180 LMEKAYAKLNGSYEALSGGTTTEGFEDFTG#GLAEWYELRKAPPNLFKIIQKALRTGSLLG 239 |
| Oncorhynchus mykiss | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+||||||||||+||||||||||#| || |+|||||||+|+||+||+| |+||| Sbjct 180 LLEKAYAKVNGCYEALSGGSTTEGFEDFTG#GNAENYDLRKPPPNMFQIIKKAIESGALLG 239 |
| Canis familiaris | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+|| |||||||+|+||||||||#|+ ||||||| | +|++|| ||||+||||| Sbjct 190 LLEKAYAKVNGSYEALSGGSTSEGFEDFTG#GVTEWYELRKAPSDLYQIILKALERGSLLG 249 |
| Danio rerio | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+||||||||||+||||||||||#|||| |||+ | |||+||+|||+ |+||| Sbjct 180 LLEKAYAKVNGCYEALSGGSTTEGFEDFTG#GIAEMYELKSAPTNLFQIIKKALDSGALLG 239 |
| Xenopus laevis | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 |||||||| ||||||||||+|+||||||||#|+ ||||++| | +|| || ||||+|||+| Sbjct 190 LLEKAYAKANGCYEALSGGSTSEGFEDFTG#GVTEWYEMKKAPKDLFNIILKALERGSLMG 249 |
| Pongo pygmaeus | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+|| |||||||+|+||||||||#|+ ||||||| | +|++|| ||||+||||| Sbjct 190 LLEKAYAKVNGSYEALSGGSTSEGFEDFTG#GVTEWYELRKAPSDLYQIILKALERGSLLG 249 |
| synthetic construct | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+|| |||||||+|+||||||||#|+ ||||||| | +|++|| ||||+||||| Sbjct 190 LLEKAYAKVNGSYEALSGGSTSEGFEDFTG#GVTEWYELRKAPSDLYQIILKALERGSLLG 249 |
| Bos taurus | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+|| |||||||+|+||||||||#|+ ||||||| | +|+ || ||||+||||| Sbjct 190 LLEKAYAKVNGSYEALSGGSTSEGFEDFTG#GVTEWYELRKAPSDLYNIILKALERGSLLG 249 |
| Sus scrofa | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+|| |||||||+|+||||||||#|+ ||||||| | +|+ || ||||+||||| Sbjct 190 LLEKAYAKVNGSYEALSGGSTSEGFEDFTG#GVTEWYELRKAPSDLYSIILKALERGSLLG 249 |
| Stizostedion vitreum vitreum | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+||||||||||+||||||||||#|||| |||++| || ||||| +|||+| Sbjct 181 LLEKAYAKLNGCYEALSGGSTTEGFEDFTG#GIAEVYELKQPDARLFHIIQKAQSRGSLMG 240 |
| Rattus norvegicus | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+|| ||||||| |+| ||||||#|+ |||+|+| | +|++|| ||||+||||| Sbjct 190 LLEKAYAKVNGSYEALSGGCTSEAFEDFTG#GVTEWYDLQKAPSDLYQIILKALERGSLLG 249 |
| Coturnix coturnix | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+|||||+||||+||||||||||#|+|| |+|++ | |+ ||+||||+||||| Sbjct 183 LLEKAYAKLNGCYESLSGGSTTEGFEDFTG#GVAEMYDLKRAPRNMGHIIRKALERGSLLG 242 |
| Danio rerio | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+||||||||||+| ||||||||#|+ | |||+| | +|| || +|+|+||||| Sbjct 185 LLEKAYAKLNGCYEALSGGSTCEGFEDFTG#GVTEMYELKKAPADLFSIIGRAIERGSLLG 244 |
| Xenopus laevis | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+|| ||+||||+||||||||||#|+|| ||||| | +| |||++|||+||||| Sbjct 183 LLEKAYAKLNGSYESLSGGSTTEGFEDFTG#GVAEMYELRKAPRDLDKIIKRALERGSLLG 242 |
| Drosophila pseudoobscura | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||++| |||| || | | |||||#|+ |||++++ ||||| |+ || |+||++| Sbjct 395 LLEKAYAKLHGSYEALKGGTTCEAMEDFTG#GVTEWYDIKEAPPNLFSIMMKAAERGSMMG 454 |
| Drosophila melanogaster | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||++| |||| || | | |||||#|+ |||++++ ||||| |+ || |+||++| Sbjct 389 LLEKAYAKLHGSYEALKGGTTCEAMEDFTG#GVTEWYDIKEAPPNLFSIMMKAAERGSMMG 448 |
| Drosophila melanogaster | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||++| |||| ||+| | |||||#|++|||+|++ | ||| |+||| |+ |++| Sbjct 195 LLEKAYAKLHGSYEALKGGSTCEAMEDFTG#GVSEWYDLKEAPGNLFTILQKAAERNSMMG 254 |
| Canis familiaris | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||+|| |||||||+| ||||||||#|+ +|+||| || ++++||+|+ ||+| Sbjct 262 LLEKAYAKLNGSYEALSGGSTVEGFEDFTG#GVTYTIQLQKPPRNLLRMLRKAVERSSLMG 321 |
| Macaca fascicularis | Query 180 LLEKAYAKINGCYEALSGGATTEGFEDFTG#GIAEWYELRKPPPNLFKIIQKALEKGSLLG 239 ||||||||++| |||| || ||| |||||#|+ |++|+| | ++ ||++||+|+|||+| Sbjct 204 LLEKAYAKLHGSYEALKGGNTTEAMEDFTG#GVTEFFEIRDAPSDMHKIMKKAIERGSLMG 263 |
[Site 18] LGCSIDITSA247-ADSEAVTYQK
Ala247  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu238 | Gly239 | Cys240 | Ser241 | Ile242 | Asp243 | Ile244 | Thr245 | Ser246 | Ala247 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala248 | Asp249 | Ser250 | Glu251 | Ala252 | Val253 | Thr254 | Tyr255 | Gln256 | Lys257 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RKPPPNLFKIIQKALEKGSLLGCSIDITSAADSEAVTYQKLVKGHAYSVTGAEEVESSGS |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 125.00 | 21 | m-calpain |
| 2 | N/A | 125.00 | 14 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 3 | Rattus norvegicus | 125.00 | 9 | calpain 2 |
| 4 | Macaca mulatta | 122.00 | 2 | PREDICTED: calpain 2, large subunit |
| 5 | Homo sapiens | 119.00 | 2 | calpain 2, large subunit |
| 6 | Homo sapiens | 118.00 | 9 | CAPN2 protein |
| 7 | Gallus gallus | 106.00 | 2 | calpain 2, (m/II) large subunit |
| 8 | Xenopus tropicalis | 103.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Oncorhynchus mykiss | 101.00 | 1 | m-calpain |
| 10 | Tetraodon nigroviridis | 96.30 | 6 | unnamed protein product |
| 11 | Gallus gallus | 94.40 | 1 | mu-calpain large subunit |
| 12 | Danio rerio | 94.00 | 5 | calpain 2, (m/II) large subunit a |
| 13 | Xenopus laevis | 91.70 | 3 | calpain |
| 14 | Danio rerio | 89.00 | 1 | calpain 1, large subunit |
| 15 | Canis familiaris | 88.20 | 17 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 16 | Sus scrofa | 87.80 | 3 | calpain 1, large subunit |
| 17 | Bos taurus | 87.80 | 3 | micromolar calcium activated neutral protease 1 |
| 18 | synthetic construct | 87.80 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 19 | Pongo pygmaeus | 87.80 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | Xenopus laevis | 85.90 | 1 | mu/m-calpain large subunit |
| 21 | Stizostedion vitreum vitreum | 85.50 | 1 | putative calpain-like protein |
| 22 | Coturnix coturnix | 80.50 | 1 | quail calpain |
| 23 | Oncorhynchus mykiss | 78.20 | 1 | gill-specific calpain |
| 24 | Rattus norvegicus | 75.90 | 1 | calpain 1, large subunit |
| 25 | Canis familiaris | 67.40 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 26 | Oryctolagus cuniculus | 58.20 | 1 | calpain 3, (p94) |
| 27 | Drosophila melanogaster | 53.90 | 5 | Calpain, without calmodulin-like domain |
| 28 | Drosophila melanogaster | 53.90 | 3 | Calpain-A CG7563-PA, isoform A |
| 29 | Drosophila pseudoobscura | 53.10 | 1 | GA20829-PA |
| 30 | Caenorhabditis elegans | 50.40 | 3 | CaLPain family member (clp-1) |
| 31 | Caenorhabditis briggsae AF16 | 50.40 | 1 | hypothetical protein CBG22945 |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 |
| N/A | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 |
| Rattus norvegicus | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 |
| Macaca mulatta | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 +||||||||||||||+||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 218 KKPPPNLFKIIQKALQKGSLLGCSIDITSA#ADSEAITYQKLVKGHAYSVTGAEEVESSGS 277 |
| Homo sapiens | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 +||||||||||||||+||||||||||||||#|||||+|+|||||||||||||||||||+|| Sbjct 218 KKPPPNLFKIIQKALQKGSLLGCSIDITSA#ADSEAITFQKLVKGHAYSVTGAEEVESNGS 277 |
| Homo sapiens | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 +||||||||||||||+||||||||||||||#|||||+|+|||||||||||||||||||+|| Sbjct 218 KKPPPNLFKIIQKALQKGSLLGCSIDITSA#ADSEAITFQKLVKGHAYSVTGAEEVESNGS 277 |
| Gallus gallus | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 +| ||||||||||||+||||||||||||||#|++|||| ||||||||||||||||| || Sbjct 218 QKAPPNLFKIIQKALQKGSLLGCSIDITSA#AETEAVTSQKLVKGHAYSVTGAEEVNFRGS 277 |
| Xenopus tropicalis | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 || |||||||||||| |||||||||||||#|++||+|+|||||||||||| ||||+ |+ Sbjct 218 RKAPPNLFKIIQKALRTGSLLGCSIDITSA#AETEAITFQKLVKGHAYSVTAAEEVQYRGN 277 |
| Oncorhynchus mykiss | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 ||||||+|+||+||+| |+|||||||||||#||||||| ||||||||||+||| || | Sbjct 218 RKPPPNMFQIIKKAIESGALLGCSIDITSA#ADSEAVTRQKLVKGHAYSLTGAVEVTYRG 276 |
| Tetraodon nigroviridis | Query 222 PNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 |+|| ||+||||+|||||||||||||#+||||+||+|||||||||||||++|| | Sbjct 228 PHLFHIIKKALERGSLLGCSIDITSA#SDSEAITYRKLVKGHAYSVTGADQVEYRG 282 |
| Gallus gallus | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 |||| +|++|| ||||+|||||||||||||# | ||||++|||||||||||||+++ | Sbjct 228 RKPPADLYQIILKALERGSLLGCSIDITSA#FDMEAVTFKKLVKGHAYSVTGAKQISYRG 286 |
| Danio rerio | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEV 272 + | |||+||+|||+ |+|||||||||||#||||||||||||||||||+||| || Sbjct 218 KSAPTNLFQIIKKALDSGALLGCSIDITSA#ADSEAVTYQKLVKGHAYSLTGATEV 272 |
| Xenopus laevis | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 +| |||||+||||||+ ||||||||||+|# |+||+| +|||||||||||||||| | Sbjct 218 KKAPPNLFQIIQKALKAESLLGCSIDITNA#YDTEAITSRKLVKGHAYSVTGAEEVLYRG 276 |
| Danio rerio | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSGS 277 +| | +|| || +|+|+|||||||||||| # | ||||++|||||||||||||||| |+ Sbjct 223 KKAPADLFSIIGRAIERGSLLGCSIDITSK#FDMEAVTFKKLVKGHAYSVTGAEEVVYRGN 282 |
| Canis familiaris | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 || | +|++|| ||||+||||||||||+| # | ||+|++|||||||||||||++| | Sbjct 228 RKAPSDLYQIILKALERGSLLGCSIDISSV#LDMEAITFKKLVKGHAYSVTGAKQVNYQG 286 |
| Sus scrofa | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 || | +|+ || ||||+||||||||||+| # | ||||++|||||||||||||++| | Sbjct 228 RKAPSDLYSIILKALERGSLLGCSIDISSV#LDMEAVTFKKLVKGHAYSVTGAKQVNYQG 286 |
| Bos taurus | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 || | +|+ || ||||+||||||||||+| # | ||||++|||||||||||||++| | Sbjct 228 RKAPSDLYNIILKALERGSLLGCSIDISSI#LDMEAVTFKKLVKGHAYSVTGAKQVNYQG 286 |
| synthetic construct | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 || | +|++|| ||||+||||||||||+| # | ||+|++|||||||||||||++| | Sbjct 228 RKAPSDLYQIILKALERGSLLGCSIDISSV#LDMEAITFKKLVKGHAYSVTGAKQVNYRG 286 |
| Pongo pygmaeus | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 || | +|++|| ||||+||||||||||+| # | ||+|++|||||||||||||++| | Sbjct 228 RKAPSDLYQIILKALERGSLLGCSIDISSV#LDMEAITFKKLVKGHAYSVTGAKQVNYRG 286 |
| Xenopus laevis | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 || | +| |||++|||+|||||||||||||# | ||||++||||||||||| +|| | Sbjct 221 RKAPRDLDKIIKRALERGSLLGCSIDITSA#FDMEAVTFKKLVKGHAYSVTALKEVNYMG 279 |
| Stizostedion vitreum vitreum | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 ++| || ||||| +|||+||||||||+#++||||| ||||||||||||| +|| | Sbjct 219 KQPDARLFHIIQKAQSRGSLMGCSIDITSS#SESEAVTSQKLVKGHAYSVTGTAQVEHQG 277 |
| Coturnix coturnix | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 ++ | |+ ||+||||+|||||| ||||||# | ||||++||||||||||| ++| | Sbjct 221 KRAPRNMGHIIRKALERGSLLGCPIDITSA#FDMEAVTFKKLVKGHAYSVTAFKDVNYRG 279 |
| Oncorhynchus mykiss | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 + | +|| |++|| ++||++||||||||+#|+||| | ||||||||+|| ||| | Sbjct 196 KSAPTDLFNIMKKAYDRGSMMGCSIDITSS#AESEAQTASGLVKGHAYSITGLEEVNYRG 254 |
| Rattus norvegicus | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 +| | +|++|| ||||+||||||||+|+ # | ||+|++ ||+||||||| |++| | Sbjct 228 QKAPSDLYQIILKALERGSLLGCSINISDI#RDLEAITFKNLVRGHAYSVTDAKQVTYQG 286 |
| Canis familiaris | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 +||| || ++++||+|+ ||+||||+| | #++ | +| + ||+||||||| +|| | Sbjct 300 QKPPRNLLRMLRKAVERSSLMGCSIEIVSE#SELETMTQKMLVRGHAYSVTDLQEVWYQG 358 |
| Oryctolagus cuniculus | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVTGAEEVESSG 276 + | +++||++||+|+| |+||||| # | ||||||||||| || | Sbjct 222 KDAPRDMYKIMRKAIERGFLMGCSIDTIVP#VQYETRMACGLVKGHAYSVTGLEETLFKG 280 |
| Drosophila melanogaster | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVT 267 ++ | ||| |+||| |+ |++||||+ # +|| | | |++|||||+| Sbjct 256 KEAPGNLFTILQKAAERNSMMGCSIEPDPN#V-TEAETPQGLIRGHAYSIT 304 |
| Drosophila melanogaster | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVT 267 ++ | ||| |+||| |+ |++||||+ # +|| | | |++|||||+| Sbjct 256 KEAPGNLFTILQKAAERNSMMGCSIEPDPN#V-TEAETPQGLIRGHAYSIT 304 |
| Drosophila pseudoobscura | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADSEAVTYQKLVKGHAYSVT 267 ++ ||||| |+ || |+||++|||++ # || | + |++|||||+| Sbjct 433 KEAPPNLFSIMMKAAERGSMMGCSLEPDPH#V-LEAETREGLIRGHAYSIT 481 |
| Caenorhabditis elegans | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADS---EAVTYQKLVKGHAYSVTGAEEVE 273 + || || +++ + | ||| ||||+ #|| || ||||||||+|| |+ Sbjct 438 KNPPRNLMQMMMRGFEMGSLFGCSIE----#ADPNVWEAKMSNGLVKGHAYSITGCRIVD 492 |
| Caenorhabditis briggsae AF16 | Query 218 RKPPPNLFKIIQKALEKGSLLGCSIDITSA#ADS---EAVTYQKLVKGHAYSVTGAEEVE 273 + || || +++ + | ||| ||||+ #|| || ||||||||+|| |+ Sbjct 433 KNPPRNLMQMMMRGFEMGSLFGCSIE----#ADPNVWEAKMSNGLVKGHAYSITGCRIVD 487 |
[Site 19] CSIDITSAAD249-SEAVTYQKLV
Asp249  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Cys240 | Ser241 | Ile242 | Asp243 | Ile244 | Thr245 | Ser246 | Ala247 | Ala248 | Asp249 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser250 | Glu251 | Ala252 | Val253 | Thr254 | Tyr255 | Gln256 | Lys257 | Leu258 | Val259 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PPPNLFKIIQKALEKGSLLGCSIDITSAADSEAVTYQKLVKGHAYSVTGAEEVESSGSLQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 125.00 | 21 | m-calpain |
| 2 | Rattus norvegicus | 125.00 | 9 | calpain 2 |
| 3 | N/A | 124.00 | 14 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 4 | Macaca mulatta | 123.00 | 2 | PREDICTED: calpain 2, large subunit |
| 5 | Homo sapiens | 120.00 | 9 | CAPN2 protein |
| 6 | Homo sapiens | 120.00 | 2 | calpain 2, large subunit |
| 7 | Gallus gallus | 107.00 | 2 | calpain 2, (m/II) large subunit |
| 8 | Xenopus tropicalis | 102.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Oncorhynchus mykiss | 97.40 | 1 | m-calpain |
| 10 | Tetraodon nigroviridis | 96.30 | 6 | unnamed protein product |
| 11 | Danio rerio | 94.00 | 5 | calpain 2, (m/II) large subunit a |
| 12 | Gallus gallus | 90.10 | 1 | mu-calpain large subunit |
| 13 | Xenopus laevis | 89.40 | 3 | calpain |
| 14 | Danio rerio | 87.40 | 1 | calpain 1, large subunit |
| 15 | Canis familiaris | 85.10 | 17 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 16 | Sus scrofa | 84.70 | 3 | calpain 1, large subunit |
| 17 | Bos taurus | 84.70 | 3 | micromolar calcium activated neutral protease 1 |
| 18 | Pongo pygmaeus | 84.70 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 19 | synthetic construct | 84.30 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Stizostedion vitreum vitreum | 84.30 | 1 | putative calpain-like protein |
| 21 | Xenopus laevis | 83.60 | 1 | mu/m-calpain large subunit |
| 22 | Coturnix coturnix | 79.00 | 1 | quail calpain |
| 23 | Oncorhynchus mykiss | 77.80 | 1 | gill-specific calpain |
| 24 | Rattus norvegicus | 73.90 | 1 | calpain 1, large subunit |
| 25 | Canis familiaris | 65.50 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 26 | Oryctolagus cuniculus | 58.20 | 1 | calpain 3, (p94) |
| 27 | Drosophila melanogaster | 53.10 | 3 | Calpain-B CG8107-PA |
| 28 | Drosophila melanogaster | 52.80 | 5 | CAA55297.1 |
| 29 | Drosophila pseudoobscura | 52.40 | 1 | GA20829-PA |
| 30 | Caenorhabditis elegans | 49.70 | 3 | CaLPain family member (clp-1) |
| 31 | Caenorhabditis briggsae AF16 | 49.70 | 1 | hypothetical protein CBG22945 |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 |
| Rattus norvegicus | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 |
| N/A | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 |
| Macaca mulatta | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 |||||||||||||+||||||||||||||||#|||+|||||||||||||||||||||||||| Sbjct 220 PPPNLFKIIQKALQKGSLLGCSIDITSAAD#SEAITYQKLVKGHAYSVTGAEEVESSGSLQ 279 |
| Homo sapiens | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 |||||||||||||+||||||||||||||||#|||+|+|||||||||||||||||||+|||| Sbjct 220 PPPNLFKIIQKALQKGSLLGCSIDITSAAD#SEAITFQKLVKGHAYSVTGAEEVESNGSLQ 279 |
| Homo sapiens | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 |||||||||||||+||||||||||||||||#|||+|+|||||||||||||||||||+|||| Sbjct 220 PPPNLFKIIQKALQKGSLLGCSIDITSAAD#SEAITFQKLVKGHAYSVTGAEEVESNGSLQ 279 |
| Gallus gallus | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 ||||||||||||+|||||||||||||||+#+|||| ||||||||||||||||| ||+| Sbjct 221 PPNLFKIIQKALQKGSLLGCSIDITSAAE#TEAVTSQKLVKGHAYSVTGAEEVNFRGSIQ 279 |
| Xenopus tropicalis | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 |||||||||||| ||||||||||||||+#+||+|+|||||||||||| ||||+ |+|+ Sbjct 221 PPNLFKIIQKALRTGSLLGCSIDITSAAE#TEAITFQKLVKGHAYSVTAAEEVQYRGNLE 279 |
| Oncorhynchus mykiss | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSG 276 ||||+|+||+||+| |+|||||||||||||#||||| ||||||||||+||| || | Sbjct 220 PPPNMFQIIKKAIESGALLGCSIDITSAAD#SEAVTRQKLVKGHAYSLTGAVEVTYRG 276 |
| Tetraodon nigroviridis | Query 222 PNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSG 276 |+|| ||+||||+|||||||||||||+|#|||+||+|||||||||||||++|| | Sbjct 228 PHLFHIIKKALERGSLLGCSIDITSASD#SEAITYRKLVKGHAYSVTGADQVEYRG 282 |
| Danio rerio | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEV 272 | |||+||+|||+ |+|||||||||||||#||||||||||||||||+||| || Sbjct 221 PTNLFQIIKKALDSGALLGCSIDITSAAD#SEAVTYQKLVKGHAYSLTGATEV 272 |
| Gallus gallus | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSG 276 || +|++|| ||||+||||||||||||| |# ||||++|||||||||||||+++ | Sbjct 230 PPADLYQIILKALERGSLLGCSIDITSAFD#MEAVTFKKLVKGHAYSVTGAKQISYRG 286 |
| Xenopus laevis | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSG 276 |||||+||||||+ ||||||||||+| |#+||+| +|||||||||||||||| | Sbjct 221 PPNLFQIIQKALKAESLLGCSIDITNAYD#TEAITSRKLVKGHAYSVTGAEEVLYRG 276 |
| Danio rerio | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSL 278 | +|| || +|+|+|||||||||||| |# ||||++|||||||||||||||| |++ Sbjct 226 PADLFSIIGRAIERGSLLGCSIDITSKFD#MEAVTFKKLVKGHAYSVTGAEEVVYRGNM 283 |
| Canis familiaris | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSL 278 | +|++|| ||||+||||||||||+| |# ||+|++|||||||||||||++| | + Sbjct 231 PSDLYQIILKALERGSLLGCSIDISSVLD#MEAITFKKLVKGHAYSVTGAKQVNYQGQM 288 |
| Sus scrofa | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSL 278 | +|+ || ||||+||||||||||+| |# ||||++|||||||||||||++| | + Sbjct 231 PSDLYSIILKALERGSLLGCSIDISSVLD#MEAVTFKKLVKGHAYSVTGAKQVNYQGQM 288 |
| Bos taurus | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSL 278 | +|+ || ||||+||||||||||+| |# ||||++|||||||||||||++| | + Sbjct 231 PSDLYNIILKALERGSLLGCSIDISSILD#MEAVTFKKLVKGHAYSVTGAKQVNYQGQM 288 |
| Pongo pygmaeus | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSL 278 | +|++|| ||||+||||||||||+| |# ||+|++|||||||||||||++| | + Sbjct 231 PSDLYQIILKALERGSLLGCSIDISSVLD#MEAITFKKLVKGHAYSVTGAKQVNYRGQM 288 |
| synthetic construct | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSL 278 | +|++|| ||||+||||||||||+| |# ||+|++|||||||||||||++| | + Sbjct 231 PSDLYQIILKALERGSLLGCSIDISSVLD#MEAITFKKLVKGHAYSVTGAKQVNYRGQV 288 |
| Stizostedion vitreum vitreum | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 | || ||||| +|||+||||||||+++#||||| ||||||||||||| +|| | + Sbjct 221 PDARLFHIIQKAQSRGSLMGCSIDITSSSE#SEAVTSQKLVKGHAYSVTGTAQVEHQGGTE 280 |
| Xenopus laevis | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSGSLQ 279 | +| |||++|||+||||||||||||| |# ||||++||||||||||| +|| | ++ Sbjct 224 PRDLDKIIKRALERGSLLGCSIDITSAFD#MEAVTFKKLVKGHAYSVTALKEVNYMGEME 282 |
| Coturnix coturnix | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSG 276 | |+ ||+||||+|||||| |||||| |# ||||++||||||||||| ++| | Sbjct 224 PRNMGHIIRKALERGSLLGCPIDITSAFD#MEAVTFKKLVKGHAYSVTAFKDVNYRG 279 |
| Oncorhynchus mykiss | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSG 276 | +|| |++|| ++||++||||||||+|+#||| | ||||||||+|| ||| | Sbjct 199 PTDLFNIMKKAYDRGSMMGCSIDITSSAE#SEAQTASGLVKGHAYSITGLEEVNYRG 254 |
| Rattus norvegicus | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSG 276 | +|++|| ||||+||||||||+|+ |# ||+|++ ||+||||||| |++| | Sbjct 231 PSDLYQIILKALERGSLLGCSINISDIRD#LEAITFKNLVRGHAYSVTDAKQVTYQG 286 |
| Canis familiaris | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSG 276 || || ++++||+|+ ||+||||+| | ++# | +| + ||+||||||| +|| | Sbjct 302 PPRNLLRMLRKAVERSSLMGCSIEIVSESE#LETMTQKMLVRGHAYSVTDLQEVWYQG 358 |
| Oryctolagus cuniculus | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVTGAEEVESSG 276 | +++||++||+|+| |+||||| # | ||||||||||| || | Sbjct 225 PRDMYKIMRKAIERGFLMGCSIDTIVPVQ#YETRMACGLVKGHAYSVTGLEETLFKG 280 |
| Drosophila melanogaster | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVT 267 ||||| |+ || |+||++|||++ # || | | |++|||||+| Sbjct 430 PPNLFSIMMKAAERGSMMGCSLEPDPHV-#LEAETPQGLIRGHAYSIT 475 |
| Drosophila melanogaster | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVT 267 | ||| |+||| |+ |++||||+ #+|| | | |++|||||+| Sbjct 236 PGNLFTILQKAAERNSMMGCSIEPDPNV-#TEAETPQGLIRGHAYSIT 281 |
| Drosophila pseudoobscura | Query 221 PPNLFKIIQKALEKGSLLGCSIDITSAAD#SEAVTYQKLVKGHAYSVT 267 ||||| |+ || |+||++|||++ # || | + |++|||||+| Sbjct 436 PPNLFSIMMKAAERGSMMGCSLEPDPHV-#LEAETREGLIRGHAYSIT 481 |
| Caenorhabditis elegans | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#S---EAVTYQKLVKGHAYSVTGAEEVE 273 || || +++ + | ||| ||||+ ||# || ||||||||+|| |+ Sbjct 440 PPRNLMQMMMRGFEMGSLFGCSIE----AD#PNVWEAKMSNGLVKGHAYSITGCRIVD 492 |
| Caenorhabditis briggsae AF16 | Query 220 PPPNLFKIIQKALEKGSLLGCSIDITSAAD#S---EAVTYQKLVKGHAYSVTGAEEVE 273 || || +++ + | ||| ||||+ ||# || ||||||||+|| |+ Sbjct 435 PPRNLMQMMMRGFEMGSLFGCSIE----AD#PNVWEAKMSNGLVKGHAYSITGCRIVD 487 |
[Site 20] SAADSEAVTY255-QKLVKGHAYS
Tyr255  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser246 | Ala247 | Ala248 | Asp249 | Ser250 | Glu251 | Ala252 | Val253 | Thr254 | Tyr255 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln256 | Lys257 | Leu258 | Val259 | Lys260 | Gly261 | His262 | Ala263 | Tyr264 | Ser265 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KIIQKALEKGSLLGCSIDITSAADSEAVTYQKLVKGHAYSVTGAEEVESSGSLQKLIRIR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 121.00 | 21 | AF497625_1 m-calpain 80 kDa large subunit |
| 2 | N/A | 120.00 | 14 | A Chain A, Crystal Structure Of M-Calpain |
| 3 | Rattus norvegicus | 120.00 | 9 | calpain 2 |
| 4 | Macaca mulatta | 118.00 | 2 | PREDICTED: calpain 2, large subunit |
| 5 | Homo sapiens | 116.00 | 9 | CAPN2 protein |
| 6 | Homo sapiens | 116.00 | 2 | calpain 2, large subunit |
| 7 | Gallus gallus | 105.00 | 2 | calpain 2, (m/II) large subunit |
| 8 | Xenopus tropicalis | 100.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Tetraodon nigroviridis | 96.30 | 7 | unnamed protein product |
| 10 | Oncorhynchus mykiss | 93.60 | 1 | m-calpain |
| 11 | Danio rerio | 93.20 | 5 | calpain 2, (m/II) large subunit a |
| 12 | Xenopus laevis | 93.20 | 3 | hypothetical protein LOC398772 |
| 13 | Xenopus laevis | 92.00 | 1 | mu/m-calpain large subunit |
| 14 | Danio rerio | 90.10 | 1 | calpain 1, large subunit |
| 15 | Stizostedion vitreum vitreum | 88.60 | 1 | putative calpain-like protein |
| 16 | Gallus gallus | 86.70 | 1 | mu-calpain large subunit |
| 17 | Canis familiaris | 86.30 | 17 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 18 | Sus scrofa | 86.30 | 3 | calpain 1, large subunit |
| 19 | Pongo pygmaeus | 86.30 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | Bos taurus | 85.90 | 4 | micromolar calcium activated neutral protease 1 |
| 21 | synthetic construct | 85.50 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 22 | Coturnix coturnix | 85.10 | 1 | quail calpain |
| 23 | Oncorhynchus mykiss | 79.70 | 1 | gill-specific calpain |
| 24 | Rattus norvegicus | 73.90 | 1 | calpain 1, large subunit |
| 25 | Canis familiaris | 65.90 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 26 | Oryctolagus cuniculus | 61.20 | 1 | calpain 3, (p94) |
| 27 | Caenorhabditis elegans | 50.80 | 7 | CaLPain family member (clp-6) |
| 28 | Caenorhabditis briggsae AF16 | 45.40 | 2 | Hypothetical protein CBG13923 |
| 29 | Drosophila melanogaster | 44.30 | 4 | CAA55297.1 |
| 30 | Drosophila melanogaster | 44.30 | 2 | Calpain-A CG7563-PB, isoform B |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 |
| N/A | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 |
| Rattus norvegicus | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 |
| Macaca mulatta | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 |||||||+|||||||||||||||||||+||#|||||||||||||||||||||||||||||| Sbjct 226 KIIQKALQKGSLLGCSIDITSAADSEAITY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 |
| Homo sapiens | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 |||||||+|||||||||||||||||||+|+#|||||||||||||||||||+|||||||||| Sbjct 226 KIIQKALQKGSLLGCSIDITSAADSEAITF#QKLVKGHAYSVTGAEEVESNGSLQKLIRIR 285 |
| Homo sapiens | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 |||||||+|||||||||||||||||||+|+#|||||||||||||||||||+|||||||||| Sbjct 226 KIIQKALQKGSLLGCSIDITSAADSEAITF#QKLVKGHAYSVTGAEEVESNGSLQKLIRIR 285 |
| Gallus gallus | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 |||||||+|||||||||||||||++|||| #||||||||||||||||| ||+||||||| Sbjct 226 KIIQKALQKGSLLGCSIDITSAAETEAVTS#QKLVKGHAYSVTGAEEVNFRGSIQKLIRIR 285 |
| Xenopus tropicalis | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 ||||||| ||||||||||||||++||+|+#|||||||||||| ||||+ |+|+|||||| Sbjct 226 KIIQKALRTGSLLGCSIDITSAAETEAITF#QKLVKGHAYSVTAAEEVQYRGNLEKLIRIR 285 |
| Tetraodon nigroviridis | Query 227 IIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 ||+||||+|||||||||||||+||||+||#+|||||||||||||++|| | +|+||| Sbjct 233 IIKKALERGSLLGCSIDITSASDSEAITY#RKLVKGHAYSVTGADQVEYRGEAVQLVRIR 291 |
| Oncorhynchus mykiss | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 +||+||+| |+|||||||||||||||||| #||||||||||+||| || | +|||||| Sbjct 226 QIIKKAIESGALLGCSIDITSAADSEAVTR#QKLVKGHAYSLTGAVEVTYRGRKEKLIRIR 285 |
| Danio rerio | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 +||+|||+ |+|||||||||||||||||||#||||||||||+||| || +||+|+| Sbjct 226 QIIKKALDSGALLGCSIDITSAADSEAVTY#QKLVKGHAYSLTGATEVNYRSRKEKLVRVR 285 |
| Xenopus laevis | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 |||++|||+||||||||||||| | ||||+#+||||||||||| +||+ | ++|||||| Sbjct 229 KIIKRALERGSLLGCSIDITSAFDMEAVTF#KKLVKGHAYSVTALKEVDYRGGMEKLIRIR 288 |
| Xenopus laevis | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 |||++|||+||||||||||||| | ||||+#+||||||||||| +|| | ++|||||| Sbjct 229 KIIKRALERGSLLGCSIDITSAFDMEAVTF#KKLVKGHAYSVTALKEVNYMGEMEKLIRIR 288 |
| Danio rerio | Query 227 IIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 || +|+|+|||||||||||| | ||||+#+|||||||||||||||| |++ ||+||| Sbjct 232 IIGRAIERGSLLGCSIDITSKFDMEAVTF#KKLVKGHAYSVTGAEEVVYRGNMTKLVRIR 290 |
| Stizostedion vitreum vitreum | Query 227 IIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 ||||| +|||+||||||||+++||||| #||||||||||||| +|| | +|||||| Sbjct 228 IIQKAQSRGSLMGCSIDITSSSESEAVTS#QKLVKGHAYSVTGTAQVEHQGGTEKLIRIR 286 |
| Gallus gallus | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 +|| ||||+||||||||||||| | ||||+#+|||||||||||||+++ | |||+| Sbjct 236 QIILKALERGSLLGCSIDITSAFDMEAVTF#KKLVKGHAYSVTGAKQISYRGQSLGLIRMR 295 |
| Canis familiaris | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 +|| ||||+||||||||||+| | ||+|+#+|||||||||||||++| | + |||+| Sbjct 236 QIILKALERGSLLGCSIDISSVLDMEAITF#KKLVKGHAYSVTGAKQVNYQGQMVSLIRMR 295 |
| Sus scrofa | Query 227 IIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 || ||||+||||||||||+| | ||||+#+|||||||||||||++| | + |||+| Sbjct 237 IILKALERGSLLGCSIDISSVLDMEAVTF#KKLVKGHAYSVTGAKQVNYQGQMVNLIRMR 295 |
| Pongo pygmaeus | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 +|| ||||+||||||||||+| | ||+|+#+|||||||||||||++| | + |||+| Sbjct 236 QIILKALERGSLLGCSIDISSVLDMEAITF#KKLVKGHAYSVTGAKQVNYRGQMVSLIRMR 295 |
| Bos taurus | Query 227 IIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 || ||||+||||||||||+| | ||||+#+|||||||||||||++| | + |||+| Sbjct 237 IILKALERGSLLGCSIDISSILDMEAVTF#KKLVKGHAYSVTGAKQVNYQGQMVNLIRMR 295 |
| synthetic construct | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 +|| ||||+||||||||||+| | ||+|+#+|||||||||||||++| | + |||+| Sbjct 236 QIILKALERGSLLGCSIDISSVLDMEAITF#KKLVKGHAYSVTGAKQVNYRGQVVSLIRMR 295 |
| Coturnix coturnix | Query 227 IIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 ||+||||+|||||| |||||| | ||||+#+||||||||||| ++| | ++||||| Sbjct 230 IIRKALERGSLLGCPIDITSAFDMEAVTF#KKLVKGHAYSVTAFKDVNYRGQQEQLIRIR 288 |
| Oncorhynchus mykiss | Query 227 IIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 |++|| ++||++||||||||+|+||| | # ||||||||+|| ||| | |||||| Sbjct 205 IMKKAYDRGSMMGCSIDITSSAESEAQTA#SGLVKGHAYSITGLEEVNYRGKKVKLIRIR 263 |
| Rattus norvegicus | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 +|| ||||+||||||||+|+ | ||+|+#+ ||+||||||| |++| | |||+| Sbjct 236 QIILKALERGSLLGCSINISDIRDLEAITF#KNLVRGHAYSVTDAKQVTYQGQRVNLIRMR 295 |
| Canis familiaris | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 ++++||+|+ ||+||||+| | ++ | +| #+ ||+||||||| +|| | + |||+| Sbjct 308 RMLRKAVERSSLMGCSIEIVSESELETMTQ#KMLVRGHAYSVTDLQEVWYQGRTETLIRVR 367 |
| Oryctolagus cuniculus | Query 226 KIIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 ||++||+|+| |+||||| | # ||||||||||| || | ||+|+| Sbjct 230 KIMRKAIERGFLMGCSIDTIVPVQYETRMA#CGLVKGHAYSVTGLEETLFKGEKVKLVRLR 289 |
| Caenorhabditis elegans | Query 227 IIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 +| | ++ ||| |||| + || # ||+|||||||| ||+ |+||| Sbjct 335 MIMKGMQMGSLFACSID-PDPREKEAQLA#NGLVRGHAYSVTGVHTVETDKEKVALLRIR 392 |
| Caenorhabditis briggsae AF16 | Query 227 IIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVTGAEEVESSGSLQKLIRIR 285 ++ + ++ ||| ||||| || # ||+|||||+| + | + |+||| Sbjct 487 MLVRGMQMGSLFGCSIDADENV-KEAQLT#NGLVRGHAYSITAIQTVNTYSGQTPLLRIR 544 |
| Drosophila melanogaster | Query 227 IIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVT 267 |+||| |+ |++||||+ +|| | #| |++|||||+| Sbjct 242 ILQKAAERNSMMGCSIEPDPNV-TEAETP#QGLIRGHAYSIT 281 |
| Drosophila melanogaster | Query 227 IIQKALEKGSLLGCSIDITSAADSEAVTY#QKLVKGHAYSVT 267 |+||| |+ |++||||+ +|| | #| |++|||||+| Sbjct 265 ILQKAAERNSMMGCSIEPDPNV-TEAETP#QGLIRGHAYSIT 304 |
[Site 21] LVKGHAYSVT267-GAEEVESSGS
Thr267  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu258 | Val259 | Lys260 | Gly261 | His262 | Ala263 | Tyr264 | Ser265 | Val266 | Thr267 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly268 | Ala269 | Glu270 | Glu271 | Val272 | Glu273 | Ser274 | Ser275 | Gly276 | Ser277 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LGCSIDITSAADSEAVTYQKLVKGHAYSVTGAEEVESSGSLQKLIRIRNPWGQVEWTGKW |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 126.00 | 22 | unnamed protein product |
| 2 | N/A | 126.00 | 15 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 3 | Rattus norvegicus | 126.00 | 12 | calpain 2 |
| 4 | Macaca mulatta | 124.00 | 2 | PREDICTED: calpain 2, large subunit |
| 5 | Homo sapiens | 121.00 | 2 | calpain 2, large subunit |
| 6 | Homo sapiens | 120.00 | 9 | CAPN2 protein |
| 7 | Gallus gallus | 112.00 | 2 | calpain 2, (m/II) large subunit |
| 8 | Xenopus tropicalis | 108.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Danio rerio | 105.00 | 5 | calpain 2, (m/II) large subunit a |
| 10 | Oncorhynchus mykiss | 105.00 | 1 | m-calpain |
| 11 | Tetraodon nigroviridis | 104.00 | 7 | unnamed protein product |
| 12 | Xenopus laevis | 101.00 | 3 | hypothetical protein LOC398772 |
| 13 | Stizostedion vitreum vitreum | 101.00 | 1 | putative calpain-like protein |
| 14 | Xenopus laevis | 101.00 | 1 | mu/m-calpain large subunit |
| 15 | Danio rerio | 101.00 | 1 | calpain 1, large subunit |
| 16 | Bos taurus | 96.70 | 5 | micromolar calcium activated neutral protease 1 |
| 17 | Canis familiaris | 96.30 | 20 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 18 | Gallus gallus | 96.30 | 1 | mu-calpain large subunit |
| 19 | Pongo pygmaeus | 95.90 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | synthetic construct | 95.50 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 21 | Coturnix coturnix | 95.50 | 1 | quail calpain |
| 22 | Sus scrofa | 95.10 | 4 | calpain 1, large subunit |
| 23 | Oncorhynchus mykiss | 94.00 | 1 | gill-specific calpain |
| 24 | Rattus norvegicus | 82.80 | 1 | calpain 1, large subunit |
| 25 | Canis familiaris | 77.40 | 1 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 26 | Oryctolagus cuniculus | 74.30 | 1 | calpain 3, (p94) |
| 27 | Ovis aries | 66.60 | 2 | skeletal muscle-specific calpain |
| 28 | Bos taurus | 63.20 | 1 | skeletal muscle-specific calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |
| N/A | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |
| Rattus norvegicus | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |
| Macaca mulatta | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||||||||||+||||||||||||||#||||||||||||||||||||||+|||||+| Sbjct 238 LGCSIDITSAADSEAITYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGEVEWTGRW 297 |
| Homo sapiens | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||||||||||+|+||||||||||||#|||||||+||||||||||||||+|||||+| Sbjct 238 LGCSIDITSAADSEAITFQKLVKGHAYSVT#GAEEVESNGSLQKLIRIRNPWGEVEWTGRW 297 |
| Homo sapiens | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||||||||||+|+||||||||||||#|||||||+||||||||||||||+|||||+| Sbjct 238 LGCSIDITSAADSEAITFQKLVKGHAYSVT#GAEEVESNGSLQKLIRIRNPWGEVEWTGRW 297 |
| Gallus gallus | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||||||++|||| ||||||||||||#||||| ||+|||||||||||+||||||| Sbjct 238 LGCSIDITSAAETEAVTSQKLVKGHAYSVT#GAEEVNFRGSIQKLIRIRNPWGEVEWTGKW 297 |
| Xenopus tropicalis | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||||||++||+|+||||||||||||# ||||+ |+|+||||||||||+||||| | Sbjct 238 LGCSIDITSAAETEAITFQKLVKGHAYSVT#AAEEVQYRGNLEKLIRIRNPWGEVEWTGAW 297 |
| Danio rerio | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 ||||||||||||||||||||||||||||+|#|| || +||+|+||||||||||| | Sbjct 238 LGCSIDITSAADSEAVTYQKLVKGHAYSLT#GATEVNYRSRKEKLVRVRNPWGQVEWTGAW 297 |
| Oncorhynchus mykiss | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 ||||||||||||||||| ||||||||||+|#|| || | +|||||||||||||||| | Sbjct 238 LGCSIDITSAADSEAVTRQKLVKGHAYSLT#GAVEVTYRGRKEKLIRIRNPWGQVEWTGAW 297 |
| Tetraodon nigroviridis | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 ||||||||||+||||+||+|||||||||||#||++|| | +|+||||||||||| | | Sbjct 244 LGCSIDITSASDSEAITYRKLVKGHAYSVT#GADQVEYRGEAVQLVRIRNPWGQVEWNGAW 303 |
| Xenopus laevis | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||||| | ||||++|||||||||||# +||+ | ++|||||||||||||||| | Sbjct 241 LGCSIDITSAFDMEAVTFKKLVKGHAYSVT#ALKEVDYRGGMEKLIRIRNPWGQVEWTGAW 300 |
| Stizostedion vitreum vitreum | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 +||||||||+++||||| ||||||||||||#| +|| | +|||||||||||||||| | Sbjct 239 MGCSIDITSSSESEAVTSQKLVKGHAYSVT#GTAQVEHQGGTEKLIRIRNPWGQVEWTGAW 298 |
| Xenopus laevis | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||||| | ||||++|||||||||||# +|| | ++|||||||||||||||| | Sbjct 241 LGCSIDITSAFDMEAVTFKKLVKGHAYSVT#ALKEVNYMGEMEKLIRIRNPWGQVEWTGAW 300 |
| Danio rerio | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 ||||||||| | ||||++|||||||||||#||||| |++ ||+|||||||+||||| | Sbjct 243 LGCSIDITSKFDMEAVTFKKLVKGHAYSVT#GAEEVVYRGNMTKLVRIRNPWGEVEWTGAW 302 |
| Bos taurus | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||+| | ||||++|||||||||||#||++| | + |||+|||||+||||| | Sbjct 248 LGCSIDISSILDMEAVTFKKLVKGHAYSVT#GAKQVNYQGQMVNLIRMRNPWGEVEWTGAW 307 |
| Canis familiaris | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||+| | ||+|++|||||||||||#||++| | + |||+|||||+||||| | Sbjct 248 LGCSIDISSVLDMEAITFKKLVKGHAYSVT#GAKQVNYQGQMVSLIRMRNPWGEVEWTGAW 307 |
| Gallus gallus | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||||| | ||||++|||||||||||#||+++ | |||+|||||+||||| | Sbjct 248 LGCSIDITSAFDMEAVTFKKLVKGHAYSVT#GAKQISYRGQSLGLIRMRNPWGEVEWTGAW 307 |
| Pongo pygmaeus | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||+| | ||+|++|||||||||||#||++| | + |||+|||||+||||| | Sbjct 248 LGCSIDISSVLDMEAITFKKLVKGHAYSVT#GAKQVNYRGQMVSLIRMRNPWGEVEWTGAW 307 |
| synthetic construct | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||+| | ||+|++|||||||||||#||++| | + |||+|||||+||||| | Sbjct 248 LGCSIDISSVLDMEAITFKKLVKGHAYSVT#GAKQVNYRGQVVSLIRMRNPWGEVEWTGAW 307 |
| Coturnix coturnix | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 ||| |||||| | ||||++|||||||||||# ++| | ++||||||||||||||| | Sbjct 241 LGCPIDITSAFDMEAVTFKKLVKGHAYSVT#AFKDVNYRGQQEQLIRIRNPWGQVEWTGAW 300 |
| Sus scrofa | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||||+| | ||||++|||||||||||#||++| | + |||+|||||+||||| | Sbjct 248 LGCSIDISSVLDMEAVTFKKLVKGHAYSVT#GAKQVNYQGQMVNLIRMRNPWGEVEWTGAW 307 |
| Oncorhynchus mykiss | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 +||||||||+|+||| | ||||||||+|#| ||| | |||||||||||||| | | Sbjct 216 MGCSIDITSSAESEAQTASGLVKGHAYSIT#GLEEVNYRGKKVKLIRIRNPWGQVEWNGAW 275 |
| Rattus norvegicus | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 |||||+|+ | ||+|++ ||+|||||||# |++| | |||+|||||+||| | | Sbjct 248 LGCSINISDIRDLEAITFKNLVRGHAYSVT#DAKQVTYQGQRVNLIRMRNPWGEVEWKGPW 307 |
| Canis familiaris | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 +||||+| | ++ | +| + ||+|||||||# +|| | + |||+|||||+||| | | Sbjct 320 MGCSIEIVSESELETMTQKMLVRGHAYSVT#DLQEVWYQGRTETLIRVRNPWGRVEWNGAW 379 |
| Oryctolagus cuniculus | Query 238 LGCSIDITSAADSEAVTYQKLVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 +||||| | ||||||||||#| || | ||+|+||||||||| | | Sbjct 242 MGCSIDTIVPVQYETRMACGLVKGHAYSVT#GLEETLFKGEKVKLVRLRNPWGQVEWNGSW 301 |
| Ovis aries | Query 258 LVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 ||||||||||#| || | ||+|+||||||||| | | Sbjct 331 LVKGHAYSVT#GLEEALYKGEKVKLVRLRNPWGQVEWNGSW 370 |
| Bos taurus | Query 258 LVKGHAYSVT#GAEEVESSGSLQKLIRIRNPWGQVEWTGKW 297 ||||||||||#| || | ||+|+||||||||| | | Sbjct 53 LVKGHAYSVT#GLEEALYKGEKVKLVRLRNPWGQVEWNGSW 92 |
[Site 22] CDSYKKWKLT359-KMDGNWRRGS
Thr359  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Cys350 | Asp351 | Ser352 | Tyr353 | Lys354 | Lys355 | Trp356 | Lys357 | Leu358 | Thr359 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys360 | Met361 | Asp362 | Gly363 | Asn364 | Trp365 | Arg366 | Arg367 | Gly368 | Ser369 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DFLRHYSRLEICNLTPDTLTCDSYKKWKLTKMDGNWRRGSTAGGCRNYPNTFWMNPQYLI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 143.00 | 16 | unnamed protein product |
| 2 | N/A | 143.00 | 10 | A Chain A, Crystal Structure Of M-Calpain |
| 3 | Rattus norvegicus | 143.00 | 9 | calpain 2 |
| 4 | Homo sapiens | 137.00 | 13 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 137.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 137.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Bos grunniens | 125.00 | 1 | calpain 2 |
| 8 | Xenopus tropicalis | 120.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Gallus gallus | 119.00 | 3 | calpain 2, (m/II) large subunit |
| 10 | Stizostedion vitreum vitreum | 112.00 | 1 | putative calpain-like protein |
| 11 | Tetraodon nigroviridis | 110.00 | 6 | unnamed protein product |
| 12 | Danio rerio | 106.00 | 5 | calpain 2, (m/II) large subunit a |
| 13 | Xenopus laevis | 102.00 | 3 | calpain 1, (mu/I) large subunit |
| 14 | Oncorhynchus mykiss | 102.00 | 1 | m-calpain |
| 15 | Canis familiaris | 99.80 | 22 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 16 | Gallus gallus | 99.80 | 1 | mu-calpain large subunit |
| 17 | Xenopus laevis | 99.00 | 1 | mu/m-calpain large subunit |
| 18 | Pongo pygmaeus | 97.80 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 19 | synthetic construct | 97.80 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Bos taurus | 97.40 | 5 | calpain 1, (mu/I) large subunit |
| 21 | Sus scrofa | 97.10 | 2 | calpain 1, large subunit |
| 22 | Coturnix coturnix | 96.30 | 1 | quail calpain |
| 23 | Danio rerio | 94.00 | 1 | calpain 1, large subunit |
| 24 | Rattus norvegicus | 94.00 | 1 | calpain 1, large subunit |
| 25 | Mus musculus | 87.40 | 1 | stomach specific calpain nCL-2 |
| 26 | Oncorhynchus mykiss | 86.70 | 1 | gill-specific calpain |
| 27 | Drosophila melanogaster | 85.10 | 3 | Calpain-B CG8107-PA |
| 28 | Macaca fascicularis | 85.10 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 29 | Drosophila pseudoobscura | 84.70 | 1 | GA20829-PA |
| 30 | Drosophila melanogaster | 84.30 | 5 | CAA55297.1 |
| 31 | Oryctolagus cuniculus | 83.20 | 1 | calpain 3, (p94) |
| 32 | Ovis aries | 81.60 | 2 | skeletal muscle-specific calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |
| N/A | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |
| Rattus norvegicus | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |
| Homo sapiens | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |||||||||||||||||||| |+|||||||#|||||||||||||||||||||||||||||| Sbjct 330 DFLRHYSRLEICNLTPDTLTSDTYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |
| Homo sapiens | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |||||||||||||||||||| |+|||||||#|||||||||||||||||||||||||||||| Sbjct 330 DFLRHYSRLEICNLTPDTLTSDTYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |
| Macaca mulatta | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |||||||||||||||||||| |+|||||||#|||||||||||||||||||||||||||||| Sbjct 330 DFLRHYSRLEICNLTPDTLTSDTYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |
| Bos grunniens | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |||||||||||||||||||| |+|||||||#|||||||||||||||||||||||||||||| Sbjct 42 DFLRHYSRLEICNLTPDTLTSDTYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 101 |
| Xenopus tropicalis | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||||+|||||||||||||| | | || ||#|||| ||||||||||||+| ||||||||+| Sbjct 330 DFLRNYSRLEICNLTPDTLASDKYSKWSLT#KMDGTWRRGSTAGGCRNFPETFWMNPQYVI 389 |
| Gallus gallus | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||||||||||||||||||| | |||| | #|+|||||||+||||||||||||| |||||| Sbjct 330 DFLRHYSRLEICNLTPDTLASDRYKKWSLL#KLDGNWRRGATAGGCRNYPNTFWTNPQYLI 389 |
| Stizostedion vitreum vitreum | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |||||| |||||||||| |+ |+ || |+#| ||+||||||||||||||+||| |||++| Sbjct 331 DFLRHYDRLEICNLTPDALSDDTVSKWALS#KFDGSWRRGSTAGGCRNYPDTFWTNPQFVI 390 |
| Tetraodon nigroviridis | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |||| |||||||||||| || | |||| |#+ + |||||||||||||||+||+|||++| Sbjct 336 DFLRQYSRLEICNLTPDALTGDEYKKWAET#EFEDTWRRGSTAGGCRNYPNSFWLNPQFVI 395 |
| Danio rerio | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||+| ||||||| ||||||| || | | + #| || ||+|||||||||+| |||||||+ | Sbjct 328 DFVRQYSRLEICTLTPDTLTSDSLKHWSVC#KFDGTWRKGSTAGGCRNHPYTFWMNPQFKI 387 |
| Xenopus laevis | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |||| +||||||||||| || |+|| |# +|+||+|||||||||+| |||+|||+ | Sbjct 340 DFLREFSRLEICNLTPDALTARRYRKWNTT#VYNGSWRKGSTAGGCRNFPATFWINPQFKI 399 |
| Oncorhynchus mykiss | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 +|||||||+||| ||||||| || | | + # ||+||+|||||||||+ |||||||++| Sbjct 328 EFLRHYSRIEICTLTPDTLTDDSVKHWSVC#NYDGSWRKGSTAGGCRNHAYTFWMNPQFVI 387 |
| Canis familiaris | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||+| ++|||||||||| | | +|| |# +| |||||||||||||| |||+|||+ | Sbjct 337 DFMREFTRLEICNLTPDALKSRSIRKWNTT#LYEGTWRRGSTAGGCRNYPATFWVNPQFKI 396 |
| Gallus gallus | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |||| ++|||||||||| | ++|| # ||+|||||||||||||| |||+|||+ | Sbjct 340 DFLREFTRLEICNLTPDALQSRKFRKWNTR#LYDGSWRRGSTAGGCRNYPATFWINPQFKI 399 |
| Xenopus laevis | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 +||| +||||||||||||| + || |# ||+|||||||||||| | |||+|||+ + Sbjct 333 EFLRQFSRLEICNLTPDTLNKEGLSKWHTT#LYDGSWRRGSTAGGCRNNPATFWINPQFKV 392 |
| Pongo pygmaeus | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||+| ++|||||||||| | + +|| |# +| |||||||||||||| |||+|||+ | Sbjct 340 DFMREFTRLEICNLTPDALKSRTIRKWNTT#LYEGTWRRGSTAGGCRNYPATFWVNPQFKI 399 |
| synthetic construct | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||+| ++|||||||||| | + +|| |# +| |||||||||||||| |||+|||+ | Sbjct 340 DFMREFTRLEICNLTPDALKSRTIRKWNTT#LYEGTWRRGSTAGGCRNYPATFWVNPQFKI 399 |
| Bos taurus | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||+| ++|||||||||| | ++ | |# +| |||||||||||||| |||+|||+ | Sbjct 340 DFMREFTRLEICNLTPDALKSQRFRNWNTT#LYEGTWRRGSTAGGCRNYPATFWVNPQFKI 399 |
| Sus scrofa | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |||| ++|||||||||| | + | |# +| |||||||||||||| |||+|||+ | Sbjct 340 DFLREFTRLEICNLTPDALKSQRVRNWNTT#LYEGTWRRGSTAGGCRNYPATFWVNPQFKI 399 |
| Coturnix coturnix | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||+| +||||||||||| || | +| # +| |||||||||||| | |||+|||+ | Sbjct 333 DFMREFSRLEICNLTPDALTKDELSRWHTQ#VFEGTWRRGSTAGGCRNNPATFWINPQFKI 392 |
| Danio rerio | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 |||| ++|||||||| | | ||| # +| |||||||||||||| ||| |||+ + Sbjct 335 DFLREFTRLEICNLTADALQASQVKKWSTA#NYNGEWRRGSTAGGCRNYPATFWNNPQFKV 394 |
| Rattus norvegicus | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||+| +++||||||||| | + + | |# +| |||||||||||||| |||+|||+ | Sbjct 340 DFIREFTKLEICNLTPDALKSRTLRNWNTT#FYEGTWRRGSTAGGCRNYPATFWVNPQFKI 399 |
| Mus musculus | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQY 387 |||+ +||||||||+||+|+ + +|| | # +| | ||||||||+||| |+| |||+ Sbjct 5 DFLKQFSRLEICNLSPDSLSSEEIQKWNLV#LFNGRWTRGSTAGGCQNYPATYWTNPQF 62 |
| Oncorhynchus mykiss | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQY 387 || +| ++||||||||+|| |+ +||++ # +||| ||||||||||+ +||| |||+ Sbjct 309 DFKANYDKIEICNLTPDSLTDDTKRKWEVN#MFEGNWIRGSTAGGCRNFIDTFWTNPQF 366 |
| Drosophila melanogaster | Query 330 DFLRHYSRLEICNLTPDTLTCD----SYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNP 385 ||| |+ |+|||||+||+|| | | +||+++# +| | | |||||||+ ||| || Sbjct 544 DFLNHFDRVEICNLSPDSLTEDQQHSSRRKWEMS#MFEGEWTSGVTAGGCRNFLETFWHNP 603 Query 386 QYLI 389 ||+| Sbjct 604 QYII 607 |
| Macaca fascicularis | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQY 387 ||+ |+++||||||| | | | + | ++# +| | || +||||||+|+||| |||| Sbjct 403 DFIYHFTKLEICNLTADALQSDKLQTWTVS#VNEGRWVRGCSAGGCRNFPDTFWTNPQY 460 |
| Drosophila pseudoobscura | Query 330 DFLRHYSRLEICNLTPDTLTCD----SYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNP 385 ||| |+ |+||||| ||+|| | | +||+++# +| | | |||||||+ ||| || Sbjct 550 DFLNHFDRVEICNLNPDSLTEDQQNSSRRKWEMS#MFEGEWTSGVTAGGCRNFLETFWHNP 609 Query 386 QYLI 389 ||+| Sbjct 610 QYVI 613 |
| Drosophila melanogaster | Query 330 DFLRHYSRLEICNLTPDTLTCDSY----KKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNP 385 ||| |+ |+|||||+||+|| | +||+++# +| | | |||||||+ +||| || Sbjct 350 DFLNHFDRVEICNLSPDSLTEDQQHSGKRKWEMS#MYEGEWTPGVTAGGCRNFLDTFWHNP 409 Query 386 QYLI 389 ||+| Sbjct 410 QYII 413 |
| Oryctolagus cuniculus | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQY 387 ||+ |+++||||||| | | | + | ++# +| | || +||||||+|+||| |||| Sbjct 335 DFVFHFTKLEICNLTADALESDKLQTWTVS#VNEGRWVRGCSAGGCRNFPDTFWTNPQY 392 |
| Ovis aries | Query 330 DFLRHYSRLEICNLTPDTLTCDSYKKWKLT#KMDGNWRRGSTAGGCRNYPNTFWMNPQY 387 ||+ |+++||||||| | | | + | ++# +| | || +||||||+|+||| |||| Sbjct 404 DFIYHFTKLEICNLTADALESDKLQTWTVS#VNEGRWVRGCSAGGCRNFPDTFWTNPQY 461 |
[Site 23] DSYKKWKLTK360-MDGNWRRGST
Lys360  Met
Met
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp351 | Ser352 | Tyr353 | Lys354 | Lys355 | Trp356 | Lys357 | Leu358 | Thr359 | Lys360 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Met361 | Asp362 | Gly363 | Asn364 | Trp365 | Arg366 | Arg367 | Gly368 | Ser369 | Thr370 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| FLRHYSRLEICNLTPDTLTCDSYKKWKLTKMDGNWRRGSTAGGCRNYPNTFWMNPQYLIK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 143.00 | 11 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Mus musculus | 142.00 | 18 | unnamed protein product |
| 3 | Rattus norvegicus | 142.00 | 9 | calpain 2 |
| 4 | Homo sapiens | 137.00 | 13 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 137.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 137.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Bos grunniens | 125.00 | 1 | calpain 2 |
| 8 | Xenopus tropicalis | 120.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Gallus gallus | 119.00 | 3 | calpain 2, (m/II) large subunit |
| 10 | Stizostedion vitreum vitreum | 111.00 | 1 | putative calpain-like protein |
| 11 | Tetraodon nigroviridis | 110.00 | 5 | unnamed protein product |
| 12 | Danio rerio | 106.00 | 5 | calpain 2, (m/II) large subunit a |
| 13 | Oncorhynchus mykiss | 103.00 | 1 | m-calpain |
| 14 | Xenopus laevis | 101.00 | 3 | calpain 1, (mu/I) large subunit |
| 15 | Canis familiaris | 98.60 | 22 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 16 | Xenopus laevis | 98.60 | 1 | mu/m-calpain large subunit |
| 17 | Gallus gallus | 97.40 | 1 | mu-calpain large subunit |
| 18 | synthetic construct | 96.70 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 19 | Pongo pygmaeus | 96.30 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | Coturnix coturnix | 96.30 | 1 | quail calpain |
| 21 | Bos taurus | 95.90 | 5 | calpain 1, (mu/I) large subunit |
| 22 | Sus scrofa | 95.50 | 2 | calpain 1, large subunit |
| 23 | Rattus norvegicus | 92.80 | 1 | calpain 1, large subunit |
| 24 | Danio rerio | 91.70 | 1 | calpain 1, large subunit |
| 25 | Oncorhynchus mykiss | 87.40 | 1 | gill-specific calpain |
| 26 | Mus musculus | 85.10 | 1 | stomach specific calpain nCL-2 |
| 27 | Macaca fascicularis | 84.30 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 28 | Drosophila melanogaster | 83.20 | 5 | calpain |
| 29 | Drosophila melanogaster | 83.20 | 3 | Calpain-B CG8107-PA |
| 30 | Drosophila pseudoobscura | 82.80 | 1 | GA20829-PA |
| 31 | Oryctolagus cuniculus | 82.80 | 1 | calpain 3, (p94) |
| 32 | Ovis aries | 81.30 | 2 | skeletal muscle-specific calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |
| Mus musculus | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |
| Rattus norvegicus | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |
| Homo sapiens | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 ||||||||||||||||||| |+||||||||#|||||||||||||||||||||||||||||| Sbjct 331 FLRHYSRLEICNLTPDTLTSDTYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |
| Homo sapiens | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 ||||||||||||||||||| |+||||||||#|||||||||||||||||||||||||||||| Sbjct 331 FLRHYSRLEICNLTPDTLTSDTYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |
| Macaca mulatta | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 ||||||||||||||||||| |+||||||||#|||||||||||||||||||||||||||||| Sbjct 331 FLRHYSRLEICNLTPDTLTSDTYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |
| Bos grunniens | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 ||||||||||||||||||| |+||||||||#|||||||||||||||||||||||||||||| Sbjct 43 FLRHYSRLEICNLTPDTLTSDTYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 102 |
| Xenopus tropicalis | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |||+|||||||||||||| | | || |||#||| ||||||||||||+| ||||||||+|| Sbjct 331 FLRNYSRLEICNLTPDTLASDKYSKWSLTK#MDGTWRRGSTAGGCRNFPETFWMNPQYVIK 390 |
| Gallus gallus | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |||||||||||||||||| | |||| | |#+|||||||+||||||||||||| ||||||| Sbjct 331 FLRHYSRLEICNLTPDTLASDRYKKWSLLK#LDGNWRRGATAGGCRNYPNTFWTNPQYLIK 390 |
| Stizostedion vitreum vitreum | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 ||||| |||||||||| |+ |+ || |+|# ||+||||||||||||||+||| |||++|+ Sbjct 332 FLRHYDRLEICNLTPDALSDDTVSKWALSK#FDGSWRRGSTAGGCRNYPDTFWTNPQFVIR 391 |
| Tetraodon nigroviridis | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 ||| |||||||||||| || | |||| |+# + |||||||||||||||+||+|||++|| Sbjct 337 FLRQYSRLEICNLTPDALTGDEYKKWAETE#FEDTWRRGSTAGGCRNYPNSFWLNPQFVIK 396 |
| Danio rerio | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |+| ||||||| ||||||| || | | + |# || ||+|||||||||+| |||||||+ || Sbjct 329 FVRQYSRLEICTLTPDTLTSDSLKHWSVCK#FDGTWRKGSTAGGCRNHPYTFWMNPQFKIK 388 |
| Oncorhynchus mykiss | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |||||||+||| ||||||| || | | + # ||+||+|||||||||+ |||||||++|+ Sbjct 329 FLRHYSRIEICTLTPDTLTDDSVKHWSVCN#YDGSWRKGSTAGGCRNHAYTFWMNPQFVIR 388 |
| Xenopus laevis | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 ||| +||||||||||| || |+|| | # +|+||+|||||||||+| |||+|||+ || Sbjct 341 FLREFSRLEICNLTPDALTARRYRKWNTTV#YNGSWRKGSTAGGCRNFPATFWINPQFKIK 400 |
| Canis familiaris | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |+| ++|||||||||| | | +|| | # +| |||||||||||||| |||+|||+ |+ Sbjct 338 FMREFTRLEICNLTPDALKSRSIRKWNTTL#YEGTWRRGSTAGGCRNYPATFWVNPQFKIR 397 |
| Xenopus laevis | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||| +||||||||||||| + || | # ||+|||||||||||| | |||+|||+ + Sbjct 334 FLRQFSRLEICNLTPDTLNKEGLSKWHTTL#YDGSWRRGSTAGGCRNNPATFWINPQFKV 392 |
| Gallus gallus | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||| ++|||||||||| | ++|| # ||+|||||||||||||| |||+|||+ | Sbjct 341 FLREFTRLEICNLTPDALQSRKFRKWNTRL#YDGSWRRGSTAGGCRNYPATFWINPQFKI 399 |
| synthetic construct | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |+| ++|||||||||| | + +|| | # +| |||||||||||||| |||+|||+ |+ Sbjct 341 FMREFTRLEICNLTPDALKSRTIRKWNTTL#YEGTWRRGSTAGGCRNYPATFWVNPQFKIR 400 |
| Pongo pygmaeus | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |+| ++|||||||||| | + +|| | # +| |||||||||||||| |||+|||+ |+ Sbjct 341 FMREFTRLEICNLTPDALKSRTIRKWNTTL#YEGTWRRGSTAGGCRNYPATFWVNPQFKIR 400 |
| Coturnix coturnix | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |+| +||||||||||| || | +| # +| |||||||||||| | |||+|||+ || Sbjct 334 FMREFSRLEICNLTPDALTKDELSRWHTQV#FEGTWRRGSTAGGCRNNPATFWINPQFKIK 393 |
| Bos taurus | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |+| ++|||||||||| | ++ | | # +| |||||||||||||| |||+|||+ |+ Sbjct 341 FMREFTRLEICNLTPDALKSQRFRNWNTTL#YEGTWRRGSTAGGCRNYPATFWVNPQFKIR 400 |
| Sus scrofa | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 ||| ++|||||||||| | + | | # +| |||||||||||||| |||+|||+ |+ Sbjct 341 FLREFTRLEICNLTPDALKSQRVRNWNTTL#YEGTWRRGSTAGGCRNYPATFWVNPQFKIR 400 |
| Rattus norvegicus | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |+| +++||||||||| | + + | | # +| |||||||||||||| |||+|||+ |+ Sbjct 341 FIREFTKLEICNLTPDALKSRTLRNWNTTF#YEGTWRRGSTAGGCRNYPATFWVNPQFKIR 400 |
| Danio rerio | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLI 389 ||| ++|||||||| | | ||| # +| |||||||||||||| ||| |||+ + Sbjct 336 FLREFTRLEICNLTADALQASQVKKWSTAN#YNGEWRRGSTAGGCRNYPATFWNNPQFKV 394 |
| Oncorhynchus mykiss | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQY 387 | +| ++||||||||+|| |+ +||++ # +||| ||||||||||+ +||| |||+ Sbjct 310 FKANYDKIEICNLTPDSLTDDTKRKWEVNM#FEGNWIRGSTAGGCRNFIDTFWTNPQF 366 |
| Mus musculus | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQY 387 ||+ +||||||||+||+|+ + +|| | # +| | ||||||||+||| |+| |||+ Sbjct 6 FLKQFSRLEICNLSPDSLSSEEIQKWNLVL#FNGRWTRGSTAGGCQNYPATYWTNPQF 62 |
| Macaca fascicularis | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |+ |+++||||||| | | | + | ++ # +| | || +||||||+|+||| |||| +| Sbjct 404 FIYHFTKLEICNLTADALQSDKLQTWTVSV#NEGRWVRGCSAGGCRNFPDTFWTNPQYRLK 463 |
| Drosophila melanogaster | Query 331 FLRHYSRLEICNLTPDTLTCD----SYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQ 386 || |+ |+|||||+||+|| | | +||+++ # +| | | |||||||+ ||| ||| Sbjct 545 FLNHFDRVEICNLSPDSLTEDQQHSSRRKWEMSM#FEGEWTSGVTAGGCRNFLETFWHNPQ 604 Query 387 YLI 389 |+| Sbjct 605 YII 607 |
| Drosophila melanogaster | Query 331 FLRHYSRLEICNLTPDTLTCD----SYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQ 386 || |+ |+|||||+||+|| | | +||+++ # +| | | |||||||+ ||| ||| Sbjct 545 FLNHFDRVEICNLSPDSLTEDQQHSSRRKWEMSM#FEGEWTSGVTAGGCRNFLETFWHNPQ 604 Query 387 YLI 389 |+| Sbjct 605 YII 607 |
| Drosophila pseudoobscura | Query 331 FLRHYSRLEICNLTPDTLTCD----SYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQ 386 || |+ |+||||| ||+|| | | +||+++ # +| | | |||||||+ ||| ||| Sbjct 551 FLNHFDRVEICNLNPDSLTEDQQNSSRRKWEMSM#FEGEWTSGVTAGGCRNFLETFWHNPQ 610 Query 387 YLI 389 |+| Sbjct 611 YVI 613 |
| Oryctolagus cuniculus | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |+ |+++||||||| | | | + | ++ # +| | || +||||||+|+||| |||| +| Sbjct 336 FVFHFTKLEICNLTADALESDKLQTWTVSV#NEGRWVRGCSAGGCRNFPDTFWTNPQYRLK 395 |
| Ovis aries | Query 331 FLRHYSRLEICNLTPDTLTCDSYKKWKLTK#MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK 390 |+ |+++||||||| | | | + | ++ # +| | || +||||||+|+||| |||| +| Sbjct 405 FIYHFTKLEICNLTADALESDKLQTWTVSV#NEGRWVRGCSAGGCRNFPDTFWTNPQYRLK 464 |
[Site 24] GGCRNYPNTF381-WMNPQYLIKL
Phe381  Trp
Trp
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly372 | Gly373 | Cys374 | Arg375 | Asn376 | Tyr377 | Pro378 | Asn379 | Thr380 | Phe381 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Trp382 | Met383 | Asn384 | Pro385 | Gln386 | Tyr387 | Leu388 | Ile389 | Lys390 | Leu391 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SYKKWKLTKMDGNWRRGSTAGGCRNYPNTFWMNPQYLIKLEEEDEDDEDGERGCTFLVGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 132.00 | 10 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 132.00 | 8 | calpain 2 |
| 3 | Mus musculus | 130.00 | 21 | calpain 2 |
| 4 | Xenopus tropicalis | 108.00 | 4 | calpain 2, (m/II) large subunit |
| 5 | Homo sapiens | 103.00 | 13 | AF261089_1 calpain large polypeptide L2 |
| 6 | Homo sapiens | 103.00 | 2 | calpain 2, large subunit |
| 7 | Macaca mulatta | 103.00 | 2 | PREDICTED: calpain 2, large subunit |
| 8 | Stizostedion vitreum vitreum | 102.00 | 1 | putative calpain-like protein |
| 9 | Bos grunniens | 100.00 | 1 | calpain 2 |
| 10 | Tetraodon nigroviridis | 93.60 | 5 | unnamed protein product |
| 11 | Gallus gallus | 93.20 | 3 | calpain 2, (m/II) large subunit |
| 12 | Xenopus laevis | 89.40 | 4 | calpain 1, (mu/I) large subunit |
| 13 | Danio rerio | 85.10 | 5 | Capn8 protein |
| 14 | Canis familiaris | 82.80 | 22 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 15 | Pongo pygmaeus | 81.60 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 16 | synthetic construct | 81.60 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 17 | Rattus norvegicus | 79.70 | 1 | calpain 1, large subunit |
| 18 | Sus scrofa | 78.60 | 1 | calpain 1, large subunit |
| 19 | Oncorhynchus mykiss | 74.30 | 1 | m-calpain |
| 20 | Bos taurus | 73.60 | 3 | calpain 1, (mu/I) large subunit |
| 21 | Gallus gallus | 72.80 | 1 | mu-calpain large subunit |
| 22 | Schistosoma japonicum | 71.60 | 2 | SJCHGC08992 protein |
| 23 | Danio rerio | 71.60 | 1 | calpain 1, large subunit |
| 24 | Drosophila melanogaster | 70.50 | 4 | CAA55297.1 |
| 25 | Drosophila melanogaster | 70.50 | 2 | Calpain-A CG7563-PB, isoform B |
| 26 | Xenopus laevis | 67.40 | 1 | mu/m-calpain large subunit |
| 27 | Coturnix coturnix | 67.00 | 1 | quail calpain |
| 28 | Oncorhynchus mykiss | 62.00 | 1 | gill-specific calpain |
| 29 | Oryctolagus cuniculus | 61.60 | 1 | calpain 3, (p94) |
| 30 | Macaca fascicularis | 61.60 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 |
| Rattus norvegicus | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 |
| Mus musculus | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 ||||||||||||||||||||||||||||||#||||||||||||||||+||||||||||||| Sbjct 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDEEDGERGCTFLVGL 411 |
| Xenopus tropicalis | Query 353 YKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 | || |||||| ||||||||||||+| ||#||||||+||||| |+| +| | ||||+||| Sbjct 353 YSKWSLTKMDGTWRRGSTAGGCRNFPETF#WMNPQYVIKLEEADDDPDDNEEGCTFIVGL 411 |
| Homo sapiens | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 +|||||||||||||||||||||||||||||#||||||||||||||||+|||| |||||||| Sbjct 352 TYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDEEDGESGCTFLVGL 411 |
| Homo sapiens | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 +|||||||||||||||||||||||||||||#||||||||||||||||+|||| |||||||| Sbjct 352 TYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDEEDGESGCTFLVGL 411 |
| Macaca mulatta | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 +|||||||||||||||||||||||||||||#||||||||||||||||+|||| |||||||| Sbjct 352 TYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDEEDGESGCTFLVGL 411 |
| Stizostedion vitreum vitreum | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 + || |+| ||+||||||||||||||+||#| |||++|+|||||+| ||| ||+|+||| Sbjct 353 TVSKWALSKFDGSWRRGSTAGGCRNYPDTF#WTNPQFVIRLEEEDDDPADGELGCSFVVGL 412 |
| Bos grunniens | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 +|||||||||||||||||||||||||||||#|||||||||||||||| |||| |||||||| Sbjct 64 TYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDQEDGESGCTFLVGL 123 |
| Tetraodon nigroviridis | Query 353 YKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 |||| |+ + |||||||||||||||+|#|+|||++||||| |+| + + ||||+||| Sbjct 359 YKKWAETEFEDTWRRGSTAGGCRNYPNSF#WLNPQFVIKLEEVDDDPGEDQEGCTFIVGL 417 |
| Gallus gallus | Query 353 YKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 |||| | |+|||||||+||||||||||||#| |||||||||||||| +| | |||||+|| Sbjct 353 YKKWSLLKLDGNWRRGATAGGCRNYPNTF#WTNPQYLIKLEEEDEDPDDPEGGCTFLIGL 411 |
| Xenopus laevis | Query 353 YKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDG-ERGCTFLVGL 411 |+|| | +|+||+|||||||||+| ||#|+|||+ ||||||| ||+ | | |||||+ | Sbjct 363 YRKWNTTVYNGSWRKGSTAGGCRNFPATF#WINPQFKIKLEEEDGDDDYGSETGCTFLLAL 422 |
| Danio rerio | Query 355 KWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 +| ++ +|||| ||||||||| | ||# |||++||||| |+| || |||||||| Sbjct 355 RWSYSQFEGNWRVGSTAGGCRNNPATF#CSNPQFMIKLEEIDDDPHDGVNGCTFLVGL 411 |
| Canis familiaris | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDED---GERGCTFL 408 | +|| | +| |||||||||||||| ||#|+|||+ |+||| |+ |+| | ||+|+ Sbjct 359 SIRKWNTTLYEGTWRRGSTAGGCRNYPATF#WVNPQFKIRLEETDDVDDDYGGRESGCSFV 418 Query 409 VGL 411 + | Sbjct 419 LAL 421 |
| Pongo pygmaeus | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGER--GCTFLV 409 + +|| | +| |||||||||||||| ||#|+|||+ |+|+| |+ |+ |+| ||+|++ Sbjct 362 TIRKWNTTLYEGTWRRGSTAGGCRNYPATF#WVNPQFKIRLDETDDPDDYGDRESGCSFVL 421 Query 410 GL 411 | Sbjct 422 AL 423 |
| synthetic construct | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGER--GCTFLV 409 + +|| | +| |||||||||||||| ||#|+|||+ |+|+| |+ |+ |+| ||+|++ Sbjct 362 TIRKWNTTLYEGTWRRGSTAGGCRNYPATF#WVNPQFKIRLDETDDPDDYGDRESGCSFVL 421 Query 410 GL 411 | Sbjct 422 AL 423 |
| Rattus norvegicus | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEE-EDEDDEDG-ERGCTFLV 409 + + | | +| |||||||||||||| ||#|+|||+ |+||| +| || | | ||+||+ Sbjct 362 TLRNWNTTFYEGTWRRGSTAGGCRNYPATF#WVNPQFKIRLEEVDDADDYDSRESGCSFLL 421 Query 410 GL 411 | Sbjct 422 AL 423 |
| Sus scrofa | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEED--EDDEDG-ERGCTFLVG 410 + | | +| |||||||||||||| ||#|+|||+ |+||| | ||| | | ||+|++ Sbjct 364 RNWNTTLYEGTWRRGSTAGGCRNYPATF#WVNPQFKIRLEETDDPEDDYGGRESGCSFVLA 423 Query 411 L 411 | Query 411 L 411 |
| Oncorhynchus mykiss | Query 352 SYKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 | | | + ||+||+|||||||||+ ||#|||||++|+|+|||+| +| | ||+|+||| Sbjct 350 SVKHWSVCNYDGSWRKGSTAGGCRNHAYTF#WMNPQFVIRLDEEDDDPDDNEVGCSFVVGL 409 |
| Bos taurus | Query 353 YKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDG----ERGCTFL 408 ++ | | +| |||||||||||||| ||#|+|||+ |+||| |+ | | | ||+|| Sbjct 363 FRNWNTTLYEGTWRRGSTAGGCRNYPATF#WVNPQFKIRLEETDDPDPDDYGGRESGCSFL 422 Query 409 VGL 411 + | Sbjct 423 LAL 425 |
| Gallus gallus | Query 353 YKKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDG---ERGCTFLV 409 ++|| ||+|||||||||||||| ||#|+|||+ | ||| |+| +| | ||+||+ Sbjct 363 FRKWNTRLYDGSWRRGSTAGGCRNYPATF#WINPQFKICLEEVDDDGDDFGGREPGCSFLL 422 Query 410 GL 411 | Sbjct 423 AL 424 |
| Schistosoma japonicum | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 ++||+| +| | | |||||| |+||||#+||||+ +++ + || |+|| | +||| Sbjct 61 RRWKITCEEGEWLRNSTAGGCTNFPNTF#YMNPQFHVQIVDPDESDDDGTG--TLVVGL 116 |
| Danio rerio | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 ||| +| |||||||||||||| ||#| |||+ + |+ | |+ |+||| | Sbjct 359 KKWSTANYNGEWRRGSTAGGCRNYPATF#WNNPQFKVALKHP---DSPGQSECSFLVAL 413 |
| Drosophila melanogaster | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 +||+++ +| | | |||||||+ +||#| ||||+| | + ||+||+|+ || +| | Sbjct 378 RKWEMSMYEGEWTPGVTAGGCRNFLDTF#WHNPQYIITLVDPDEEDEEGQ--CTVIVAL 433 |
| Drosophila melanogaster | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 +||+++ +| | | |||||||+ +||#| ||||+| | + ||+||+|+ || +| | Sbjct 401 RKWEMSMYEGEWTPGVTAGGCRNFLDTF#WHNPQYIITLVDPDEEDEEGQ--CTVIVAL 456 |
| Xenopus laevis | Query 355 KWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 || | ||+|||||||||||| | ||#|+|||+ + | |||+| +| | |+||| | Sbjct 358 KWHTTLYDGSWRRGSTAGGCRNNPATF#WINPQFKVTLLEEDDDPDDNELACSFLVAL 414 |
| Coturnix coturnix | Query 355 KWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 +| +| |||||||||||| | ||#|+|||+ ||| |||+| | | |+||| | Sbjct 358 RWHTQVFEGTWRRGSTAGGCRNNPATF#WINPQFKIKLLEEDDDPGDDEVACSFLVAL 414 |
| Oncorhynchus mykiss | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 +||++ +||| ||||||||||+ +||#| |||+ + | |++++ |+ ++ | Sbjct 333 RKWEVNMFEGNWIRGSTAGGCRNFIDTF#WTNPQFKLTLLHADDNNDK----CSVVIAL 386 |
| Oryctolagus cuniculus | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 + | ++ +| | || +||||||+|+||#| |||| +|| |||+| || | |+||| | Sbjct 359 QTWTVSVNEGRWVRGCSAGGCRNFPDTF#WTNPQYRLKLLEEDDDPEDSEVICSFLVAL 416 |
| Macaca fascicularis | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTF#WMNPQYLIKLEEEDEDDEDGERGCTFLVGL 411 + | ++ +| | || +||||||+|+||#| |||| +|| |||+| +| | |+||| | Sbjct 427 QTWTVSVNEGRWVRGCSAGGCRNFPDTF#WTNPQYRLKLLEEDDDPDDSEVICSFLVAL 484 |
[Site 25] CRNYPNTFWM383-NPQYLIKLEE
Met383  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Cys374 | Arg375 | Asn376 | Tyr377 | Pro378 | Asn379 | Thr380 | Phe381 | Trp382 | Met383 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn384 | Pro385 | Gln386 | Tyr387 | Leu388 | Ile389 | Lys390 | Leu391 | Glu392 | Glu393 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| KKWKLTKMDGNWRRGSTAGGCRNYPNTFWMNPQYLIKLEEEDEDDEDGERGCTFLVGLIQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 132.00 | 10 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 131.00 | 8 | calpain 2 |
| 3 | Mus musculus | 130.00 | 21 | unnamed protein product |
| 4 | Homo sapiens | 127.00 | 13 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 127.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 127.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Bos grunniens | 123.00 | 1 | calpain 2 |
| 8 | Xenopus tropicalis | 109.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Stizostedion vitreum vitreum | 106.00 | 1 | putative calpain-like protein |
| 10 | Tetraodon nigroviridis | 95.90 | 5 | unnamed protein product |
| 11 | Gallus gallus | 94.40 | 3 | calpain 2, (m/II) large subunit |
| 12 | Xenopus laevis | 89.70 | 4 | calpain 1, (mu/I) large subunit |
| 13 | Danio rerio | 87.80 | 5 | Capn8 protein |
| 14 | Canis familiaris | 84.30 | 22 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 15 | Pongo pygmaeus | 84.30 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 16 | synthetic construct | 84.30 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 17 | Rattus norvegicus | 82.40 | 1 | calpain 1, large subunit |
| 18 | Sus scrofa | 81.60 | 1 | calpain 1, large subunit |
| 19 | Oncorhynchus mykiss | 77.00 | 1 | m-calpain |
| 20 | Bos taurus | 75.10 | 3 | calpain 1, (mu/I) large subunit |
| 21 | Danio rerio | 74.70 | 1 | calpain 1, large subunit |
| 22 | Schistosoma japonicum | 74.30 | 2 | SJCHGC08992 protein |
| 23 | Gallus gallus | 73.90 | 1 | mu-calpain large subunit |
| 24 | Drosophila melanogaster | 73.20 | 4 | CAA55297.1 |
| 25 | Drosophila melanogaster | 72.80 | 2 | Calpain-A CG7563-PB, isoform B |
| 26 | Xenopus laevis | 70.10 | 1 | mu/m-calpain large subunit |
| 27 | Coturnix coturnix | 69.30 | 1 | quail calpain |
| 28 | Macaca fascicularis | 64.30 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 29 | Oncorhynchus mykiss | 64.30 | 1 | gill-specific calpain |
| 30 | Oryctolagus cuniculus | 63.90 | 1 | calpain 3, (p94) |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 |
| Rattus norvegicus | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 |
| Mus musculus | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 ||||||||||||||||||||||||||||||#||||||||||||||+||||||||||||||| Sbjct 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDEEDGERGCTFLVGLIQ 413 |
| Homo sapiens | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 ||||||||||||||||||||||||||||||#||||||||||||||+|||| |||||||||| Sbjct 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDEEDGESGCTFLVGLIQ 413 |
| Homo sapiens | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 ||||||||||||||||||||||||||||||#||||||||||||||+|||| |||||||||| Sbjct 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDEEDGESGCTFLVGLIQ 413 |
| Macaca mulatta | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 ||||||||||||||||||||||||||||||#||||||||||||||+|||| |||||||||| Sbjct 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDEEDGESGCTFLVGLIQ 413 |
| Bos grunniens | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 ||||||||||||||||||||||||||||||#|||||||||||||| |||| |||||||||| Sbjct 66 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDQEDGESGCTFLVGLIQ 125 |
| Xenopus tropicalis | Query 355 KWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 || |||||| ||||||||||||+| ||||#||||+||||| |+| +| | ||||+||||| Sbjct 355 KWSLTKMDGTWRRGSTAGGCRNFPETFWM#NPQYVIKLEEADDDPDDNEEGCTFIVGLIQ 413 |
| Stizostedion vitreum vitreum | Query 355 KWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 || |+| ||+||||||||||||||+||| #|||++|+|||||+| ||| ||+|+||||| Sbjct 356 KWALSKFDGSWRRGSTAGGCRNYPDTFWT#NPQFVIRLEEEDDDPADGELGCSFVVGLIQ 414 |
| Tetraodon nigroviridis | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 ||| |+ + |||||||||||||||+||+#|||++||||| |+| + + ||||+|||+| Sbjct 360 KKWAETEFEDTWRRGSTAGGCRNYPNSFWL#NPQFVIKLEEVDDDPGEDQEGCTFIVGLMQ 419 |
| Gallus gallus | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 ||| | |+|||||||+||||||||||||| #|||||||||||||| +| | |||||+|||| Sbjct 354 KKWSLLKLDGNWRRGATAGGCRNYPNTFWT#NPQYLIKLEEEDEDPDDPEGGCTFLIGLIQ 413 |
| Xenopus laevis | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDG-ERGCTFLVGLI 412 +|| | +|+||+|||||||||+| |||+#|||+ ||||||| ||+ | | |||||+ |+ Sbjct 364 RKWNTTVYNGSWRKGSTAGGCRNFPATFWI#NPQFKIKLEEEDGDDDYGSETGCTFLLALM 423 Query 413 Q 413 Query 413 Q 413 |
| Danio rerio | Query 355 KWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 +| ++ +|||| ||||||||| | || #|||++||||| |+| || ||||||||+| Sbjct 355 RWSYSQFEGNWRVGSTAGGCRNNPATFCS#NPQFMIKLEEIDDDPHDGVNGCTFLVGLMQ 413 |
| Canis familiaris | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDED---GERGCTFLVG 410 +|| | +| |||||||||||||| |||+#|||+ |+||| |+ |+| | ||+|++ Sbjct 361 RKWNTTLYEGTWRRGSTAGGCRNYPATFWV#NPQFKIRLEETDDVDDDYGGRESGCSFVLA 420 Query 411 LIQ 413 |+| Sbjct 421 LMQ 423 |
| Pongo pygmaeus | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGER--GCTFLVGL 411 +|| | +| |||||||||||||| |||+#|||+ |+|+| |+ |+ |+| ||+|++ | Sbjct 364 RKWNTTLYEGTWRRGSTAGGCRNYPATFWV#NPQFKIRLDETDDPDDYGDRESGCSFVLAL 423 Query 412 IQ 413 +| Sbjct 424 MQ 425 |
| synthetic construct | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGER--GCTFLVGL 411 +|| | +| |||||||||||||| |||+#|||+ |+|+| |+ |+ |+| ||+|++ | Sbjct 364 RKWNTTLYEGTWRRGSTAGGCRNYPATFWV#NPQFKIRLDETDDPDDYGDRESGCSFVLAL 423 Query 412 IQ 413 +| Sbjct 424 MQ 425 |
| Rattus norvegicus | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEE-EDEDDEDG-ERGCTFLVGL 411 + | | +| |||||||||||||| |||+#|||+ |+||| +| || | | ||+||+ | Sbjct 364 RNWNTTFYEGTWRRGSTAGGCRNYPATFWV#NPQFKIRLEEVDDADDYDSRESGCSFLLAL 423 Query 412 IQ 413 +| Sbjct 424 MQ 425 |
| Sus scrofa | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEED--EDDEDG-ERGCTFLVG 410 + | | +| |||||||||||||| |||+#|||+ |+||| | ||| | | ||+|++ Sbjct 364 RNWNTTLYEGTWRRGSTAGGCRNYPATFWV#NPQFKIRLEETDDPEDDYGGRESGCSFVLA 423 Query 411 LIQ 413 |+| Sbjct 424 LMQ 426 |
| Oncorhynchus mykiss | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 | | + ||+||+|||||||||+ ||||#|||++|+|+|||+| +| | ||+|+||||| Sbjct 352 KHWSVCNYDGSWRKGSTAGGCRNHAYTFWM#NPQFVIRLDEEDDDPDDNEVGCSFVVGLIQ 411 |
| Bos taurus | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDG----ERGCTFLV 409 + | | +| |||||||||||||| |||+#|||+ |+||| |+ | | | ||+||+ Sbjct 364 RNWNTTLYEGTWRRGSTAGGCRNYPATFWV#NPQFKIRLEETDDPDPDDYGGRESGCSFLL 423 Query 410 GLIQ 413 |+| Sbjct 424 ALMQ 427 |
| Danio rerio | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 ||| +| |||||||||||||| ||| #|||+ + |+ | |+ |+||| |+| Sbjct 359 KKWSTANYNGEWRRGSTAGGCRNYPATFWN#NPQFKVALKHP---DSPGQSECSFLVALMQ 415 |
| Schistosoma japonicum | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 ++||+| +| | | |||||| |+||||+|#|||+ +++ + || |+|| | +|||+| Sbjct 61 RRWKITCEEGEWLRNSTAGGCTNFPNTFYM#NPQFHVQIVDPDESDDDGTG--TLVVGLMQ 118 |
| Gallus gallus | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDG---ERGCTFLVG 410 +|| ||+|||||||||||||| |||+#|||+ | ||| |+| +| | ||+||+ Sbjct 364 RKWNTRLYDGSWRRGSTAGGCRNYPATFWI#NPQFKICLEEVDDDGDDFGGREPGCSFLLA 423 Query 411 LIQ 413 |+| Sbjct 424 LMQ 426 |
| Drosophila melanogaster | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 +||+++ +| | | |||||||+ +||| #||||+| | + ||+||+|+ || +| |+| Sbjct 378 RKWEMSMYEGEWTPGVTAGGCRNFLDTFWH#NPQYIITLVDPDEEDEEGQ--CTVIVALMQ 435 |
| Drosophila melanogaster | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 +||+++ +| | | |||||||+ +||| #||||+| | + ||+||+|+ || +| |+| Sbjct 401 RKWEMSMYEGEWTPGVTAGGCRNFLDTFWH#NPQYIITLVDPDEEDEEGQ--CTVIVALMQ 458 |
| Xenopus laevis | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 || | ||+|||||||||||| | |||+#|||+ + | |||+| +| | |+||| |+| Sbjct 357 SKWHTTLYDGSWRRGSTAGGCRNNPATFWI#NPQFKVTLLEEDDDPDDNELACSFLVALMQ 416 |
| Coturnix coturnix | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 +| +| |||||||||||| | |||+#|||+ ||| |||+| | | |+||| |+| Sbjct 357 SRWHTQVFEGTWRRGSTAGGCRNNPATFWI#NPQFKIKLLEEDDDPGDDEVACSFLVALMQ 416 |
| Macaca fascicularis | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 + | ++ +| | || +||||||+|+||| #|||| +|| |||+| +| | |+||| |+| Sbjct 427 QTWTVSVNEGRWVRGCSAGGCRNFPDTFWT#NPQYRLKLLEEDDDPDDSEVICSFLVALMQ 486 |
| Oncorhynchus mykiss | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 +||++ +||| ||||||||||+ +||| #|||+ + | |++++ |+ ++ |+| Sbjct 333 RKWEVNMFEGNWIRGSTAGGCRNFIDTFWT#NPQFKLTLLHADDNNDK----CSVVIALMQ 388 |
| Oryctolagus cuniculus | Query 354 KKWKLTKMDGNWRRGSTAGGCRNYPNTFWM#NPQYLIKLEEEDEDDEDGERGCTFLVGLIQ 413 + | ++ +| | || +||||||+|+||| #|||| +|| |||+| || | |+||| |+| Sbjct 359 QTWTVSVNEGRWVRGCSAGGCRNFPDTFWT#NPQYRLKLLEEDDDPEDSEVICSFLVALMQ 418 |
[Site 26] PNTFWMNPQY387-LIKLEEEDED
Tyr387  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro378 | Asn379 | Thr380 | Phe381 | Trp382 | Met383 | Asn384 | Pro385 | Gln386 | Tyr387 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu388 | Ile389 | Lys390 | Leu391 | Glu392 | Glu393 | Glu394 | Asp395 | Glu396 | Asp397 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LTKMDGNWRRGSTAGGCRNYPNTFWMNPQYLIKLEEEDEDDEDGERGCTFLVGLIQKHRR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 132.00 | 11 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 132.00 | 9 | calpain 2 |
| 3 | Mus musculus | 130.00 | 20 | unnamed protein product |
| 4 | Homo sapiens | 128.00 | 13 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 128.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 128.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Bos grunniens | 115.00 | 1 | calpain 2 |
| 8 | Xenopus tropicalis | 110.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Stizostedion vitreum vitreum | 106.00 | 1 | putative calpain-like protein |
| 10 | Gallus gallus | 97.10 | 3 | calpain 2, (m/II) large subunit |
| 11 | Tetraodon nigroviridis | 94.40 | 5 | unnamed protein product |
| 12 | Xenopus laevis | 89.70 | 4 | calpain 1, (mu/I) large subunit |
| 13 | Danio rerio | 87.00 | 5 | Capn8 protein |
| 14 | Rattus norvegicus | 84.30 | 1 | calpain 1, large subunit |
| 15 | Pongo pygmaeus | 84.30 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 16 | synthetic construct | 84.30 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 17 | Canis familiaris | 84.00 | 17 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 18 | Sus scrofa | 83.20 | 2 | calpain 1, large subunit |
| 19 | Gallus gallus | 79.70 | 1 | mu-calpain large subunit |
| 20 | Oncorhynchus mykiss | 78.60 | 1 | m-calpain |
| 21 | Bos taurus | 76.60 | 5 | calpain 1, (mu/I) large subunit |
| 22 | Coturnix coturnix | 75.90 | 1 | quail calpain |
| 23 | Danio rerio | 73.90 | 1 | calpain 1, large subunit |
| 24 | Drosophila melanogaster | 73.20 | 4 | CAA55297.1 |
| 25 | Drosophila melanogaster | 73.20 | 2 | Calpain-A CG7563-PB, isoform B |
| 26 | Xenopus laevis | 71.60 | 1 | mu/m-calpain large subunit |
| 27 | Schistosoma japonicum | 69.30 | 2 | unknown |
| 28 | Macaca fascicularis | 67.40 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 29 | Oryctolagus cuniculus | 67.00 | 1 | calpain 3, (p94) |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 |
| Rattus norvegicus | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 |
| Mus musculus | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 ||||||||||||||||||||||||||||||#||||||||||+||||||||||||||||||| Sbjct 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDEEDGERGCTFLVGLIQKHRR 417 |
| Homo sapiens | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 ||||||||||||||||||||||||||||||#||||||||||+|||| |||||||||||||| Sbjct 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDEEDGESGCTFLVGLIQKHRR 417 |
| Homo sapiens | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 ||||||||||||||||||||||||||||||#||||||||||+|||| |||||||||||||| Sbjct 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDEEDGESGCTFLVGLIQKHRR 417 |
| Macaca mulatta | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 ||||||||||||||||||||||||||||||#||||||||||+|||| |||||||||||||| Sbjct 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDEEDGESGCTFLVGLIQKHRR 417 |
| Bos grunniens | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQ 413 ||||||||||||||||||||||||||||||#|||||||||| |||| |||||||||| Sbjct 70 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDQEDGESGCTFLVGLIQ 125 |
| Xenopus tropicalis | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 |||||| ||||||||||||+| ||||||||#+||||| |+| +| | ||||+||||||+|| Sbjct 358 LTKMDGTWRRGSTAGGCRNFPETFWMNPQY#VIKLEEADDDPDDNEEGCTFIVGLIQKNRR 417 |
| Stizostedion vitreum vitreum | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 |+| ||+||||||||||||||+||| |||+#+|+|||||+| ||| ||+|+||||||+|| Sbjct 359 LSKFDGSWRRGSTAGGCRNYPDTFWTNPQF#VIRLEEEDDDPADGELGCSFVVGLIQKNRR 418 |
| Gallus gallus | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 | |+|||||||+||||||||||||| ||||#|||||||||| +| | |||||+|||||||| Sbjct 358 LLKLDGNWRRGATAGGCRNYPNTFWTNPQY#LIKLEEEDEDPDDPEGGCTFLIGLIQKHRR 417 |
| Tetraodon nigroviridis | Query 359 TKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 |+ + |||||||||||||||+||+|||+#+||||| |+| + + ||||+|||+||+|| Sbjct 365 TEFEDTWRRGSTAGGCRNYPNSFWLNPQF#VIKLEEVDDDPGEDQEGCTFIVGLMQKNRR 423 |
| Xenopus laevis | Query 359 TKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDG-ERGCTFLVGLIQKHRR 417 | +|+||+|||||||||+| |||+|||+# ||||||| ||+ | | |||||+ |+||++| Sbjct 369 TVYNGSWRKGSTAGGCRNFPATFWINPQF#KIKLEEEDGDDDYGSETGCTFLLALMQKNKR 428 |
| Danio rerio | Query 359 TKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 ++ +|||| ||||||||| | || |||+#+||||| |+| || ||||||||+|| | Sbjct 359 SQFEGNWRVGSTAGGCRNNPATFCSNPQF#MIKLEEIDDDPHDGVNGCTFLVGLMQKDGR 417 |
| Rattus norvegicus | Query 359 TKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEE-EDEDDEDG-ERGCTFLVGLIQKH 415 | +| |||||||||||||| |||+|||+# |+||| +| || | | ||+||+ |+||| Sbjct 369 TFYEGTWRRGSTAGGCRNYPATFWVNPQF#KIRLEEVDDADDYDSRESGCSFLLALMQKH 427 |
| Pongo pygmaeus | Query 359 TKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGER--GCTFLVGLIQKH 415 | +| |||||||||||||| |||+|||+# |+|+| |+ |+ |+| ||+|++ |+||| Sbjct 369 TLYEGTWRRGSTAGGCRNYPATFWVNPQF#KIRLDETDDPDDYGDRESGCSFVLALMQKH 427 |
| synthetic construct | Query 359 TKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGER--GCTFLVGLIQKH 415 | +| |||||||||||||| |||+|||+# |+|+| |+ |+ |+| ||+|++ |+||| Sbjct 369 TLYEGTWRRGSTAGGCRNYPATFWVNPQF#KIRLDETDDPDDYGDRESGCSFVLALMQKH 427 |
| Canis familiaris | Query 359 TKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDG---ERGCTFLVGLIQKH 415 | +| |||||||||||||| |||+|||+# |+||| |+ |+| | ||+|++ |+||| Sbjct 366 TLYEGTWRRGSTAGGCRNYPATFWVNPQF#KIRLEETDDVDDDYGGRESGCSFVLALMQKH 425 |
| Sus scrofa | Query 359 TKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEED--EDDEDG-ERGCTFLVGLIQKH 415 | +| |||||||||||||| |||+|||+# |+||| | ||| | | ||+|++ |+||| Sbjct 369 TLYEGTWRRGSTAGGCRNYPATFWVNPQF#KIRLEETDDPEDDYGGRESGCSFVLALMQKH 428 |
| Gallus gallus | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDG---ERGCTFLVGLIQKHRR 417 ||+|||||||||||||| |||+|||+# | ||| |+| +| | ||+||+ |+||||| Sbjct 372 DGSWRRGSTAGGCRNYPATFWINPQF#KICLEEVDDDGDDFGGREPGCSFLLALMQKHRR 430 |
| Oncorhynchus mykiss | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 ||+||+|||||||||+ |||||||+#+|+|+|||+| +| | ||+|+||||||+|| Sbjct 360 DGSWRKGSTAGGCRNHAYTFWMNPQF#VIRLDEEDDDPDDNEVGCSFVVGLIQKNRR 415 |
| Bos taurus | Query 359 TKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDG----ERGCTFLVGLIQK 414 | +| |||||||||||||| |||+|||+# |+||| |+ | | | ||+||+ |+|| Sbjct 369 TLYEGTWRRGSTAGGCRNYPATFWVNPQF#KIRLEETDDPDPDDYGGRESGCSFLLALMQK 428 Query 415 H 415 | Query 415 H 415 |
| Coturnix coturnix | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 +| |||||||||||| | |||+|||+# ||| |||+| | | |+||| |+||||| Sbjct 364 FEGTWRRGSTAGGCRNNPATFWINPQF#KIKLLEEDDDPGDDEVACSFLVALMQKHRR 420 |
| Danio rerio | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 +| |||||||||||||| ||| |||+# + |+ | |+ |+||| |+|| || Sbjct 367 NGEWRRGSTAGGCRNYPATFWNNPQF#KVALKHP---DSPGQSECSFLVALMQKDRR 419 |
| Drosophila melanogaster | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 ++ +| | | |||||||+ +||| ||||#+| | + ||+||+|+ || +| |+||+|| Sbjct 382 MSMYEGEWTPGVTAGGCRNFLDTFWHNPQY#IITLVDPDEEDEEGQ--CTVIVALMQKNRR 439 |
| Drosophila melanogaster | Query 358 LTKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 ++ +| | | |||||||+ +||| ||||#+| | + ||+||+|+ || +| |+||+|| Sbjct 405 MSMYEGEWTPGVTAGGCRNFLDTFWHNPQY#IITLVDPDEEDEEGQ--CTVIVALMQKNRR 462 |
| Xenopus laevis | Query 359 TKMDGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 | ||+|||||||||||| | |||+|||+# + | |||+| +| | |+||| |+||+|| Sbjct 362 TLYDGSWRRGSTAGGCRNNPATFWINPQF#KVTLLEEDDDPDDNELACSFLVALMQKNRR 420 |
| Schistosoma japonicum | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 +| |+| +||||||+ +|||+||||#++ +|+ ||+|| | | +|||+||+|| Sbjct 16 EGCWKRRVSAGGCRNFLDTFWINPQY#VVSVEDPDENDE--ENKGTLIVGLMQKNRR 69 |
| Macaca fascicularis | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 +| | || +||||||+|+||| ||||# +|| |||+| +| | |+||| |+||+|| Sbjct 435 EGRWVRGCSAGGCRNFPDTFWTNPQY#RLKLLEEDDDPDDSEVICSFLVALMQKNRR 490 |
| Oryctolagus cuniculus | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQY#LIKLEEEDEDDEDGERGCTFLVGLIQKHRR 417 +| | || +||||||+|+||| ||||# +|| |||+| || | |+||| |+||+|| Sbjct 367 EGRWVRGCSAGGCRNFPDTFWTNPQY#RLKLLEEDDDPEDSEVICSFLVALMQKNRR 422 |
[Site 27] FWMNPQYLIK390-LEEEDEDDED
Lys390  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe381 | Trp382 | Met383 | Asn384 | Pro385 | Gln386 | Tyr387 | Leu388 | Ile389 | Lys390 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu391 | Glu392 | Glu393 | Glu394 | Asp395 | Glu396 | Asp397 | Asp398 | Glu399 | Asp400 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MDGNWRRGSTAGGCRNYPNTFWMNPQYLIKLEEEDEDDEDGERGCTFLVGLIQKHRRRQR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 132.00 | 13 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 132.00 | 8 | calpain 2 |
| 3 | Mus musculus | 130.00 | 20 | unnamed protein product |
| 4 | Homo sapiens | 128.00 | 13 | AF261089_1 calpain large polypeptide L2 |
| 5 | Macaca mulatta | 128.00 | 2 | PREDICTED: calpain 2, large subunit |
| 6 | Homo sapiens | 128.00 | 2 | calpain 2, large subunit |
| 7 | Xenopus tropicalis | 110.00 | 4 | calpain 2, (m/II) large subunit |
| 8 | Bos grunniens | 110.00 | 1 | calpain 2 |
| 9 | Stizostedion vitreum vitreum | 105.00 | 1 | putative calpain-like protein |
| 10 | Gallus gallus | 98.60 | 3 | calpain 2, (m/II) large subunit |
| 11 | Tetraodon nigroviridis | 95.10 | 5 | unnamed protein product |
| 12 | Xenopus laevis | 92.40 | 4 | calpain 1, (mu/I) large subunit |
| 13 | Danio rerio | 87.80 | 5 | Capn8 protein |
| 14 | Canis familiaris | 83.60 | 19 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 15 | Gallus gallus | 83.60 | 1 | mu-calpain large subunit |
| 16 | Rattus norvegicus | 83.60 | 1 | calpain 1, large subunit |
| 17 | Pongo pygmaeus | 83.20 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 18 | synthetic construct | 83.20 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 19 | Sus scrofa | 82.40 | 2 | calpain 1, large subunit |
| 20 | Oncorhynchus mykiss | 81.30 | 1 | m-calpain |
| 21 | Coturnix coturnix | 80.10 | 1 | quail calpain |
| 22 | Danio rerio | 76.60 | 1 | calpain 1, large subunit |
| 23 | Bos taurus | 75.90 | 4 | calpain 1, (mu/I) large subunit |
| 24 | Drosophila melanogaster | 74.70 | 4 | CAA55297.1 |
| 25 | Drosophila melanogaster | 74.70 | 2 | Calpain-A CG7563-PB, isoform B |
| 26 | Xenopus laevis | 73.20 | 1 | mu/m-calpain large subunit |
| 27 | Schistosoma japonicum | 71.60 | 1 | unknown |
| 28 | Macaca fascicularis | 69.30 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 29 | Ovis aries | 67.80 | 1 | skeletal muscle-specific calpain p94 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 |
| Rattus norvegicus | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 |
| Mus musculus | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDEEDGERGCTFLVGLIQKHRRRQR 420 |
| Homo sapiens | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 ||||||||||||||||||||||||||||||#|||||||+|||| ||||||||||||||||| Sbjct 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDEEDGESGCTFLVGLIQKHRRRQR 420 |
| Macaca mulatta | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 ||||||||||||||||||||||||||||||#|||||||+|||| ||||||||||||||||| Sbjct 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDEEDGESGCTFLVGLIQKHRRRQR 420 |
| Homo sapiens | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 ||||||||||||||||||||||||||||||#|||||||+|||| ||||||||||||||||| Sbjct 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDEEDGESGCTFLVGLIQKHRRRQR 420 |
| Xenopus tropicalis | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 ||| ||||||||||||+| ||||||||+||#||| |+| +| | ||||+||||||+||+|| Sbjct 361 MDGTWRRGSTAGGCRNFPETFWMNPQYVIK#LEEADDDPDDNEEGCTFIVGLIQKNRRKQR 420 |
| Bos grunniens | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQ 413 ||||||||||||||||||||||||||||||#||||||| |||| |||||||||| Sbjct 73 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDQEDGESGCTFLVGLIQ 125 |
| Stizostedion vitreum vitreum | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 ||+||||||||||||||+||| |||++|+#|||||+| ||| ||+|+||||||+||| | Sbjct 362 FDGSWRRGSTAGGCRNYPDTFWTNPQFVIR#LEEEDDDPADGELGCSFVVGLIQKNRRRAR 421 |
| Gallus gallus | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 +|||||||+||||||||||||| |||||||#||||||| +| | |||||+||||||||+|| Sbjct 361 LDGNWRRGATAGGCRNYPNTFWTNPQYLIK#LEEEDEDPDDPEGGCTFLIGLIQKHRRKQR 420 |
| Tetraodon nigroviridis | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 + |||||||||||||||+||+|||++||#||| |+| + + ||||+|||+||+||| | Sbjct 367 FEDTWRRGSTAGGCRNYPNSFWLNPQFVIK#LEEVDDDPGEDQEGCTFIVGLMQKNRRRLR 426 |
| Xenopus laevis | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDG-ERGCTFLVGLIQKHRRRQR 420 +|+||+|||||||||+| |||+|||+ ||#||||| ||+ | | |||||+ |+||++|++| Sbjct 372 NGSWRKGSTAGGCRNFPATFWINPQFKIK#LEEEDGDDDYGSETGCTFLLALMQKNKRKER 431 |
| Danio rerio | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 +|||| ||||||||| | || |||++||#||| |+| || ||||||||+|| |+ + Sbjct 362 EGNWRVGSTAGGCRNNPATFCSNPQFMIK#LEEIDDDPHDGVNGCTFLVGLMQKDGRKDK 420 |
| Canis familiaris | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDG---ERGCTFLVGLIQKH 415 +| |||||||||||||| |||+|||+ |+#||| |+ |+| | ||+|++ |+||| Sbjct 369 EGTWRRGSTAGGCRNYPATFWVNPQFKIR#LEETDDVDDDYGGRESGCSFVLALMQKH 425 |
| Gallus gallus | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDG---ERGCTFLVGLIQKHRRR 418 ||+|||||||||||||| |||+|||+ | #||| |+| +| | ||+||+ |+|||||| Sbjct 372 DGSWRRGSTAGGCRNYPATFWINPQFKIC#LEEVDDDGDDFGGREPGCSFLLALMQKHRRR 431 Query 419 QR 420 +| Sbjct 432 ER 433 |
| Rattus norvegicus | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEE-EDEDDEDG-ERGCTFLVGLIQKH 415 +| |||||||||||||| |||+|||+ |+#||| +| || | | ||+||+ |+||| Sbjct 372 EGTWRRGSTAGGCRNYPATFWVNPQFKIR#LEEVDDADDYDSRESGCSFLLALMQKH 427 |
| Pongo pygmaeus | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGER--GCTFLVGLIQKH 415 +| |||||||||||||| |||+|||+ |+#|+| |+ |+ |+| ||+|++ |+||| Sbjct 372 EGTWRRGSTAGGCRNYPATFWVNPQFKIR#LDETDDPDDYGDRESGCSFVLALMQKH 427 |
| synthetic construct | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGER--GCTFLVGLIQKH 415 +| |||||||||||||| |||+|||+ |+#|+| |+ |+ |+| ||+|++ |+||| Sbjct 372 EGTWRRGSTAGGCRNYPATFWVNPQFKIR#LDETDDPDDYGDRESGCSFVLALMQKH 427 |
| Sus scrofa | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEED--EDDEDG-ERGCTFLVGLIQKH 415 +| |||||||||||||| |||+|||+ |+#||| | ||| | | ||+|++ |+||| Sbjct 372 EGTWRRGSTAGGCRNYPATFWVNPQFKIR#LEETDDPEDDYGGRESGCSFVLALMQKH 428 |
| Oncorhynchus mykiss | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 ||+||+|||||||||+ |||||||++|+#|+|||+| +| | ||+|+||||||+||| | Sbjct 360 DGSWRKGSTAGGCRNHAYTFWMNPQFVIR#LDEEDDDPDDNEVGCSFVVGLIQKNRRRLR 418 |
| Coturnix coturnix | Query 361 MDGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 +| |||||||||||| | |||+|||+ ||#| |||+| | | |+||| |+||||||+| Sbjct 364 FEGTWRRGSTAGGCRNNPATFWINPQFKIK#LLEEDDDPGDDEVACSFLVALMQKHRRRER 423 |
| Danio rerio | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 +| |||||||||||||| ||| |||+ + #|+ | |+ |+||| |+|| ||++| Sbjct 367 NGEWRRGSTAGGCRNYPATFWNNPQFKVA#LKHP---DSPGQSECSFLVALMQKDRRKKR 422 |
| Bos taurus | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDG----ERGCTFLVGLIQKH 415 +| |||||||||||||| |||+|||+ |+#||| |+ | | | ||+||+ |+||| Sbjct 372 EGTWRRGSTAGGCRNYPATFWVNPQFKIR#LEETDDPDPDDYGGRESGCSFLLALMQKH 429 |
| Drosophila melanogaster | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 +| | | |||||||+ +||| ||||+| #| + ||+||+|+ || +| |+||+|| +| Sbjct 386 EGEWTPGVTAGGCRNFLDTFWHNPQYIIT#LVDPDEEDEEGQ--CTVIVALMQKNRRSKR 442 |
| Drosophila melanogaster | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 +| | | |||||||+ +||| ||||+| #| + ||+||+|+ || +| |+||+|| +| Sbjct 409 EGEWTPGVTAGGCRNFLDTFWHNPQYIIT#LVDPDEEDEEGQ--CTVIVALMQKNRRSKR 465 |
| Xenopus laevis | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 ||+|||||||||||| | |||+|||+ + #| |||+| +| | |+||| |+||+|| +| Sbjct 365 DGSWRRGSTAGGCRNNPATFWINPQFKVT#LLEEDDDPDDNELACSFLVALMQKNRRNER 423 |
| Schistosoma japonicum | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 +| |+| +||||||+ +|||+||||++ #+|+ ||+|| | | +|||+||+||+ | Sbjct 16 EGCWKRRVSAGGCRNFLDTFWINPQYVVS#VEDPDENDE--ENKGTLIVGLMQKNRRKMR 72 |
| Macaca fascicularis | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 +| | || +||||||+|+||| |||| +|#| |||+| +| | |+||| |+||+||+ | Sbjct 435 EGRWVRGCSAGGCRNFPDTFWTNPQYRLK#LLEEDDDPDDSEVICSFLVALMQKNRRKDR 493 |
| Ovis aries | Query 362 DGNWRRGSTAGGCRNYPNTFWMNPQYLIK#LEEEDEDDEDGERGCTFLVGLIQKHRRRQR 420 + | || +||||||+|+||| |||| +|#| |||+| +| | |+||| |+||+||+ | Sbjct 149 EARWVRGCSAGGCRNFPDTFWTNPQYRLK#LLEEDDDPDDSEVICSFLVALMQKNRRKDR 207 |
[Site 28] MNPQYLIKLE392-EEDEDDEDGE
Glu392  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Met383 | Asn384 | Pro385 | Gln386 | Tyr387 | Leu388 | Ile389 | Lys390 | Leu391 | Glu392 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu393 | Glu394 | Asp395 | Glu396 | Asp397 | Asp398 | Glu399 | Asp400 | Gly401 | Glu402 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GNWRRGSTAGGCRNYPNTFWMNPQYLIKLEEEDEDDEDGERGCTFLVGLIQKHRRRQRKM |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 132.00 | 13 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 132.00 | 9 | calpain 2 |
| 3 | Mus musculus | 130.00 | 20 | m-calpain large subunit |
| 4 | Homo sapiens | 128.00 | 13 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 128.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 128.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus tropicalis | 109.00 | 4 | calpain 2, (m/II) large subunit |
| 8 | Stizostedion vitreum vitreum | 106.00 | 1 | putative calpain-like protein |
| 9 | Bos grunniens | 106.00 | 1 | calpain 2 |
| 10 | Tetraodon nigroviridis | 99.40 | 5 | unnamed protein product |
| 11 | Gallus gallus | 99.00 | 3 | calpain 2, (m/II) large subunit |
| 12 | Xenopus laevis | 92.80 | 4 | calpain 1, (mu/I) large subunit |
| 13 | Danio rerio | 87.80 | 5 | Capn8 protein |
| 14 | Rattus norvegicus | 83.60 | 1 | calpain 1, large subunit |
| 15 | Canis familiaris | 83.20 | 19 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 16 | synthetic construct | 83.20 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 17 | Pongo pygmaeus | 83.20 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 18 | Sus scrofa | 82.00 | 2 | calpain 1, large subunit |
| 19 | Gallus gallus | 82.00 | 1 | mu-calpain large subunit |
| 20 | Oncorhynchus mykiss | 81.30 | 1 | m-calpain |
| 21 | Coturnix coturnix | 80.10 | 1 | quail calpain |
| 22 | Danio rerio | 77.00 | 1 | calpain 1, large subunit |
| 23 | Drosophila melanogaster | 76.60 | 4 | CAA55297.1 |
| 24 | Drosophila melanogaster | 76.60 | 2 | Calpain-A CG7563-PB, isoform B |
| 25 | Bos taurus | 75.50 | 5 | calpain 1, (mu/I) large subunit |
| 26 | Xenopus laevis | 74.30 | 1 | mu/m-calpain large subunit |
| 27 | Schistosoma japonicum | 71.60 | 1 | unknown |
| 28 | Macaca fascicularis | 71.60 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 29 | Ovis aries | 69.70 | 1 | skeletal muscle-specific calpain p94 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 |
| Rattus norvegicus | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 |
| Mus musculus | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 ||||||||||||||||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDEEDGERGCTFLVGLIQKHRRRQRKM 422 |
| Homo sapiens | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 ||||||||||||||||||||||||||||||#|||||+|||| ||||||||||||||||||| Sbjct 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDEEDGESGCTFLVGLIQKHRRRQRKM 422 |
| Homo sapiens | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 ||||||||||||||||||||||||||||||#|||||+|||| ||||||||||||||||||| Sbjct 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDEEDGESGCTFLVGLIQKHRRRQRKM 422 |
| Macaca mulatta | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 ||||||||||||||||||||||||||||||#|||||+|||| ||||||||||||||||||| Sbjct 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDEEDGESGCTFLVGLIQKHRRRQRKM 422 |
| Xenopus tropicalis | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 | ||||||||||||+| ||||||||+||||#| |+| +| | ||||+||||||+||+|||| Sbjct 363 GTWRRGSTAGGCRNFPETFWMNPQYVIKLE#EADDDPDDNEEGCTFIVGLIQKNRRKQRKM 422 |
| Stizostedion vitreum vitreum | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 |+||||||||||||||+||| |||++|+||#|||+| ||| ||+|+||||||+||| ||+ Sbjct 364 GSWRRGSTAGGCRNYPDTFWTNPQFVIRLE#EEDDDPADGELGCSFVVGLIQKNRRRARKL 423 |
| Bos grunniens | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQ 413 ||||||||||||||||||||||||||||||#||||| |||| |||||||||| Sbjct 75 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDQEDGESGCTFLVGLIQ 125 |
| Tetraodon nigroviridis | Query 364 NWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 |||||||||||||||+||+|||++||||#| |+| + + ||||+|||+||+||| ||| Sbjct 370 TWRRGSTAGGCRNYPNSFWLNPQFVIKLE#EVDDDPGEDQEGCTFIVGLMQKNRRRLRKM 428 |
| Gallus gallus | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 ||||||+||||||||||||| |||||||||#||||| +| | |||||+||||||||+|||| Sbjct 363 GNWRRGATAGGCRNYPNTFWTNPQYLIKLE#EEDEDPDDPEGGCTFLIGLIQKHRRKQRKM 422 |
| Xenopus laevis | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDG-ERGCTFLVGLIQKHRRRQRK 421 |+||+|||||||||+| |||+|||+ ||||#||| ||+ | | |||||+ |+||++|++|+ Sbjct 373 GSWRKGSTAGGCRNFPATFWINPQFKIKLE#EEDGDDDYGSETGCTFLLALMQKNKRKERR 432 Query 422 M 422 Sbjct 433 F 433 |
| Danio rerio | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRK 421 |||| ||||||||| | || |||++||||#| |+| || ||||||||+|| |+ ++ Sbjct 363 GNWRVGSTAGGCRNNPATFCSNPQFMIKLE#EIDDDPHDGVNGCTFLVGLMQKDGRKDKR 421 |
| Rattus norvegicus | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#E-EDEDDEDG-ERGCTFLVGLIQKH 415 | |||||||||||||| |||+|||+ |+||#| +| || | | ||+||+ |+||| Sbjct 373 GTWRRGSTAGGCRNYPATFWVNPQFKIRLE#EVDDADDYDSRESGCSFLLALMQKH 427 |
| Canis familiaris | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDG---ERGCTFLVGLIQKH 415 | |||||||||||||| |||+|||+ |+||#| |+ |+| | ||+|++ |+||| Sbjct 370 GTWRRGSTAGGCRNYPATFWVNPQFKIRLE#ETDDVDDDYGGRESGCSFVLALMQKH 425 |
| synthetic construct | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGER--GCTFLVGLIQKH 415 | |||||||||||||| |||+|||+ |+|+#| |+ |+ |+| ||+|++ |+||| Sbjct 373 GTWRRGSTAGGCRNYPATFWVNPQFKIRLD#ETDDPDDYGDRESGCSFVLALMQKH 427 |
| Pongo pygmaeus | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGER--GCTFLVGLIQKH 415 | |||||||||||||| |||+|||+ |+|+#| |+ |+ |+| ||+|++ |+||| Sbjct 373 GTWRRGSTAGGCRNYPATFWVNPQFKIRLD#ETDDPDDYGDRESGCSFVLALMQKH 427 |
| Sus scrofa | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EED--EDDEDG-ERGCTFLVGLIQKH 415 | |||||||||||||| |||+|||+ |+||#| | ||| | | ||+|++ |+||| Sbjct 373 GTWRRGSTAGGCRNYPATFWVNPQFKIRLE#ETDDPEDDYGGRESGCSFVLALMQKH 428 |
| Gallus gallus | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDG---ERGCTFLVGLIQKHRRRQ 419 |+|||||||||||||| |||+|||+ | ||#| |+| +| | ||+||+ |+||||||+ Sbjct 373 GSWRRGSTAGGCRNYPATFWINPQFKICLE#EVDDDGDDFGGREPGCSFLLALMQKHRRRE 432 Query 420 RK 421 |+ Sbjct 433 RR 434 |
| Oncorhynchus mykiss | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 |+||+|||||||||+ |||||||++|+|+#|||+| +| | ||+|+||||||+||| ||+ Sbjct 361 GSWRKGSTAGGCRNHAYTFWMNPQFVIRLD#EEDDDPDDNEVGCSFVVGLIQKNRRRLRKV 420 |
| Coturnix coturnix | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 | |||||||||||| | |||+|||+ ||| #|||+| | | |+||| |+||||||+|++ Sbjct 366 GTWRRGSTAGGCRNNPATFWINPQFKIKLL#EEDDDPGDDEVACSFLVALMQKHRRRERRV 425 |
| Danio rerio | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRK 421 | |||||||||||||| ||| |||+ + |+# | |+ |+||| |+|| ||++|+ Sbjct 368 GEWRRGSTAGGCRNYPATFWNNPQFKVALK#HP---DSPGQSECSFLVALMQKDRRKKRR 423 |
| Drosophila melanogaster | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 | | | |||||||+ +||| ||||+| | #+ ||+||+|+ || +| |+||+|| +| | Sbjct 387 GEWTPGVTAGGCRNFLDTFWHNPQYIITLV#DPDEEDEEGQ--CTVIVALMQKNRRSKRNM 444 |
| Drosophila melanogaster | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 | | | |||||||+ +||| ||||+| | #+ ||+||+|+ || +| |+||+|| +| | Sbjct 410 GEWTPGVTAGGCRNFLDTFWHNPQYIITLV#DPDEEDEEGQ--CTVIVALMQKNRRSKRNM 467 |
| Bos taurus | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDG----ERGCTFLVGLIQKH 415 | |||||||||||||| |||+|||+ |+||#| |+ | | | ||+||+ |+||| Sbjct 373 GTWRRGSTAGGCRNYPATFWVNPQFKIRLE#ETDDPDPDDYGGRESGCSFLLALMQKH 429 |
| Xenopus laevis | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRK 421 |+|||||||||||| | |||+|||+ + | #|||+| +| | |+||| |+||+|| +|| Sbjct 366 GSWRRGSTAGGCRNNPATFWINPQFKVTLL#EEDDDPDDNELACSFLVALMQKNRRNERK 424 |
| Schistosoma japonicum | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRK 421 | |+| +||||||+ +|||+||||++ +|#+ ||+|| | | +|||+||+||+ |+ Sbjct 17 GCWKRRVSAGGCRNFLDTFWINPQYVVSVE#DPDENDE--ENKGTLIVGLMQKNRRKMRQ 73 |
| Macaca fascicularis | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 | | || +||||||+|+||| |||| +|| #|||+| +| | |+||| |+||+||+ ||+ Sbjct 436 GRWVRGCSAGGCRNFPDTFWTNPQYRLKLL#EEDDDPDDSEVICSFLVALMQKNRRKDRKL 495 |
| Ovis aries | Query 363 GNWRRGSTAGGCRNYPNTFWMNPQYLIKLE#EEDEDDEDGERGCTFLVGLIQKHRRRQRKM 422 | || +||||||+|+||| |||| +|| #|||+| +| | |+||| |+||+||+ ||+ Sbjct 150 ARWVRGCSAGGCRNFPDTFWTNPQYRLKLL#EEDDDPDDSEVICSFLVALMQKNRRKDRKL 209 |
[Site 29] DEDGERGCTF407-LVGLIQKHRR
Phe407  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp398 | Glu399 | Asp400 | Gly401 | Glu402 | Arg403 | Gly404 | Cys405 | Thr406 | Phe407 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu408 | Val409 | Gly410 | Leu411 | Ile412 | Gln413 | Lys414 | His415 | Arg416 | Arg417 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PNTFWMNPQYLIKLEEEDEDDEDGERGCTFLVGLIQKHRRRQRKMGEDMHTIGFGIYEVP |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 131.00 | 15 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 131.00 | 12 | calpain 2 |
| 3 | Mus musculus | 129.00 | 20 | unnamed protein product |
| 4 | Homo sapiens | 127.00 | 11 | AF261089_1 calpain large polypeptide L2 |
| 5 | Macaca mulatta | 127.00 | 2 | PREDICTED: calpain 2, large subunit |
| 6 | Homo sapiens | 127.00 | 2 | calpain 2, large subunit |
| 7 | Xenopus tropicalis | 112.00 | 4 | calpain 2, (m/II) large subunit |
| 8 | Stizostedion vitreum vitreum | 105.00 | 1 | putative calpain-like protein |
| 9 | Gallus gallus | 97.80 | 3 | calpain 2, (m/II) large subunit |
| 10 | Tetraodon nigroviridis | 97.40 | 6 | unnamed protein product |
| 11 | Xenopus laevis | 90.50 | 4 | calpain 1, (mu/I) large subunit |
| 12 | Oncorhynchus mykiss | 84.70 | 1 | m-calpain |
| 13 | Danio rerio | 82.80 | 5 | Capn8 protein |
| 14 | Coturnix coturnix | 79.70 | 1 | quail calpain |
| 15 | Gallus gallus | 75.90 | 1 | mu-calpain large subunit |
| 16 | Xenopus laevis | 73.90 | 1 | mu/m-calpain large subunit |
| 17 | Danio rerio | 71.60 | 1 | calpain 1, large subunit |
| 18 | synthetic construct | 71.20 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 19 | Pongo pygmaeus | 71.20 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | Canis familiaris | 70.90 | 21 | PREDICTED: similar to calpain 3 isoform c isoform |
| 21 | Bos taurus | 70.90 | 5 | lens-specific calpain Lp82 |
| 22 | Ovis aries | 70.90 | 3 | skeletal muscle-specific calpain |
| 23 | Sus scrofa | 70.90 | 3 | calpain 3 |
| 24 | Macaca fascicularis | 70.90 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 25 | Rattus norvegicus | 70.50 | 1 | calpain 1, large subunit |
| 26 | Oryctolagus cuniculus | 70.50 | 1 | calpain 3, (p94) |
| 27 | Schistosoma japonicum | 69.30 | 1 | unknown |
| 28 | Drosophila melanogaster | 68.20 | 4 | CAA55297.1 |
| 29 | Drosophila melanogaster | 68.20 | 2 | Calpain-A CG7563-PB, isoform B |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |
| Rattus norvegicus | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |
| Mus musculus | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 ||||||||||||||||||||+|||||||||#|||||||||||||||||||||||||||||| Sbjct 378 PNTFWMNPQYLIKLEEEDEDEEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |
| Homo sapiens | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 ||||||||||||||||||||+|||| ||||#|||||||||||||||||||||||||||||| Sbjct 378 PNTFWMNPQYLIKLEEEDEDEEDGESGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |
| Macaca mulatta | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 ||||||||||||||||||||+|||| ||||#|||||||||||||||||||||||||||||| Sbjct 378 PNTFWMNPQYLIKLEEEDEDEEDGESGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |
| Homo sapiens | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 ||||||||||||||||||||+|||| ||||#|||||||||||||||||||||||||||||| Sbjct 378 PNTFWMNPQYLIKLEEEDEDEEDGESGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |
| Xenopus tropicalis | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 | ||||||||+||||| |+| +| | ||||#+||||||+||+||||||||||||| ||||| Sbjct 378 PETFWMNPQYVIKLEEADDDPDDNEEGCTF#IVGLIQKNRRKQRKMGEDMHTIGFAIYEVP 437 |
| Stizostedion vitreum vitreum | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |+||| |||++|+|||||+| ||| ||+|#+||||||+||| ||+||||||+|| ||+|| Sbjct 379 PDTFWTNPQFVIRLEEEDDDPADGELGCSF#VVGLIQKNRRRARKLGEDMHTVGFAIYQVP 438 |
| Gallus gallus | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 ||||| |||||||||||||| +| | ||||#|+||||||||+||||||||||||| ||||| Sbjct 378 PNTFWTNPQYLIKLEEEDEDPDDPEGGCTF#LIGLIQKHRRKQRKMGEDMHTIGFAIYEVP 437 |
| Tetraodon nigroviridis | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 ||+||+|||++||||| |+| + + ||||#+|||+||+||| ||||||| |||| |||+| Sbjct 384 PNSFWLNPQFVIKLEEVDDDPGEDQEGCTF#IVGLMQKNRRRLRKMGEDMETIGFAIYELP 443 |
| Xenopus laevis | Query 378 PNTFWMNPQYLIKLEEEDEDDEDG-ERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEV 436 | |||+|||+ ||||||| ||+ | | ||||#|+ |+||++|++|+ |+|| |||| +||| Sbjct 388 PATFWINPQFKIKLEEEDGDDDYGSETGCTF#LLALMQKNKRKERRFGKDMETIGFAVYEV 447 Query 437 P 437 | Query 437 P 437 |
| Oncorhynchus mykiss | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |||||||++|+|+|||+| +| | ||+|#+||||||+||| ||+||||||||| ||||| Sbjct 378 TFWMNPQFVIRLDEEDDDPDDNEVGCSF#VVGLIQKNRRRLRKVGEDMHTIGFAIYEVP 435 |
| Danio rerio | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 | || |||++||||| |+| || ||||#||||+|| |+ ++ | |++|||| ||+|| Sbjct 378 PATFCSNPQFMIKLEEIDDDPHDGVNGCTF#LVGLMQKDGRKDKRFGRDLNTIGFAIYKVP 437 |
| Coturnix coturnix | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 | |||+|||+ ||| |||+| | | |+|#|| |+||||||+|++| ||||||| +|||| Sbjct 381 PATFWINPQFKIKLLEEDDDPGDDEVACSF#LVALMQKHRRRERRVGGDMHTIGFAVYEVP 440 |
| Gallus gallus | Query 378 PNTFWMNPQYLIKLEE---EDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIY 434 | |||+|||+ | ||| + +| | ||+|#|+ |+||||||+|+ |+|| |||| +| Sbjct 388 PATFWINPQFKICLEEVDDDGDDFGGREPGCSF#LLALMQKHRRRERRYGKDMETIGFAVY 447 Query 435 EVP 437 ||| Query 435 EVP 437 |
| Xenopus laevis | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 | |||+|||+ + | |||+| +| | |+|#|| |+||+|| +|| |+||||||| +|||| Sbjct 381 PATFWINPQFKVTLLEEDDDPDDNELACSF#LVALMQKNRRNERKAGQDMHTIGFAVYEVP 440 |
| Danio rerio | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 | ||| |||+ + |+ | |+ |+|#|| |+|| ||++|+ |+|| |||| ||||| Sbjct 383 PATFWNNPQFKVALKHPDSP---GQSECSF#LVALMQKDRRKKRREGQDMETIGFAIYEVP 439 |
| synthetic construct | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGER--GCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYE 435 | |||+|||+ |+|+| |+ |+ |+| ||+|#++ |+||||||+|+ | || |||| +|| Sbjct 388 PATFWVNPQFKIRLDETDDPDDYGDRESGCSF#VLALMQKHRRRERRFGRDMETIGFAVYE 447 Query 436 VP 437 || Query 436 VP 437 |
| Pongo pygmaeus | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGER--GCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYE 435 | |||+|||+ |+|+| |+ |+ |+| ||+|#++ |+||||||+|+ | || |||| +|| Sbjct 388 PATFWVNPQFKIRLDETDDPDDYGDRESGCSF#VLALMQKHRRRERRFGRDMETIGFAVYE 447 Query 436 VP 437 || Query 436 VP 437 |
| Canis familiaris | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |+||| |||| +|| |||+| +| | |+|#|| |+||+||+ ||+| ++ |||| ||||| Sbjct 383 PDTFWTNPQYRLKLLEEDDDPDDSEVICSF#LVALMQKNRRKDRKLGANLFTIGFAIYEVP 442 |
| Bos taurus | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |+||| |||| +|| |||+| +| | |+|#|| |+||+||+ ||+| ++ |||| ||||| Sbjct 383 PDTFWTNPQYRLKLLEEDDDPDDSEVICSF#LVALMQKNRRKDRKLGANLFTIGFAIYEVP 442 |
| Ovis aries | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |+||| |||| +|| |||+| +| | |+|#|| |+||+||+ ||+| ++ |||| ||||| Sbjct 452 PDTFWTNPQYRLKLLEEDDDPDDSEVICSF#LVALMQKNRRKDRKLGANLFTIGFAIYEVP 511 |
| Sus scrofa | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |+||| |||| +|| |||+| +| | |+|#|| |+||+||+ ||+| ++ |||| ||||| Sbjct 451 PDTFWTNPQYRLKLLEEDDDPDDSEVICSF#LVALMQKNRRKDRKLGANLFTIGFAIYEVP 510 |
| Macaca fascicularis | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |+||| |||| +|| |||+| +| | |+|#|| |+||+||+ ||+| ++ |||| ||||| Sbjct 451 PDTFWTNPQYRLKLLEEDDDPDDSEVICSF#LVALMQKNRRKDRKLGANLFTIGFAIYEVP 510 |
| Rattus norvegicus | Query 378 PNTFWMNPQYLIKLEE-EDEDDEDG-ERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYE 435 | |||+|||+ |+||| +| || | | ||+|#|+ |+||||||+|+ | || |||| +|+ Sbjct 388 PATFWVNPQFKIRLEEVDDADDYDSRESGCSF#LLALMQKHRRRERRFGRDMETIGFAVYQ 447 Query 436 VP 437 || Query 436 VP 437 |
| Oryctolagus cuniculus | Query 378 PNTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |+||| |||| +|| |||+| || | |+|#|| |+||+||+ ||+| ++ |||| ||||| Sbjct 383 PDTFWTNPQYRLKLLEEDDDPEDSEVICSF#LVALMQKNRRKDRKLGANLFTIGFAIYEVP 442 |
| Schistosoma japonicum | Query 379 NTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 +|||+||||++ +|+ ||+|| | | #+|||+||+||+ |+ | |+ |||+ +|++| Sbjct 33 DTFWINPQYVVSVEDPDENDE--ENKGTL#IVGLMQKNRRKMRQEGADLLTIGYAVYKLP 89 |
| Drosophila melanogaster | Query 379 NTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEV 436 +||| ||||+| | + ||+||+|+ || #+| |+||+|| +| || + |||| || + Sbjct 403 DTFWHNPQYIITLVDPDEEDEEGQ--CTV#IVALMQKNRRSKRNMGMECLTIGFAIYSL 458 |
| Drosophila melanogaster | Query 379 NTFWMNPQYLIKLEEEDEDDEDGERGCTF#LVGLIQKHRRRQRKMGEDMHTIGFGIYEV 436 +||| ||||+| | + ||+||+|+ || #+| |+||+|| +| || + |||| || + Sbjct 426 DTFWHNPQYIITLVDPDEEDEEGQ--CTV#IVALMQKNRRSKRNMGMECLTIGFAIYSL 481 |
[Site 30] DGERGCTFLV409-GLIQKHRRRQ
Val409  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp400 | Gly401 | Glu402 | Arg403 | Gly404 | Cys405 | Thr406 | Phe407 | Leu408 | Val409 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly410 | Leu411 | Ile412 | Gln413 | Lys414 | His415 | Arg416 | Arg417 | Arg418 | Gln419 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TFWMNPQYLIKLEEEDEDDEDGERGCTFLVGLIQKHRRRQRKMGEDMHTIGFGIYEVPEE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 129.00 | 15 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 129.00 | 12 | calpain 2 |
| 3 | Mus musculus | 127.00 | 20 | m-calpain large subunit |
| 4 | Homo sapiens | 125.00 | 11 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 125.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 125.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Xenopus tropicalis | 108.00 | 4 | calpain 2, (m/II) large subunit |
| 8 | Stizostedion vitreum vitreum | 102.00 | 1 | putative calpain-like protein |
| 9 | Tetraodon nigroviridis | 94.70 | 6 | unnamed protein product |
| 10 | Gallus gallus | 94.40 | 3 | calpain 2, (m/II) large subunit |
| 11 | Xenopus laevis | 89.00 | 4 | calpain 1, (mu/I) large subunit |
| 12 | Oncorhynchus mykiss | 85.10 | 1 | m-calpain |
| 13 | Danio rerio | 82.00 | 5 | Capn8 protein |
| 14 | Coturnix coturnix | 80.90 | 1 | quail calpain |
| 15 | Gallus gallus | 74.30 | 1 | mu-calpain large subunit |
| 16 | Xenopus laevis | 73.60 | 1 | mu/m-calpain large subunit |
| 17 | Ovis aries | 71.60 | 3 | muscle specific calpain 3 |
| 18 | Schistosoma japonicum | 70.90 | 1 | unknown |
| 19 | Canis familiaris | 70.50 | 21 | PREDICTED: similar to Calpain-3 (Calpain L3) (Cal |
| 20 | Bos taurus | 70.50 | 5 | skeletal muscle-specific calpain p94 |
| 21 | synthetic construct | 70.50 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 22 | Danio rerio | 70.50 | 1 | calpain 1, large subunit |
| 23 | Pongo pygmaeus | 70.50 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 24 | Sus scrofa | 70.10 | 3 | calpain 3 |
| 25 | Macaca fascicularis | 70.10 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 26 | Oryctolagus cuniculus | 69.70 | 1 | calpain 3, (p94) |
| 27 | Rattus norvegicus | 69.30 | 1 | calpain 1, large subunit |
| 28 | Drosophila melanogaster | 68.20 | 4 | CAA55297.1 |
| 29 | Drosophila melanogaster | 68.20 | 2 | Calpain-A CG7563-PB, isoform B |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 |
| Rattus norvegicus | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 |
| Mus musculus | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||||||||||||||||||+|||||||||||#|||||||||||||||||||||||||||||| Sbjct 380 TFWMNPQYLIKLEEEDEDEEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 |
| Homo sapiens | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||||||||||||||||||+|||| ||||||#|||||||||||||||||||||||||||||| Sbjct 380 TFWMNPQYLIKLEEEDEDEEDGESGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 |
| Homo sapiens | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||||||||||||||||||+|||| ||||||#|||||||||||||||||||||||||||||| Sbjct 380 TFWMNPQYLIKLEEEDEDEEDGESGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 |
| Macaca mulatta | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||||||||||||||||||+|||| ||||||#|||||||||||||||||||||||||||||| Sbjct 380 TFWMNPQYLIKLEEEDEDEEDGESGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 |
| Xenopus tropicalis | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||||||||+||||| |+| +| | ||||+|#|||||+||+||||||||||||| ||||| + Sbjct 380 TFWMNPQYVIKLEEADDDPDDNEEGCTFIV#GLIQKNRRKQRKMGEDMHTIGFAIYEVPPQ 439 |
| Stizostedion vitreum vitreum | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||| |||++|+|||||+| ||| ||+|+|#|||||+||| ||+||||||+|| ||+||+| Sbjct 381 TFWTNPQFVIRLEEEDDDPADGELGCSFVV#GLIQKNRRRARKLGEDMHTVGFAIYQVPDE 440 |
| Tetraodon nigroviridis | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 +||+|||++||||| |+| + + ||||+|#||+||+||| ||||||| |||| |||+||| Sbjct 386 SFWLNPQFVIKLEEVDDDPGEDQEGCTFIV#GLMQKNRRRLRKMGEDMETIGFAIYELPEE 445 |
| Gallus gallus | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||| |||||||||||||| +| | |||||+#||||||||+||||||||||||| ||||| | Sbjct 380 TFWTNPQYLIKLEEEDEDPDDPEGGCTFLI#GLIQKHRRKQRKMGEDMHTIGFAIYEVPPE 439 |
| Xenopus laevis | Query 380 TFWMNPQYLIKLEEEDEDDEDG-ERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPE 438 |||+|||+ ||||||| ||+ | | |||||+# |+||++|++|+ |+|| |||| +|||| Sbjct 390 TFWINPQFKIKLEEEDGDDDYGSETGCTFLL#ALMQKNKRKERRFGKDMETIGFAVYEVPR 449 Query 439 E 439 | Query 439 E 439 |
| Oncorhynchus mykiss | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 |||||||++|+|+|||+| +| | ||+|+|#|||||+||| ||+||||||||| ||||| + Sbjct 378 TFWMNPQFVIRLDEEDDDPDDNEVGCSFVV#GLIQKNRRRLRKVGEDMHTIGFAIYEVPSQ 437 |
| Danio rerio | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 || |||++||||| |+| || ||||||#||+|| |+ ++ | |++|||| ||+||+| Sbjct 380 TFCSNPQFMIKLEEIDDDPHDGVNGCTFLV#GLMQKDGRKDKRFGRDLNTIGFAIYKVPDE 439 |
| Coturnix coturnix | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 |||+|||+ ||| |||+| | | |+|||# |+||||||+|++| ||||||| +|||||| Sbjct 383 TFWINPQFKIKLLEEDDDPGDDEVACSFLV#ALMQKHRRRERRVGGDMHTIGFAVYEVPEE 442 |
| Gallus gallus | Query 380 TFWMNPQYLIKLEE---EDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEV 436 |||+|||+ | ||| + +| | ||+||+# |+||||||+|+ |+|| |||| +||| Sbjct 390 TFWINPQFKICLEEVDDDGDDFGGREPGCSFLL#ALMQKHRRRERRYGKDMETIGFAVYEV 449 Query 437 PEE 439 | | Sbjct 450 PPE 452 |
| Xenopus laevis | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 |||+|||+ + | |||+| +| | |+|||# |+||+|| +|| |+||||||| +||||+| Sbjct 383 TFWINPQFKVTLLEEDDDPDDNELACSFLV#ALMQKNRRNERKAGQDMHTIGFAVYEVPDE 442 |
| Ovis aries | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||| |||| +|| |||+| +| | |+|||# |+||+||+ ||+| ++ |||| |||||+| Sbjct 2 TFWTNPQYRLKLLEEDDDPDDSEVICSFLV#ALMQKNRRKDRKLGANLFTIGFAIYEVPKE 61 |
| Schistosoma japonicum | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 |||+||||++ +|+ ||+|| | | +|#||+||+||+ |+ | |+ |||+ +|++||+ Sbjct 34 TFWINPQYVVSVEDPDENDE--ENKGTLIV#GLMQKNRRKMRQEGADLLTIGYAVYKLPEQ 91 |
| Canis familiaris | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||| |||| +|| |||+| +| | |+|||# |+||+||+ ||+| ++ |||| |||||+| Sbjct 407 TFWTNPQYRLKLLEEDDDPDDSEVICSFLV#ALMQKNRRKDRKLGANLFTIGFAIYEVPKE 466 |
| Bos taurus | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||| |||| +|| |||+| +| | |+|||# |+||+||+ ||+| ++ |||| |||||+| Sbjct 167 TFWTNPQYRLKLLEEDDDPDDSEVICSFLV#ALMQKNRRKDRKLGANLFTIGFAIYEVPKE 226 |
| synthetic construct | Query 380 TFWMNPQYLIKLEEEDEDDEDGER--GCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |||+|||+ |+|+| |+ |+ |+| ||+|++# |+||||||+|+ | || |||| +|||| Sbjct 390 TFWVNPQFKIRLDETDDPDDYGDRESGCSFVL#ALMQKHRRRERRFGRDMETIGFAVYEVP 449 Query 438 EE 439 | Sbjct 450 PE 451 |
| Danio rerio | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||| |||+ + |+ | |+ |+|||# |+|| ||++|+ |+|| |||| ||||| | Sbjct 385 TFWNNPQFKVALKHPDSP---GQSECSFLV#ALMQKDRRKKRREGQDMETIGFAIYEVPRE 441 |
| Pongo pygmaeus | Query 380 TFWMNPQYLIKLEEEDEDDEDGER--GCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |||+|||+ |+|+| |+ |+ |+| ||+|++# |+||||||+|+ | || |||| +|||| Sbjct 390 TFWVNPQFKIRLDETDDPDDYGDRESGCSFVL#ALMQKHRRRERRFGRDMETIGFAVYEVP 449 Query 438 EE 439 | Sbjct 450 PE 451 |
| Sus scrofa | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||| |||| +|| |||+| +| | |+|||# |+||+||+ ||+| ++ |||| |||||+| Sbjct 453 TFWTNPQYRLKLLEEDDDPDDSEVICSFLV#ALMQKNRRKDRKLGANLFTIGFAIYEVPKE 512 |
| Macaca fascicularis | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||| |||| +|| |||+| +| | |+|||# |+||+||+ ||+| ++ |||| |||||+| Sbjct 453 TFWTNPQYRLKLLEEDDDPDDSEVICSFLV#ALMQKNRRKDRKLGANLFTIGFAIYEVPKE 512 |
| Oryctolagus cuniculus | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||| |||| +|| |||+| || | |+|||# |+||+||+ ||+| ++ |||| |||||+| Sbjct 385 TFWTNPQYRLKLLEEDDDPEDSEVICSFLV#ALMQKNRRKDRKLGANLFTIGFAIYEVPKE 444 |
| Rattus norvegicus | Query 380 TFWMNPQYLIKLEE-EDEDDEDG-ERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEVP 437 |||+|||+ |+||| +| || | | ||+||+# |+||||||+|+ | || |||| +|+|| Sbjct 390 TFWVNPQFKIRLEEVDDADDYDSRESGCSFLL#ALMQKHRRRERRFGRDMETIGFAVYQVP 449 Query 438 EE 439 | Sbjct 450 RE 451 |
| Drosophila melanogaster | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEV 436 ||| ||||+| | + ||+||+|+ || +|# |+||+|| +| || + |||| || + Sbjct 404 TFWHNPQYIITLVDPDEEDEEGQ--CTVIV#ALMQKNRRSKRNMGMECLTIGFAIYSL 458 |
| Drosophila melanogaster | Query 380 TFWMNPQYLIKLEEEDEDDEDGERGCTFLV#GLIQKHRRRQRKMGEDMHTIGFGIYEV 436 ||| ||||+| | + ||+||+|+ || +|# |+||+|| +| || + |||| || + Sbjct 427 TFWHNPQYIITLVDPDEEDEEGQ--CTVIV#ALMQKNRRSKRNMGMECLTIGFAIYSL 481 |
[Site 31] GERGCTFLVG410-LIQKHRRRQR
Gly410  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly401 | Glu402 | Arg403 | Gly404 | Cys405 | Thr406 | Phe407 | Leu408 | Val409 | Gly410 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu411 | Ile412 | Gln413 | Lys414 | His415 | Arg416 | Arg417 | Arg418 | Gln419 | Arg420 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| FWMNPQYLIKLEEEDEDDEDGERGCTFLVGLIQKHRRRQRKMGEDMHTIGFGIYEVPEEL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 128.00 | 15 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 128.00 | 12 | calpain 2 |
| 3 | Mus musculus | 126.00 | 20 | m-calpain large subunit |
| 4 | Homo sapiens | 124.00 | 10 | AF261089_1 calpain large polypeptide L2 |
| 5 | Macaca mulatta | 124.00 | 2 | PREDICTED: calpain 2, large subunit |
| 6 | Homo sapiens | 124.00 | 2 | calpain 2, large subunit |
| 7 | Xenopus tropicalis | 108.00 | 4 | calpain 2, (m/II) large subunit |
| 8 | Stizostedion vitreum vitreum | 101.00 | 1 | putative calpain-like protein |
| 9 | Tetraodon nigroviridis | 94.40 | 6 | unnamed protein product |
| 10 | Gallus gallus | 92.80 | 3 | calpain 2, (m/II) large subunit |
| 11 | Xenopus laevis | 87.40 | 4 | calpain 1, (mu/I) large subunit |
| 12 | Oncorhynchus mykiss | 84.00 | 1 | m-calpain |
| 13 | Danio rerio | 82.00 | 5 | Capn8 protein |
| 14 | Coturnix coturnix | 78.60 | 1 | quail calpain |
| 15 | Gallus gallus | 72.40 | 1 | mu-calpain large subunit |
| 16 | Xenopus laevis | 72.00 | 1 | mu/m-calpain large subunit |
| 17 | synthetic construct | 70.10 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 18 | Pongo pygmaeus | 70.10 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 19 | Canis familiaris | 69.30 | 21 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 20 | Ovis aries | 69.30 | 4 | muscle specific calpain 3 |
| 21 | Schistosoma japonicum | 69.30 | 1 | unknown |
| 22 | Bos taurus | 68.90 | 5 | skeletal muscle-specific calpain p94 |
| 23 | Rattus norvegicus | 68.90 | 1 | calpain 1, large subunit |
| 24 | Danio rerio | 68.90 | 1 | calpain 1, large subunit |
| 25 | Sus scrofa | 68.60 | 3 | calpain 1, large subunit |
| 26 | Macaca fascicularis | 68.60 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 27 | Oryctolagus cuniculus | 68.20 | 1 | calpain 3, (p94) |
| 28 | Drosophila melanogaster | 66.20 | 4 | CAA55297.1 |
| 29 | Drosophila melanogaster | 66.20 | 2 | Calpain-A CG7563-PB, isoform B |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 |
| Rattus norvegicus | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 |
| Mus musculus | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 |||||||||||||||||+||||||||||||#|||||||||||||||||||||||||||||| Sbjct 381 FWMNPQYLIKLEEEDEDEEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 |
| Homo sapiens | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 |||||||||||||||||+|||| |||||||#|||||||||||||||||||||||||||||| Sbjct 381 FWMNPQYLIKLEEEDEDEEDGESGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 |
| Macaca mulatta | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 |||||||||||||||||+|||| |||||||#|||||||||||||||||||||||||||||| Sbjct 381 FWMNPQYLIKLEEEDEDEEDGESGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 |
| Homo sapiens | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 |||||||||||||||||+|||| |||||||#|||||||||||||||||||||||||||||| Sbjct 381 FWMNPQYLIKLEEEDEDEEDGESGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 |
| Xenopus tropicalis | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 |||||||+||||| |+| +| | ||||+||#||||+||+||||||||||||| ||||| +| Sbjct 381 FWMNPQYVIKLEEADDDPDDNEEGCTFIVG#LIQKNRRKQRKMGEDMHTIGFAIYEVPPQL 440 |
| Stizostedion vitreum vitreum | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 || |||++|+|||||+| ||| ||+|+||#||||+||| ||+||||||+|| ||+||+| Sbjct 382 FWTNPQFVIRLEEEDDDPADGELGCSFVVG#LIQKNRRRARKLGEDMHTVGFAIYQVPDEF 441 |
| Tetraodon nigroviridis | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||+|||++||||| |+| + + ||||+||#|+||+||| ||||||| |||| |||+||| Sbjct 387 FWLNPQFVIKLEEVDDDPGEDQEGCTFIVG#LMQKNRRRLRKMGEDMETIGFAIYELPEE 445 |
| Gallus gallus | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 || |||||||||||||| +| | |||||+|#|||||||+||||||||||||| ||||| | Sbjct 381 FWTNPQYLIKLEEEDEDPDDPEGGCTFLIG#LIQKHRRKQRKMGEDMHTIGFAIYEVPPEF 440 |
| Xenopus laevis | Query 381 FWMNPQYLIKLEEEDEDDEDG-ERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||+|||+ ||||||| ||+ | | |||||+ #|+||++|++|+ |+|| |||| +|||| | Sbjct 391 FWINPQFKIKLEEEDGDDDYGSETGCTFLLA#LMQKNKRKERRFGKDMETIGFAVYEVPRE 450 Query 440 L 440 Sbjct 451 F 451 |
| Oncorhynchus mykiss | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 ||||||++|+|+|||+| +| | ||+|+||#||||+||| ||+||||||||| ||||| + Sbjct 379 FWMNPQFVIRLDEEDDDPDDNEVGCSFVVG#LIQKNRRRLRKVGEDMHTIGFAIYEVPSQF 438 |
| Danio rerio | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 | |||++||||| |+| || |||||||#|+|| |+ ++ | |++|||| ||+||+|| Sbjct 381 FCSNPQFMIKLEEIDDDPHDGVNGCTFLVG#LMQKDGRKDKRFGRDLNTIGFAIYKVPDEL 440 |
| Coturnix coturnix | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||+|||+ ||| |||+| | | |+||| #|+||||||+|++| ||||||| +|||||| Sbjct 384 FWINPQFKIKLLEEDDDPGDDEVACSFLVA#LMQKHRRRERRVGGDMHTIGFAVYEVPEE 442 |
| Gallus gallus | Query 381 FWMNPQYLIKLEE---EDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVP 437 ||+|||+ | ||| + +| | ||+||+ #|+||||||+|+ |+|| |||| +|||| Sbjct 391 FWINPQFKICLEEVDDDGDDFGGREPGCSFLLA#LMQKHRRRERRYGKDMETIGFAVYEVP 450 Query 438 EE 439 | Sbjct 451 PE 452 |
| Xenopus laevis | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 ||+|||+ + | |||+| +| | |+||| #|+||+|| +|| |+||||||| +||||+| Sbjct 384 FWINPQFKVTLLEEDDDPDDNELACSFLVA#LMQKNRRNERKAGQDMHTIGFAVYEVPDEF 443 |
| synthetic construct | Query 381 FWMNPQYLIKLEEEDEDDEDGER--GCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPE 438 ||+|||+ |+|+| |+ |+ |+| ||+|++ #|+||||||+|+ | || |||| +|||| Sbjct 391 FWVNPQFKIRLDETDDPDDYGDRESGCSFVLA#LMQKHRRRERRFGRDMETIGFAVYEVPP 450 Query 439 EL 440 || Query 439 EL 440 |
| Pongo pygmaeus | Query 381 FWMNPQYLIKLEEEDEDDEDGER--GCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPE 438 ||+|||+ |+|+| |+ |+ |+| ||+|++ #|+||||||+|+ | || |||| +|||| Sbjct 391 FWVNPQFKIRLDETDDPDDYGDRESGCSFVLA#LMQKHRRRERRFGRDMETIGFAVYEVPP 450 Query 439 EL 440 || Query 439 EL 440 |
| Canis familiaris | Query 381 FWMNPQYLIKLEEEDEDDED---GERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVP 437 ||+|||+ |+||| |+ |+| | ||+|++ #|+||||||+|+ | || |||| +|||| Sbjct 388 FWVNPQFKIRLEETDDVDDDYGGRESGCSFVLA#LMQKHRRRERRFGRDMETIGFAVYEVP 447 Query 438 EEL 440 || Sbjct 448 PEL 450 |
| Ovis aries | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 || |||| +|| |||+| +| | |+||| #|+||+||+ ||+| ++ |||| |||||+| Sbjct 3 FWTNPQYRLKLLEEDDDPDDSEVICSFLVA#LMQKNRRKDRKLGANLFTIGFAIYEVPKE 61 |
| Schistosoma japonicum | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEE 439 ||+||||++ +|+ ||+|| | | +||#|+||+||+ |+ | |+ |||+ +|++||+ Sbjct 35 FWINPQYVVSVEDPDENDE--ENKGTLIVG#LMQKNRRKMRQEGADLLTIGYAVYKLPEQ 91 |
| Bos taurus | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 || |||| +|| |||+| +| | |+||| #|+||+||+ ||+| ++ |||| |||||+|+ Sbjct 168 FWTNPQYRLKLLEEDDDPDDSEVICSFLVA#LMQKNRRKDRKLGANLFTIGFAIYEVPKEM 227 |
| Rattus norvegicus | Query 381 FWMNPQYLIKLEE-EDEDDEDG-ERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPE 438 ||+|||+ |+||| +| || | | ||+||+ #|+||||||+|+ | || |||| +|+|| Sbjct 391 FWVNPQFKIRLEEVDDADDYDSRESGCSFLLA#LMQKHRRRERRFGRDMETIGFAVYQVPR 450 Query 439 EL 440 || Query 439 EL 440 |
| Danio rerio | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 || |||+ + |+ | |+ |+||| #|+|| ||++|+ |+|| |||| ||||| | Sbjct 386 FWNNPQFKVALKHPDSP---GQSECSFLVA#LMQKDRRKKRREGQDMETIGFAIYEVPREF 442 |
| Sus scrofa | Query 381 FWMNPQYLIKLEEED--EDDEDG-ERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVP 437 ||+|||+ |+||| | ||| | | ||+|++ #|+||||||+|+ | || |||| +|||| Sbjct 391 FWVNPQFKIRLEETDDPEDDYGGRESGCSFVLA#LMQKHRRRERRFGRDMETIGFAVYEVP 450 Query 438 EEL 440 || Sbjct 451 PEL 453 |
| Macaca fascicularis | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 || |||| +|| |||+| +| | |+||| #|+||+||+ ||+| ++ |||| |||||+|+ Sbjct 454 FWTNPQYRLKLLEEDDDPDDSEVICSFLVA#LMQKNRRKDRKLGANLFTIGFAIYEVPKEM 513 |
| Oryctolagus cuniculus | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEVPEEL 440 || |||| +|| |||+| || | |+||| #|+||+||+ ||+| ++ |||| |||||+|+ Sbjct 386 FWTNPQYRLKLLEEDDDPEDSEVICSFLVA#LMQKNRRKDRKLGANLFTIGFAIYEVPKEM 445 |
| Drosophila melanogaster | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEV 436 || ||||+| | + ||+||+|+ || +| #|+||+|| +| || + |||| || + Sbjct 405 FWHNPQYIITLVDPDEEDEEGQ--CTVIVA#LMQKNRRSKRNMGMECLTIGFAIYSL 458 |
| Drosophila melanogaster | Query 381 FWMNPQYLIKLEEEDEDDEDGERGCTFLVG#LIQKHRRRQRKMGEDMHTIGFGIYEV 436 || ||||+| | + ||+||+|+ || +| #|+||+|| +| || + |||| || + Sbjct 428 FWHNPQYIITLVDPDEEDEEGQ--CTVIVA#LMQKNRRSKRNMGMECLTIGFAIYSL 481 |
[Site 32] KHRRRQRKMG423-EDMHTIGFGI
Gly423  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys414 | His415 | Arg416 | Arg417 | Arg418 | Gln419 | Arg420 | Lys421 | Met422 | Gly423 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu424 | Asp425 | Met426 | His427 | Thr428 | Ile429 | Gly430 | Phe431 | Gly432 | Ile433 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EDEDDEDGERGCTFLVGLIQKHRRRQRKMGEDMHTIGFGIYEVPEELTGQTNIHLSKNFF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 129.00 | 16 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 129.00 | 12 | calpain 2 |
| 3 | Mus musculus | 125.00 | 20 | calpain 2 |
| 4 | Homo sapiens | 108.00 | 13 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 108.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 108.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Gallus gallus | 101.00 | 3 | calpain 2, (m/II) large subunit |
| 8 | Xenopus tropicalis | 95.50 | 4 | calpain 2, (m/II) large subunit |
| 9 | Stizostedion vitreum vitreum | 94.70 | 1 | putative calpain-like protein |
| 10 | Tetraodon nigroviridis | 89.70 | 6 | unnamed protein product |
| 11 | Oncorhynchus mykiss | 84.00 | 1 | m-calpain |
| 12 | Coturnix coturnix | 81.30 | 1 | quail calpain |
| 13 | Xenopus laevis | 80.50 | 4 | calpain 1, (mu/I) large subunit |
| 14 | Danio rerio | 78.20 | 5 | calpain 2, (m/II) large subunit a |
| 15 | Xenopus laevis | 77.40 | 1 | mu/m-calpain large subunit |
| 16 | Gallus gallus | 75.90 | 1 | mu-calpain large subunit |
| 17 | Danio rerio | 75.10 | 1 | calpain 1, large subunit |
| 18 | Ovis aries | 65.90 | 4 | calpain 1 |
| 19 | synthetic construct | 65.90 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 65.90 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Canis familiaris | 65.50 | 20 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 22 | Bos taurus | 64.30 | 5 | calpain 3 |
| 23 | Sus scrofa | 64.30 | 3 | calpain 3 |
| 24 | Macaca fascicularis | 63.90 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 25 | Oryctolagus cuniculus | 63.20 | 1 | calpain 3, (p94) |
| 26 | Rattus norvegicus | 62.80 | 1 | calpain 1, large subunit |
| 27 | Schistosoma japonicum | 54.70 | 1 | unknown |
| 28 | Sus scrofa domestica | 49.30 | 1 | calpain large polypeptide L3 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 394 EDEDDEDGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 394 EDEDDEDGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 |
| Rattus norvegicus | Query 394 EDEDDEDGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 394 EDEDDEDGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 |
| Mus musculus | Query 394 EDEDDEDGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 ||||+|||||||||||||||||||||||||#||||||||||||||||||||||||| |||| Sbjct 394 EDEDEEDGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLGKNFF 453 |
| Homo sapiens | Query 404 GCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 ||||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 404 GCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELSGQTNIHLSKNFF 453 |
| Homo sapiens | Query 404 GCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 ||||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 404 GCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELSGQTNIHLSKNFF 453 |
| Macaca mulatta | Query 404 GCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 ||||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 404 GCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELSGQTNIHLSKNFF 453 |
| Gallus gallus | Query 404 GCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 |||||+||||||||+|||||#|||||||| ||||| | +|||||||||||| Sbjct 404 GCTFLIGLIQKHRRKQRKMG#EDMHTIGFAIYEVPPEFSGQTNIHLSKNFF 453 |
| Xenopus tropicalis | Query 395 DEDDEDGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNF 452 |+| +| | ||||+||||||+||+|||||#|||||||| ||||| +|||| +| ||+++ Sbjct 395 DDDPDDNEEGCTFIVGLIQKNRRKQRKMG#EDMHTIGFAIYEVPPQLTGQKDIRLSRDY 452 |
| Stizostedion vitreum vitreum | Query 394 EDEDDEDGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 ||+| ||| ||+|+||||||+||| ||+|#|||||+|| ||+||+| | |+||+||+| Sbjct 395 EDDDPADGELGCSFVVGLIQKNRRRARKLG#EDMHTVGFAIYQVPDEFRDQRNVHLNKNYF 454 |
| Tetraodon nigroviridis | Query 394 EDEDDEDGE--RGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKN 451 |+ ||+ || ||||+|||+||+||| ||||#||| |||| |||+||| +| |+|| || Sbjct 398 EEVDDDPGEDQEGCTFIVGLMQKNRRRLRKMG#EDMETIGFAIYELPEEYSGLKNVHLKKN 457 Query 452 FF 453 || Query 452 FF 453 |
| Oncorhynchus mykiss | Query 404 GCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 ||+|+||||||+||| ||+|#|||||||| ||||| + || +|| ||+| Sbjct 402 GCSFVVGLIQKNRRRLRKVG#EDMHTIGFAIYEVPSQFHGQREVHLDKNYF 451 |
| Coturnix coturnix | Query 404 GCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 |+||| |+||||||+|++|# ||||||| +|||||| | |+|| |+|| Sbjct 407 ACSFLVALMQKHRRRERRVG#GDMHTIGFAVYEVPEEAQGSQNVHLKKDFF 456 |
| Xenopus laevis | Query 394 EDEDDEDG-ERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNF 452 || ||+ | | |||||+ |+||++|++|+ |#+|| |||| +|||| | ||+ +|| ++| Sbjct 404 EDGDDDYGSETGCTFLLALMQKNKRKERRFG#KDMETIGFAVYEVPREFLGQSGVHLKRDF 463 Query 453 F 453 | Query 453 F 453 |
| Danio rerio | Query 404 GCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 ||+ ++|||||+||+ || |#|||||||+ ||||| + || +|| ||+| Sbjct 402 GCSVVIGLIQKNRRKMRKAG#EDMHTIGYAIYEVPSQFHGQKEVHLDKNYF 451 |
| Xenopus laevis | Query 404 GCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 |+||| |+||+|| +|| |#+||||||| +||||+| | |+|| |+|| Sbjct 407 ACSFLVALMQKNRRNERKAG#QDMHTIGFAVYEVPDEFKGCQNVHLKKDFF 456 |
| Gallus gallus | Query 402 ERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 | ||+||+ |+||||||+|+ |#+|| |||| +|||| | | + +|| ++|| Sbjct 415 EPGCSFLLALMQKHRRRERRYG#KDMETIGFAVYEVPPEHVGHSGVHLQRDFF 466 |
| Danio rerio | Query 398 DEDGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 | |+ |+||| |+|| ||++|+ |#+|| |||| ||||| | ||+ +|| ++|| Sbjct 400 DSPGQSECSFLVALMQKDRRKKRREG#QDMETIGFAIYEVPREFLGQSGVHLKRDFF 455 |
| Ovis aries | Query 402 ERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 | ||+||+ |+||||||+|+ |# || |||| +|||| || || +|| ++|| Sbjct 28 ESGCSFLLALMQKHRRRERRFG#RDMETIGFAVYEVPPELVGQPAVHLKRDFF 79 |
| synthetic construct | Query 394 EDEDDE-DGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNF 452 +| || | | ||+|++ |+||||||+|+ |# || |||| +|||| || || +|| ++| Sbjct 405 DDPDDYGDRESGCSFVLALMQKHRRRERRFG#RDMETIGFAVYEVPPELVGQPAVHLKRDF 464 Query 453 F 453 | Query 453 F 453 |
| Pongo pygmaeus | Query 394 EDEDDE-DGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNF 452 +| || | | ||+|++ |+||||||+|+ |# || |||| +|||| || || +|| ++| Sbjct 405 DDPDDYGDRESGCSFVLALMQKHRRRERRFG#RDMETIGFAVYEVPPELAGQPAVHLKRDF 464 Query 453 F 453 | Query 453 F 453 |
| Canis familiaris | Query 394 EDEDDEDG--ERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKN 451 +| ||+ | | ||+|++ |+||||||+|+ |# || |||| +|||| || || +|| ++ Sbjct 402 DDVDDDYGGRESGCSFVLALMQKHRRRERRFG#RDMETIGFAVYEVPPELVGQPAVHLKRD 461 Query 452 FF 453 || Query 452 FF 453 |
| Bos taurus | Query 405 CTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 |+||| |+||+||+ ||+|# ++ |||| |||||+|+ | || |+|| Sbjct 479 CSFLVALMQKNRRKDRKLG#ANLFTIGFAIYEVPKEMHGNKQ-HLQKDFF 526 |
| Sus scrofa | Query 405 CTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 |+||| |+||+||+ ||+|# ++ |||| |||||+|+ | || |+|| Sbjct 478 CSFLVALMQKNRRKDRKLG#ANLFTIGFAIYEVPKEMHGNKQ-HLQKDFF 525 |
| Macaca fascicularis | Query 405 CTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 |+||| |+||+||+ ||+|# ++ |||| |||||+|+ | || |+|| Sbjct 478 CSFLVALMQKNRRKDRKLG#ANLFTIGFAIYEVPKEMHGNRQ-HLQKDFF 525 |
| Oryctolagus cuniculus | Query 405 CTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNFF 453 |+||| |+||+||+ ||+|# ++ |||| |||||+|+ | || | || Sbjct 410 CSFLVALMQKNRRKDRKLG#ANLFTIGFAIYEVPKEMHGNKQ-HLQKEFF 457 |
| Rattus norvegicus | Query 394 EDEDDEDG-ERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSKNF 452 +| || | | ||+||+ |+||||||+|+ |# || |||| +|+|| || || +|| ++| Sbjct 405 DDADDYDSRESGCSFLLALMQKHRRRERRFG#RDMETIGFAVYQVPRELAGQP-VHLKRDF 463 Query 453 F 453 | Query 453 F 453 |
| Schistosoma japonicum | Query 394 EDEDDEDGERGCTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYEVPEELTGQTNIHLSK 450 || |+ | | | +|||+||+||+ |+ |# |+ |||+ +|++||+ | ++ | Sbjct 46 EDPDENDEENKGTLIVGLMQKNRRKMRQEG#ADLLTIGYAVYKLPEQHQGTLDMKFFK 102 |
| Sus scrofa domestica | Query 405 CTFLVGLIQKHRRRQRKMG#EDMHTIGFGIYE 435 |+||| |+||+||+ ||+|# ++ |||| ||| Sbjct 5 CSFLVALMQKNRRKDRKLG#ANLFTIGFAIYE 35 |
[Site 33] LTGQTNIHLS449-KNFFLTTRAR
Ser449  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu440 | Thr441 | Gly442 | Gln443 | Thr444 | Asn445 | Ile446 | His447 | Leu448 | Ser449 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys450 | Asn451 | Phe452 | Phe453 | Leu454 | Thr455 | Thr456 | Arg457 | Ala458 | Arg459 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RKMGEDMHTIGFGIYEVPEELTGQTNIHLSKNFFLTTRARERSDTFINLREVLNRFKLPP |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 130.00 | 15 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 130.00 | 10 | calpain 2 |
| 3 | Mus musculus | 129.00 | 20 | m-calpain |
| 4 | Homo sapiens | 127.00 | 13 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 127.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 127.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Gallus gallus | 111.00 | 3 | calpain 2, (m/II) large subunit |
| 8 | Xenopus tropicalis | 100.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Tetraodon nigroviridis | 94.40 | 6 | unnamed protein product |
| 10 | Danio rerio | 93.20 | 5 | calpain 2, (m/II) large subunit a |
| 11 | Oncorhynchus mykiss | 93.20 | 1 | m-calpain |
| 12 | Coturnix coturnix | 90.10 | 1 | quail calpain |
| 13 | Stizostedion vitreum vitreum | 89.40 | 1 | putative calpain-like protein |
| 14 | Xenopus laevis | 84.30 | 1 | mu/m-calpain large subunit |
| 15 | Xenopus laevis | 83.20 | 3 | calpain 1, (mu/I) large subunit |
| 16 | Danio rerio | 81.30 | 1 | calpain 1, large subunit |
| 17 | Canis familiaris | 79.70 | 21 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 18 | Gallus gallus | 79.70 | 1 | mu-calpain large subunit |
| 19 | Pongo pygmaeus | 79.30 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | synthetic construct | 79.30 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 21 | Bos taurus | 78.60 | 5 | calpain 1, (mu/I) large subunit |
| 22 | Ovis aries | 78.60 | 3 | calpain 1 |
| 23 | Sus scrofa | 75.50 | 4 | calpain 1, large subunit |
| 24 | Oryctolagus cuniculus | 73.20 | 1 | calpain 3, (p94) |
| 25 | Macaca fascicularis | 73.20 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 26 | Rattus norvegicus | 72.40 | 1 | calpain 1, large subunit |
| 27 | Canis familiaris | 62.00 | 1 | PREDICTED: similar to Calpain-9 (Digestive tract- |
| 28 | Oncorhynchus mykiss | 59.30 | 1 | gill-specific calpain |
| 29 | Rattus sp. | 52.80 | 1 | calpain II |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 |
| Rattus norvegicus | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 |
| Mus musculus | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 ||||||||||||||||||||||||||||| #|||||||||||||||||||||||||||||| Sbjct 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLG#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 |
| Homo sapiens | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 |||||||||||||||||||||+||||||||#|||||| ||||||||||||||||||||||| Sbjct 420 RKMGEDMHTIGFGIYEVPEELSGQTNIHLS#KNFFLTNRARERSDTFINLREVLNRFKLPP 479 |
| Homo sapiens | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 |||||||||||||||||||||+||||||||#|||||| ||||||||||||||||||||||| Sbjct 420 RKMGEDMHTIGFGIYEVPEELSGQTNIHLS#KNFFLTNRARERSDTFINLREVLNRFKLPP 479 |
| Macaca mulatta | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 |||||||||||||||||||||+||||||||#|||||| ||||||||||||||||||||||| Sbjct 420 RKMGEDMHTIGFGIYEVPEELSGQTNIHLS#KNFFLTNRARERSDTFINLREVLNRFKLPP 479 |
| Gallus gallus | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 |||||||||||| ||||| | +||||||||#|||||| +|||+|+||||||||||||||| Sbjct 420 RKMGEDMHTIGFAIYEVPPEFSGQTNIHLS#KNFFLTNKAREKSNTFINLREVLNRFKLPA 479 |
| Xenopus tropicalis | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 |||||||||||| ||||| +|||| +| ||#+++ | + ||+|||+||||||||||+||| Sbjct 420 RKMGEDMHTIGFAIYEVPPQLTGQKDIRLS#RDYILRNKIREKSDTYINLREVLNRFRLPP 479 |
| Tetraodon nigroviridis | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 ||||||| |||| |||+||| +| |+|| #|||||++ | ||+|||||||| +|| ||| Sbjct 426 RKMGEDMETIGFAIYELPEEYSGLKNVHLK#KNFFLSSYATARSETFINLREVCSRFCLPP 485 |
| Danio rerio | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 || ||||||||+ ||||| + || +|| #||+||| + ||+|||||||| |||||| Sbjct 418 RKAGEDMHTIGYAIYEVPSQFHGQKEVHLD#KNYFLTHAQKARSETFINLREVSTRFKLPP 477 |
| Oncorhynchus mykiss | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 ||+||||||||| ||||| + || +|| #||+||| + ||+||+||||| |||+|| Sbjct 418 RKVGEDMHTIGFAIYEVPSQFHGQREVHLD#KNYFLTHAQKARSETFVNLREVSTRFKMPP 477 |
| Coturnix coturnix | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 |++| ||||||| +|||||| | |+|| #|+||| ++| ||+|||||||| |+ +||| Sbjct 423 RRVGGDMHTIGFAVYEVPEEAQGSQNVHLK#KDFFLRNQSRARSETFINLREVSNQIRLPP 482 |
| Stizostedion vitreum vitreum | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 ||+||||||+|| ||+||+| | |+||+#||+|| + ||+|||||||| +|| ||| Sbjct 421 RKLGEDMHTVGFAIYQVPDEFRDQRNVHLN#KNYFLRHASAARSETFINLREVCSRFCLPP 480 |
| Xenopus laevis | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 || |+||||||| +||||+| | |+|| #|+||| ++ ||+|||||||| + ||| Sbjct 423 RKAGQDMHTIGFAVYEVPDEFKGCQNVHLK#KDFFLRHQSCARSETFINLREVSTKIHLPP 482 |
| Xenopus laevis | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 |+ |+|| |||| +|||| | ||+ +|| #++||| +| ||+ ||||||| | |||| Sbjct 431 RRFGKDMETIGFAVYEVPREFLGQSGVHLK#RDFFLQNASRARSEQFINLREVSTRHKLPP 490 |
| Danio rerio | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLP 478 |+ |+|| |||| ||||| | ||+ +|| #++|||| + ||+ ||||||| +||+|| Sbjct 422 RREGQDMETIGFAIYEVPREFLGQSGVHLK#RDFFLTHASSARSELFINLREVSSRFRLP 480 |
| Canis familiaris | Query 425 DMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 || |||| +|||| || || +|| #++||| +| ||+ ||||||| ||+||| Sbjct 435 DMETIGFAVYEVPPELVGQPAVHLK#RDFFLANSSRARSEQFINLREVSTRFRLPP 489 |
| Gallus gallus | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 |+ |+|| |||| +|||| | | + +|| #++|||+ +| ||+ ||||||| | +||| Sbjct 433 RRYGKDMETIGFAVYEVPPEHVGHSGVHLQ#RDFFLSNASRARSEQFINLREVSTRLRLPP 492 |
| Pongo pygmaeus | Query 425 DMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 || |||| +|||| || || +|| #++||| +| ||+ ||||||| ||+||| Sbjct 437 DMETIGFAVYEVPPELAGQPAVHLK#RDFFLANASRARSEQFINLREVSTRFRLPP 491 |
| synthetic construct | Query 425 DMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 || |||| +|||| || || +|| #++||| +| ||+ ||||||| ||+||| Sbjct 437 DMETIGFAVYEVPPELVGQPAVHLK#RDFFLANASRARSEQFINLREVSTRFRLPP 491 |
| Bos taurus | Query 425 DMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 || |||| +|||| || || +|| #++|||+ +| ||+ ||||||| ||+||| Sbjct 439 DMETIGFAVYEVPPELMGQPAVHLK#RDFFLSNASRARSEQFINLREVSTRFRLPP 493 |
| Ovis aries | Query 425 DMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 || |||| +|||| || || +|| #++||| +| ||+ ||||||| ||+||| Sbjct 51 DMETIGFAVYEVPPELVGQPAVHLK#RDFFLANASRARSEQFINLREVSTRFRLPP 105 |
| Sus scrofa | Query 425 DMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 || |||| +|||| || || +|| #++||| +| ||+ ||||||| ||+||| Sbjct 438 DMETIGFAVYEVPPELVGQP-VHLK#RDFFLANASRARSEQFINLREVSTRFRLPP 491 |
| Oryctolagus cuniculus | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 ||+| ++ |||| |||||+|+ | || #| ||| ++ || |+||+||| ||+||| Sbjct 425 RKLGANLFTIGFAIYEVPKEMHGNKQ-HLQ#KEFFLYNASKARSKTYINMREVSQRFRLPP 483 |
| Macaca fascicularis | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 ||+| ++ |||| |||||+|+ | || #|+||| +| || |+||+||| ||+||| Sbjct 493 RKLGANLFTIGFAIYEVPKEMHGNRQ-HLQ#KDFFLYNASRARSKTYINMREVSQRFRLPP 551 |
| Rattus norvegicus | Query 425 DMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 || |||| +|+|| || || +|| #++||| +| +|+ ||||||| || +||| Sbjct 437 DMETIGFAVYQVPRELAGQP-VHLK#RDFFLANASRAQSEHFINLREVSNRIRLPP 490 |
| Canis familiaris | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 ++ | || |||+ ||+ | + + ||+#|+|| ++ || |||||||| +|||||| Sbjct 237 KRFGADMLTIGYAIYQSPSK-----DEHLN#KDFFRYHASQARSKTFINLREVSDRFKLPP 291 |
| Oncorhynchus mykiss | Query 420 RKMGEDMHTIGFGIYEVPEELTGQTNIHLS#KNFFLTTRARERSDTFINLREVLNRFKLPP 479 || | |+ |||| +|| | + || #|+|| ++ || |+||+||+ || ||| Sbjct 395 RKEGLDLQTIGFALYEAPAD-----EDHLG#KDFFRYNGSKARSKTYINVREISERFTLPP 449 |
| Rattus sp. | Query 455 TTRARERSDTFINLREVLNRFKLPP 479 ||||||||||||||||||||||||| Query 455 TTRARERSDTFINLREVLNRFKLPP 479 |
[Site 34] FFLTTRARER461-SDTFINLREV
Arg461  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe452 | Phe453 | Leu454 | Thr455 | Thr456 | Arg457 | Ala458 | Arg459 | Glu460 | Arg461 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser462 | Asp463 | Thr464 | Phe465 | Ile466 | Asn467 | Leu468 | Arg469 | Glu470 | Val471 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GIYEVPEELTGQTNIHLSKNFFLTTRARERSDTFINLREVLNRFKLPPGEYVLVPSTFEP |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Rattus norvegicus | 130.00 | 10 | calpain 2 |
| 2 | N/A | 129.00 | 15 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 3 | Mus musculus | 128.00 | 20 | m-calpain |
| 4 | Homo sapiens | 127.00 | 14 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 126.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 126.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Gallus gallus | 112.00 | 3 | calpain 2, (m/II) large subunit |
| 8 | Xenopus tropicalis | 97.80 | 4 | calpain 2, (m/II) large subunit |
| 9 | Tetraodon nigroviridis | 94.70 | 6 | unnamed protein product |
| 10 | Coturnix coturnix | 94.40 | 1 | quail calpain |
| 11 | Danio rerio | 93.60 | 5 | calpain 1, (mu/I) large subunit a |
| 12 | Oncorhynchus mykiss | 93.20 | 1 | m-calpain |
| 13 | Canis familiaris | 91.70 | 22 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 14 | Stizostedion vitreum vitreum | 89.40 | 1 | putative calpain-like protein |
| 15 | Xenopus laevis | 89.00 | 3 | calpain 1, (mu/I) large subunit |
| 16 | Danio rerio | 88.20 | 1 | calpain 1, large subunit |
| 17 | Bos taurus | 87.80 | 5 | calpain 1, (mu/I) large subunit |
| 18 | synthetic construct | 87.80 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 19 | Pongo pygmaeus | 87.80 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | Ovis aries | 87.40 | 3 | calpain 1 |
| 21 | Sus scrofa | 87.00 | 4 | calpain 1, large subunit |
| 22 | Xenopus laevis | 86.30 | 1 | mu/m-calpain large subunit |
| 23 | Gallus gallus | 85.90 | 1 | mu-calpain large subunit |
| 24 | Rattus norvegicus | 83.60 | 1 | calpain 1, large subunit |
| 25 | Macaca fascicularis | 80.10 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 26 | Oryctolagus cuniculus | 79.30 | 1 | calpain 3, (p94) |
| 27 | Rattus sp. | 76.30 | 2 | calpain II |
| 28 | Canis familiaris | 72.80 | 1 | PREDICTED: similar to Calpain-9 (Digestive tract- |
| 29 | Caenorhabditis elegans | 63.90 | 1 | CaLPain family member (clp-1) |
| 30 | Oncorhynchus mykiss | 63.90 | 1 | gill-specific calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| Rattus norvegicus | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 |
| N/A | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 |
| Mus musculus | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 ||||||||||||||||| ||||||||||||#|||||||||||||||||||||||||||||| Sbjct 432 GIYEVPEELTGQTNIHLGKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 |
| Homo sapiens | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 |||||||||+|||||||||||||| |||||#|||||||||||||||||||||+|||||||| Sbjct 432 GIYEVPEELSGQTNIHLSKNFFLTNRARER#SDTFINLREVLNRFKLPPGEYILVPSTFEP 491 |
| Homo sapiens | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 |||||||||+|||||||||||||| |||||#|||||||||||||||||||||+|||||||| Sbjct 432 GIYEVPEELSGQTNIHLSKNFFLTNRARER#SDTFINLREVLNRFKLPPGEYILVPSTFEP 491 |
| Macaca mulatta | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 |||||||||+|||||||||||||| |||||#|||||||||||||||||||||+|||||||| Sbjct 432 GIYEVPEELSGQTNIHLSKNFFLTNRARER#SDTFINLREVLNRFKLPPGEYILVPSTFEP 491 |
| Gallus gallus | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 ||||| | +|||||||||||||| +|||+#|+||||||||||||||| |||++||||||| Sbjct 432 AIYEVPPEFSGQTNIHLSKNFFLTNKAREK#SNTFINLREVLNRFKLPAGEYIIVPSTFEP 491 |
| Xenopus tropicalis | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 ||||| +|||| +| ||+++ | + ||+#|||+||||||||||+|||||||++|||||| Sbjct 432 AIYEVPPQLTGQKDIRLSRDYILRNKIREK#SDTYINLREVLNRFRLPPGEYVILPSTFEP 491 |
| Tetraodon nigroviridis | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 |||+||| +| |+|| |||||++ | |#|+|||||||| +|| ||||||++||||||| Sbjct 438 AIYELPEEYSGLKNVHLKKNFFLSSYATAR#SETFINLREVCSRFCLPPGEYLIVPSTFEP 497 |
| Coturnix coturnix | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +|||||| | |+|| |+||| ++| |#|+|||||||| |+ +||||||++||||||| Sbjct 435 AVYEVPEEAQGSQNVHLKKDFFLRNQSRAR#SETFINLREVSNQIRLPPGEYIVVPSTFEP 494 |
| Danio rerio | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 ||||||| || |+|| |+|||+ + |#|+|||||||| | +||||||++||||||| Sbjct 435 AIYEVPEEYTGCQNVHLKKDFFLSNSSAAR#SETFINLREVSTRLRLPPGEYIIVPSTFEP 494 |
| Oncorhynchus mykiss | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 ||||| + || +|| ||+||| + |#|+||+||||| |||+|||||++||||||| Sbjct 430 AIYEVPSQFHGQREVHLDKNYFLTHAQKAR#SETFVNLREVSTRFKMPPGEYLIVPSTFEP 489 |
| Canis familiaris | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +|||| || || +|| ++||| +| |#|+ ||||||| ||+|||||||+||||||| Sbjct 442 AVYEVPPELVGQPAVHLKRDFFLANSSRAR#SEQFINLREVSTRFRLPPGEYVVVPSTFEP 501 |
| Stizostedion vitreum vitreum | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 ||+||+| | |+||+||+|| + |#|+|||||||| +|| ||||||++||||||| Sbjct 433 AIYQVPDEFRDQRNVHLNKNYFLRHASAAR#SETFINLREVCSRFCLPPGEYLIVPSTFEP 492 |
| Xenopus laevis | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +|||| | ||+ +|| ++||| +| |#|+ ||||||| | |||||||++||||||| Sbjct 443 AVYEVPREFLGQSGVHLKRDFFLQNASRAR#SEQFINLREVSTRHKLPPGEYIVVPSTFEP 502 |
| Danio rerio | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 ||||| | ||+ +|| ++|||| + |#|+ ||||||| +||+|| |||++||||||| Sbjct 434 AIYEVPREFLGQSGVHLKRDFFLTHASSAR#SELFINLREVSSRFRLPAGEYIIVPSTFEP 493 |
| Bos taurus | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +|||| || || +|| ++|||+ +| |#|+ ||||||| ||+|||||||+||||||| Sbjct 446 AVYEVPPELMGQPAVHLKRDFFLSNASRAR#SEQFINLREVSTRFRLPPGEYVVVPSTFEP 505 |
| synthetic construct | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +|||| || || +|| ++||| +| |#|+ ||||||| ||+|||||||+||||||| Sbjct 444 AVYEVPPELVGQPAVHLKRDFFLANASRAR#SEQFINLREVSTRFRLPPGEYVVVPSTFEP 503 |
| Pongo pygmaeus | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +|||| || || +|| ++||| +| |#|+ ||||||| ||+|||||||+||||||| Sbjct 444 AVYEVPPELAGQPAVHLKRDFFLANASRAR#SEQFINLREVSTRFRLPPGEYVVVPSTFEP 503 |
| Ovis aries | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +|||| || || +|| ++||| +| |#|+ ||||||| ||+|||||||+||||||| Sbjct 58 AVYEVPPELVGQPAVHLKRDFFLANASRAR#SEQFINLREVSTRFRLPPGEYVVVPSTFEP 117 |
| Sus scrofa | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +|||| || || +|| ++||| +| |#|+ ||||||| ||+|||||||+||||||| Sbjct 445 AVYEVPPELVGQP-VHLKRDFFLANASRAR#SEQFINLREVSTRFRLPPGEYVVVPSTFEP 503 |
| Xenopus laevis | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +||||+| | |+|| |+||| ++ |#|+|||||||| + ||||||++||||||| Sbjct 435 AVYEVPDEFKGCQNVHLKKDFFLRHQSCAR#SETFINLREVSTKIHLPPGEYIIVPSTFEP 494 |
| Gallus gallus | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +|||| | | + +|| ++|||+ +| |#|+ ||||||| | +||||||++||||||| Sbjct 445 AVYEVPPEHVGHSGVHLQRDFFLSNASRAR#SEQFINLREVSTRLRLPPGEYIVVPSTFEP 504 |
| Rattus norvegicus | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +|+|| || || +|| ++||| +| +#|+ ||||||| || +||||||++||||||| Sbjct 444 AVYQVPRELAGQP-VHLKRDFFLANASRAQ#SEHFINLREVSNRIRLPPGEYIVVPSTFEP 502 |
| Macaca fascicularis | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 |||||+|+ | || |+||| +| |#| |+||+||| ||+||| |||+||||+|| Sbjct 505 AIYEVPKEMHGNRQ-HLQKDFFLYNASRAR#SKTYINMREVSQRFRLPPSEYVIVPSTYEP 563 |
| Oryctolagus cuniculus | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 |||||+|+ | || | ||| ++ |#| |+||+||| ||+||| |||+||||+|| Sbjct 437 AIYEVPKEMHGNKQ-HLQKEFFLYNASKAR#SKTYINMREVSQRFRLPPSEYVIVPSTYEP 495 |
| Rattus sp. | Query 455 TTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 |||||||#|||||||||||||||||||||||||||||| Query 455 TTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 |
| Canis familiaris | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 ||+ | + + ||+|+|| ++ |#| |||||||| +|||||||||+|+|||||| Sbjct 249 AIYQSPSK-----DEHLNKDFFRYHASQAR#SKTFINLREVSDRFKLPPGEYILIPSTFEP 303 |
| Caenorhabditis elegans | Query 442 GQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 | ||| || ++ |#| ||||||+ ||++||| ||+||||||| Sbjct 710 GNNRGRLSKQFFAANKSAMR#SAAFINLREMTGRFRVPPGNYVVVPSTFEP 759 |
| Oncorhynchus mykiss | Query 432 GIYEVPEELTGQTNIHLSKNFFLTTRARER#SDTFINLREVLNRFKLPPGEYVLVPSTFEP 491 +|| | + || |+|| ++ |#| |+||+||+ || |||| |+|||+||+| Sbjct 407 ALYEAPAD-----EDHLGKDFFRYNGSKAR#SKTYINVREISERFTLPPGNYLLVPTTFQP 461 |
[Site 35] FLTTRARERS462-DTFINLREVL
Ser462  Asp
Asp
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe453 | Leu454 | Thr455 | Thr456 | Arg457 | Ala458 | Arg459 | Glu460 | Arg461 | Ser462 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asp463 | Thr464 | Phe465 | Ile466 | Asn467 | Leu468 | Arg469 | Glu470 | Val471 | Leu472 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| IYEVPEELTGQTNIHLSKNFFLTTRARERSDTFINLREVLNRFKLPPGEYVLVPSTFEPH |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 131.00 | 15 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 131.00 | 10 | calpain 2 |
| 3 | Mus musculus | 129.00 | 20 | m-calpain large subunit |
| 4 | Homo sapiens | 125.00 | 14 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 125.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 125.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Gallus gallus | 112.00 | 3 | calpain 2, (m/II) large subunit |
| 8 | Xenopus tropicalis | 101.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Coturnix coturnix | 97.40 | 1 | quail calpain |
| 10 | Danio rerio | 96.30 | 5 | calpain 2, (m/II) large subunit a |
| 11 | Oncorhynchus mykiss | 96.30 | 1 | m-calpain |
| 12 | Tetraodon nigroviridis | 95.10 | 6 | unnamed protein product |
| 13 | Stizostedion vitreum vitreum | 92.40 | 1 | putative calpain-like protein |
| 14 | Canis familiaris | 92.00 | 22 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 15 | Bos taurus | 91.70 | 5 | calpain 1, (mu/I) large subunit |
| 16 | Pongo pygmaeus | 91.70 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 17 | synthetic construct | 91.70 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 18 | Xenopus laevis | 90.50 | 3 | hypothetical protein LOC398772 |
| 19 | Xenopus laevis | 89.70 | 1 | mu/m-calpain large subunit |
| 20 | Danio rerio | 88.60 | 1 | calpain 1, large subunit |
| 21 | Sus scrofa | 87.80 | 4 | calpain 1, large subunit |
| 22 | Ovis aries | 87.40 | 3 | calpain 1 |
| 23 | Gallus gallus | 86.70 | 1 | mu-calpain large subunit |
| 24 | Rattus norvegicus | 84.00 | 1 | calpain 1, large subunit |
| 25 | Macaca fascicularis | 82.40 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 26 | Oryctolagus cuniculus | 82.00 | 1 | calpain 3, (p94) |
| 27 | Rattus sp. | 79.30 | 2 | calpain II |
| 28 | Canis familiaris | 75.10 | 1 | PREDICTED: similar to Calpain-9 (Digestive tract- |
| 29 | Oncorhynchus mykiss | 67.00 | 1 | gill-specific calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 |
| Rattus norvegicus | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 |
| Mus musculus | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 |||||||||||||||| |||||||||||||#|||||||||||||||||||||||||||||| Sbjct 433 IYEVPEELTGQTNIHLGKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 |
| Homo sapiens | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||||||||+|||||||||||||| ||||||#||||||||||||||||||||+||||||||+ Sbjct 433 IYEVPEELSGQTNIHLSKNFFLTNRARERS#DTFINLREVLNRFKLPPGEYILVPSTFEPN 492 |
| Homo sapiens | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||||||||+|||||||||||||| ||||||#||||||||||||||||||||+||||||||+ Sbjct 433 IYEVPEELSGQTNIHLSKNFFLTNRARERS#DTFINLREVLNRFKLPPGEYILVPSTFEPN 492 |
| Macaca mulatta | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||||||||+|||||||||||||| ||||||#||||||||||||||||||||+||||||||+ Sbjct 433 IYEVPEELSGQTNIHLSKNFFLTNRARERS#DTFINLREVLNRFKLPPGEYILVPSTFEPN 492 |
| Gallus gallus | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||||| | +|||||||||||||| +|||+|#+||||||||||||||| |||++|||||||+ Sbjct 433 IYEVPPEFSGQTNIHLSKNFFLTNKAREKS#NTFINLREVLNRFKLPAGEYIIVPSTFEPN 492 |
| Xenopus tropicalis | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||||| +|||| +| ||+++ | + ||+|#||+||||||||||+|||||||++||||||| Sbjct 433 IYEVPPQLTGQKDIRLSRDYILRNKIREKS#DTYINLREVLNRFRLPPGEYVILPSTFEPH 492 |
| Coturnix coturnix | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +|||||| | |+|| |+||| ++| ||#+|||||||| |+ +||||||++|||||||| Sbjct 436 VYEVPEEAQGSQNVHLKKDFFLRNQSRARS#ETFINLREVSNQIRLPPGEYIVVPSTFEPH 495 |
| Danio rerio | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||||| + || +|| ||+||| + ||#+|||||||| |||||||||++|||||+|| Sbjct 431 IYEVPSQFHGQKEVHLDKNYFLTHAQKARS#ETFINLREVSTRFKLPPGEYLIVPSTFDPH 490 |
| Oncorhynchus mykiss | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||||| + || +|| ||+||| + ||#+||+||||| |||+|||||++|||||||| Sbjct 431 IYEVPSQFHGQREVHLDKNYFLTHAQKARS#ETFVNLREVSTRFKMPPGEYLIVPSTFEPH 490 |
| Tetraodon nigroviridis | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 |||+||| +| |+|| |||||++ | ||#+|||||||| +|| ||||||++|||||||+ Sbjct 439 IYELPEEYSGLKNVHLKKNFFLSSYATARS#ETFINLREVCSRFCLPPGEYLIVPSTFEPN 498 |
| Stizostedion vitreum vitreum | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||+||+| | |+||+||+|| + ||#+|||||||| +|| ||||||++|||||||| Sbjct 434 IYQVPDEFRDQRNVHLNKNYFLRHASAARS#ETFINLREVCSRFCLPPGEYLIVPSTFEPH 493 |
| Canis familiaris | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +|||| || || +|| ++||| +| ||#+ ||||||| ||+|||||||+|||||||+ Sbjct 443 VYEVPPELVGQPAVHLKRDFFLANSSRARS#EQFINLREVSTRFRLPPGEYVVVPSTFEPN 502 |
| Bos taurus | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +|||| || || +|| ++|||+ +| ||#+ ||||||| ||+|||||||+|||||||+ Sbjct 447 VYEVPPELMGQPAVHLKRDFFLSNASRARS#EQFINLREVSTRFRLPPGEYVVVPSTFEPN 506 |
| Pongo pygmaeus | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +|||| || || +|| ++||| +| ||#+ ||||||| ||+|||||||+|||||||+ Sbjct 445 VYEVPPELAGQPAVHLKRDFFLANASRARS#EQFINLREVSTRFRLPPGEYVVVPSTFEPN 504 |
| synthetic construct | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +|||| || || +|| ++||| +| ||#+ ||||||| ||+|||||||+|||||||+ Sbjct 445 VYEVPPELVGQPAVHLKRDFFLANASRARS#EQFINLREVSTRFRLPPGEYVVVPSTFEPN 504 |
| Xenopus laevis | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +||||+| | |+|| |+||| ++ ||#+|||||||| + +||||||++|||||||| Sbjct 436 VYEVPDEFKGCQNVHLKKDFFLRHQSCARS#ETFINLREVSTQIRLPPGEYIIVPSTFEPH 495 |
| Xenopus laevis | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +||||+| | |+|| |+||| ++ ||#+|||||||| + ||||||++|||||||| Sbjct 436 VYEVPDEFKGCQNVHLKKDFFLRHQSCARS#ETFINLREVSTKIHLPPGEYIIVPSTFEPH 495 |
| Danio rerio | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||||| | ||+ +|| ++|||| + ||#+ ||||||| +||+|| |||++|||||||+ Sbjct 435 IYEVPREFLGQSGVHLKRDFFLTHASSARS#ELFINLREVSSRFRLPAGEYIIVPSTFEPN 494 |
| Sus scrofa | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +|||| || || +|| ++||| +| ||#+ ||||||| ||+|||||||+|||||||+ Sbjct 446 VYEVPPELVGQP-VHLKRDFFLANASRARS#EQFINLREVSTRFRLPPGEYVVVPSTFEPN 504 |
| Ovis aries | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +|||| || || +|| ++||| +| ||#+ ||||||| ||+|||||||+|||||||+ Sbjct 59 VYEVPPELVGQPAVHLKRDFFLANASRARS#EQFINLREVSTRFRLPPGEYVVVPSTFEPN 118 |
| Gallus gallus | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +|||| | | + +|| ++|||+ +| ||#+ ||||||| | +||||||++|||||||+ Sbjct 446 VYEVPPEHVGHSGVHLQRDFFLSNASRARS#EQFINLREVSTRLRLPPGEYIVVPSTFEPN 505 |
| Rattus norvegicus | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +|+|| || || +|| ++||| +| +|#+ ||||||| || +||||||++|||||||+ Sbjct 445 VYQVPRELAGQP-VHLKRDFFLANASRAQS#EHFINLREVSNRIRLPPGEYIVVPSTFEPN 503 |
| Macaca fascicularis | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 |||||+|+ | || |+||| +| ||# |+||+||| ||+||| |||+||||+||| Sbjct 506 IYEVPKEMHGNRQ-HLQKDFFLYNASRARS#KTYINMREVSQRFRLPPSEYVIVPSTYEPH 564 |
| Oryctolagus cuniculus | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 |||||+|+ | || | ||| ++ ||# |+||+||| ||+||| |||+||||+||| Sbjct 438 IYEVPKEMHGNKQ-HLQKEFFLYNASKARS#KTYINMREVSQRFRLPPSEYVIVPSTYEPH 496 |
| Rattus sp. | Query 455 TTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||||||||#|||||||||||||||||||||||||||||| Query 455 TTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 |
| Canis familiaris | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 ||+ | + + ||+|+|| ++ ||# |||||||| +|||||||||+|+||||||| Sbjct 250 IYQSPSK-----DEHLNKDFFRYHASQARS#KTFINLREVSDRFKLPPGEYILIPSTFEPH 304 |
| Oncorhynchus mykiss | Query 433 IYEVPEELTGQTNIHLSKNFFLTTRARERS#DTFINLREVLNRFKLPPGEYVLVPSTFEPH 492 +|| | + || |+|| ++ ||# |+||+||+ || |||| |+|||+||+|| Sbjct 408 LYEAPAD-----EDHLGKDFFRYNGSKARS#KTYINVREISERFTLPPGNYLLVPTTFQPH 462 |
[Site 36] RERSDTFINL468-REVLNRFKLP
Leu468  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg459 | Glu460 | Arg461 | Ser462 | Asp463 | Thr464 | Phe465 | Ile466 | Asn467 | Leu468 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg469 | Glu470 | Val471 | Leu472 | Asn473 | Arg474 | Phe475 | Lys476 | Leu477 | Pro478 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ELTGQTNIHLSKNFFLTTRARERSDTFINLREVLNRFKLPPGEYVLVPSTFEPHKNGDFC |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 133.00 | 15 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 133.00 | 11 | calpain 2 |
| 3 | Mus musculus | 130.00 | 20 | calpain 2 |
| 4 | Homo sapiens | 125.00 | 12 | AF261089_1 calpain large polypeptide L2 |
| 5 | Homo sapiens | 125.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 125.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Gallus gallus | 109.00 | 3 | calpain 2, (m/II) large subunit |
| 8 | Xenopus tropicalis | 102.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Danio rerio | 99.80 | 5 | calpain 2, (m/II) large subunit a |
| 10 | Tetraodon nigroviridis | 97.40 | 6 | unnamed protein product |
| 11 | Oncorhynchus mykiss | 96.70 | 1 | m-calpain |
| 12 | Stizostedion vitreum vitreum | 96.70 | 1 | putative calpain-like protein |
| 13 | Coturnix coturnix | 91.70 | 1 | quail calpain |
| 14 | Canis familiaris | 91.30 | 22 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 15 | synthetic construct | 90.90 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 16 | Bos taurus | 90.50 | 5 | calpain 1, (mu/I) large subunit |
| 17 | Pongo pygmaeus | 90.50 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 18 | Ovis aries | 87.80 | 3 | calpain 1 |
| 19 | Sus scrofa | 87.00 | 4 | calpain 1, large subunit |
| 20 | Xenopus laevis | 85.90 | 3 | hypothetical protein LOC398772 |
| 21 | Xenopus laevis | 85.10 | 1 | mu/m-calpain large subunit |
| 22 | Gallus gallus | 84.30 | 1 | mu-calpain large subunit |
| 23 | Danio rerio | 83.60 | 1 | calpain 1, large subunit |
| 24 | Canis familiaris | 81.60 | 2 | PREDICTED: similar to Calpain-9 (Digestive tract- |
| 25 | Rattus norvegicus | 81.30 | 1 | calpain 1, large subunit |
| 26 | Rattus sp. | 79.70 | 2 | calpain II |
| 27 | Macaca fascicularis | 77.80 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 28 | Oryctolagus cuniculus | 76.60 | 1 | calpain 3, (p94) |
| 29 | Oncorhynchus mykiss | 70.10 | 1 | gill-specific calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 |
| Rattus norvegicus | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 |
| Mus musculus | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 |||||||||| |||||||||||||||||||#|||||||||||||||||||||||||+|||| Sbjct 439 ELTGQTNIHLGKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKDGDFC 498 |
| Homo sapiens | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 ||+|||||||||||||| ||||||||||||#||||||||||||||+||||||||+|+|||| Sbjct 439 ELSGQTNIHLSKNFFLTNRARERSDTFINL#REVLNRFKLPPGEYILVPSTFEPNKDGDFC 498 |
| Homo sapiens | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 ||+|||||||||||||| ||||||||||||#||||||||||||||+||||||||+|+|||| Sbjct 439 ELSGQTNIHLSKNFFLTNRARERSDTFINL#REVLNRFKLPPGEYILVPSTFEPNKDGDFC 498 |
| Macaca mulatta | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 ||+|||||||||||||| ||||||||||||#||||||||||||||+||||||||+|+|||| Sbjct 439 ELSGQTNIHLSKNFFLTNRARERSDTFINL#REVLNRFKLPPGEYILVPSTFEPNKDGDFC 498 |
| Gallus gallus | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 | +|||||||||||||| +|||+|+|||||#|||||||||| |||++|||||||+ ||||| Sbjct 439 EFSGQTNIHLSKNFFLTNKAREKSNTFINL#REVLNRFKLPAGEYIIVPSTFEPNLNGDFC 498 |
| Xenopus tropicalis | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 +|||| +| ||+++ | + ||+|||+|||#|||||||+|||||||++|||||||| |||| Sbjct 439 QLTGQKDIRLSRDYILRNKIREKSDTYINL#REVLNRFRLPPGEYVILPSTFEPHKVGDFC 498 |
| Danio rerio | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 + || +|| ||+||| + ||+|||||#||| |||||||||++|||||+|||||||| Sbjct 437 QFHGQKEVHLDKNYFLTHAQKARSETFINL#REVSTRFKLPPGEYLIVPSTFDPHKNGDFC 496 |
| Tetraodon nigroviridis | Query 442 GQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 || +|| |||||+ + ||+|||||#||| +|||||||||+++||||||| ||||| Sbjct 439 GQREVHLDKNFFLSHAQKARSETFINL#REVCSRFKLPPGEYLIIPSTFEPHLNGDFC 495 |
| Oncorhynchus mykiss | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 + || +|| ||+||| + ||+||+||#||| |||+|||||++|||||||| ||||| Sbjct 437 QFHGQREVHLDKNYFLTHAQKARSETFVNL#REVSTRFKMPPGEYLIVPSTFEPHLNGDFC 496 |
| Stizostedion vitreum vitreum | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 | | |+||+||+|| + ||+|||||#||| +|| ||||||++|||||||||||||| Sbjct 440 EFRDQRNVHLNKNYFLRHASAARSETFINL#REVCSRFCLPPGEYLIVPSTFEPHKNGDFC 499 |
| Coturnix coturnix | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 | | |+|| |+||| ++| ||+|||||#||| |+ +||||||++||||||||| || Sbjct 442 EAQGSQNVHLKKDFFLRNQSRARSETFINL#REVSNQIRLPPGEYIVVPSTFEPHKEADF 500 |
| Canis familiaris | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 || || +|| ++||| +| ||+ ||||#||| ||+|||||||+|||||||+| ||| Sbjct 449 ELVGQPAVHLKRDFFLANSSRARSEQFINL#REVSTRFRLPPGEYVVVPSTFEPNKEGDF 507 |
| synthetic construct | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 || || +|| ++||| +| ||+ ||||#||| ||+|||||||+|||||||+| ||| Sbjct 451 ELVGQPAVHLKRDFFLANASRARSEQFINL#REVSTRFRLPPGEYVVVPSTFEPNKEGDF 509 |
| Bos taurus | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 || || +|| ++|||+ +| ||+ ||||#||| ||+|||||||+|||||||+| ||| Sbjct 453 ELMGQPAVHLKRDFFLSNASRARSEQFINL#REVSTRFRLPPGEYVVVPSTFEPNKEGDF 511 |
| Pongo pygmaeus | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 || || +|| ++||| +| ||+ ||||#||| ||+|||||||+|||||||+| ||| Sbjct 451 ELAGQPAVHLKRDFFLANASRARSEQFINL#REVSTRFRLPPGEYVVVPSTFEPNKEGDF 509 |
| Ovis aries | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 || || +|| ++||| +| ||+ ||||#||| ||+|||||||+|||||||+| ||| Sbjct 65 ELVGQPAVHLKRDFFLANASRARSEQFINL#REVSTRFRLPPGEYVVVPSTFEPNKEGDFV 124 |
| Sus scrofa | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 || || +|| ++||| +| ||+ ||||#||| ||+|||||||+|||||||+| ||| Sbjct 452 ELVGQP-VHLKRDFFLANASRARSEQFINL#REVSTRFRLPPGEYVVVPSTFEPNKEGDF 509 |
| Xenopus laevis | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 | | |+|| |+||| ++ ||+|||||#||| + +||||||++||||||||| || Sbjct 442 EFKGCQNVHLKKDFFLRHQSCARSETFINL#REVSTQIRLPPGEYIIVPSTFEPHKEADF 500 |
| Xenopus laevis | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 | | |+|| |+||| ++ ||+|||||#||| + ||||||++||||||||| || Sbjct 442 EFKGCQNVHLKKDFFLRHQSCARSETFINL#REVSTKIHLPPGEYIIVPSTFEPHKEADF 500 |
| Gallus gallus | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 | | + +|| ++|||+ +| ||+ ||||#||| | +||||||++|||||||++ ||| Sbjct 452 EHVGHSGVHLQRDFFLSNASRARSEQFINL#REVSTRLRLPPGEYIVVPSTFEPNREGDF 510 |
| Danio rerio | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 | ||+ +|| ++|||| + ||+ ||||#||| +||+|| |||++|||||||+| || Sbjct 441 EFLGQSGVHLKRDFFLTHASSARSELFINL#REVSSRFRLPAGEYIIVPSTFEPNKEADF 499 |
| Canis familiaris | Query 447 HLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDFC 498 ||+|+|| ++ || |||||#||| +|||||||||+|+|||||||+ ||| Sbjct 259 HLNKDFFRYHASQARSKTFINL#REVSDRFKLPPGEYILIPSTFEPHQEADFC 310 |
| Rattus norvegicus | Query 439 ELTGQTNIHLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 || || +|| ++||| +| +|+ ||||#||| || +||||||++|||||||+| ||| Sbjct 451 ELAGQP-VHLKRDFFLANASRAQSEHFINL#REVSNRIRLPPGEYIVVPSTFEPNKEGDF 508 |
| Rattus sp. | Query 455 TTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPH 492 ||||||||||||||#|||||||||||||||||||||||| Query 455 TTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPH 492 |
| Macaca fascicularis | Query 447 HLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 || |+||| +| || |+||+#||| ||+||| |||+||||+|||+ |+| Sbjct 519 HLQKDFFLYNASRARSKTYINM#REVSQRFRLPPSEYVIVPSTYEPHQEGEF 569 |
| Oryctolagus cuniculus | Query 447 HLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 || | ||| ++ || |+||+#||| ||+||| |||+||||+|||+ |+| Sbjct 451 HLQKEFFLYNASKARSKTYINM#REVSQRFRLPPSEYVIVPSTYEPHQEGEF 501 |
| Oncorhynchus mykiss | Query 447 HLSKNFFLTTRARERSDTFINL#REVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 || |+|| ++ || |+||+#||+ || |||| |+|||+||+|| || Sbjct 417 HLGKDFFRYNGSKARSKTYINV#REISERFTLPPGNYLLVPTTFQPHDEADF 467 |
[Site 37] ERSDTFINLR469-EVLNRFKLPP
Arg469  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu460 | Arg461 | Ser462 | Asp463 | Thr464 | Phe465 | Ile466 | Asn467 | Leu468 | Arg469 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu470 | Val471 | Leu472 | Asn473 | Arg474 | Phe475 | Lys476 | Leu477 | Pro478 | Pro479 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LTGQTNIHLSKNFFLTTRARERSDTFINLREVLNRFKLPPGEYVLVPSTFEPHKNGDFCI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 134.00 | 15 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 134.00 | 11 | calpain 2 |
| 3 | Mus musculus | 131.00 | 20 | unnamed protein product |
| 4 | Homo sapiens | 126.00 | 12 | AF261089_1 calpain large polypeptide L2 |
| 5 | Macaca mulatta | 126.00 | 2 | PREDICTED: calpain 2, large subunit |
| 6 | Homo sapiens | 125.00 | 2 | calpain 2, large subunit |
| 7 | Gallus gallus | 108.00 | 3 | calpain 2, (m/II) large subunit |
| 8 | Xenopus tropicalis | 102.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Danio rerio | 101.00 | 5 | calpain 2, (m/II) large subunit a |
| 10 | Tetraodon nigroviridis | 100.00 | 7 | unnamed protein product |
| 11 | Oncorhynchus mykiss | 98.60 | 1 | m-calpain |
| 12 | Stizostedion vitreum vitreum | 97.40 | 1 | putative calpain-like protein |
| 13 | Coturnix coturnix | 91.30 | 1 | quail calpain |
| 14 | Canis familiaris | 90.10 | 22 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 15 | Bos taurus | 89.70 | 5 | calpain 1, (mu/I) large subunit |
| 16 | Pongo pygmaeus | 89.70 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 17 | synthetic construct | 89.70 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 18 | Ovis aries | 87.00 | 3 | calpain 1 |
| 19 | Sus scrofa | 86.30 | 4 | calpain 1, large subunit |
| 20 | Xenopus laevis | 85.10 | 3 | hypothetical protein LOC398772 |
| 21 | Canis familiaris | 85.10 | 1 | PREDICTED: similar to Calpain-9 (Digestive tract- |
| 22 | Gallus gallus | 84.70 | 1 | mu-calpain large subunit |
| 23 | Xenopus laevis | 83.20 | 1 | mu/m-calpain large subunit |
| 24 | Danio rerio | 82.80 | 1 | calpain 1, large subunit |
| 25 | Rattus norvegicus | 82.40 | 1 | calpain 1, large subunit |
| 26 | Rattus sp. | 79.70 | 2 | calpain II |
| 27 | Macaca fascicularis | 78.60 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 28 | Oryctolagus cuniculus | 77.00 | 1 | calpain 3, (p94) |
| 29 | Oncorhynchus mykiss | 72.00 | 1 | gill-specific calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 |
| Rattus norvegicus | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 |
| Mus musculus | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 ||||||||| ||||||||||||||||||||#||||||||||||||||||||||||+||||| Sbjct 440 LTGQTNIHLGKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKDGDFCI 499 |
| Homo sapiens | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 |+|||||||||||||| |||||||||||||#|||||||||||||+||||||||+|+||||| Sbjct 440 LSGQTNIHLSKNFFLTNRARERSDTFINLR#EVLNRFKLPPGEYILVPSTFEPNKDGDFCI 499 |
| Macaca mulatta | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 |+|||||||||||||| |||||||||||||#|||||||||||||+||||||||+|+||||| Sbjct 440 LSGQTNIHLSKNFFLTNRARERSDTFINLR#EVLNRFKLPPGEYILVPSTFEPNKDGDFCI 499 |
| Homo sapiens | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 |+|||||||||||||| |||||||||||||#|||||||||||||+||||||||+|+||||| Sbjct 440 LSGQTNIHLSKNFFLTNRARERSDTFINLR#EVLNRFKLPPGEYILVPSTFEPNKDGDFCI 499 |
| Gallus gallus | Query 441 TGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 +|||||||||||||| +|||+|+||||||#||||||||| |||++|||||||+ |||||+ Sbjct 441 SGQTNIHLSKNFFLTNKAREKSNTFINLR#EVLNRFKLPAGEYIIVPSTFEPNLNGDFCL 499 |
| Xenopus tropicalis | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 |||| +| ||+++ | + ||+|||+||||#||||||+|||||||++|||||||| ||||+ Sbjct 440 LTGQKDIRLSRDYILRNKIREKSDTYINLR#EVLNRFRLPPGEYVILPSTFEPHKVGDFCL 499 |
| Danio rerio | Query 442 GQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 || +|| ||+||| + ||+||||||#|| |||||||||++|||||+||||||||+ Sbjct 440 GQKEVHLDKNYFLTHAQKARSETFINLR#EVSTRFKLPPGEYLIVPSTFDPHKNGDFCV 497 |
| Tetraodon nigroviridis | Query 442 GQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 || +|| |||||+ + ||+||||||#|| +|||||||||+++||||||| |||||| Sbjct 439 GQREVHLDKNFFLSHAQKARSETFINLR#EVCSRFKLPPGEYLIIPSTFEPHLNGDFCI 496 |
| Oncorhynchus mykiss | Query 442 GQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 || +|| ||+||| + ||+||+|||#|| |||+|||||++|||||||| |||||| Sbjct 440 GQREVHLDKNYFLTHAQKARSETFVNLR#EVSTRFKMPPGEYLIVPSTFEPHLNGDFCI 497 |
| Stizostedion vitreum vitreum | Query 443 QTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 | |+||+||+|| + ||+||||||#|| +|| ||||||++||||||||||||||+ Sbjct 444 QRNVHLNKNYFLRHASAARSETFINLR#EVCSRFCLPPGEYLIVPSTFEPHKNGDFCV 500 |
| Coturnix coturnix | Query 442 GQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 | |+|| |+||| ++| ||+||||||#|| |+ +||||||++||||||||| || + Sbjct 445 GSQNVHLKKDFFLRNQSRARSETFINLR#EVSNQIRLPPGEYIVVPSTFEPHKEADFIL 502 |
| Canis familiaris | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 | || +|| ++||| +| ||+ |||||#|| ||+|||||||+|||||||+| ||| + Sbjct 450 LVGQPAVHLKRDFFLANSSRARSEQFINLR#EVSTRFRLPPGEYVVVPSTFEPNKEGDFVL 509 |
| Bos taurus | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 | || +|| ++|||+ +| ||+ |||||#|| ||+|||||||+|||||||+| ||| + Sbjct 454 LMGQPAVHLKRDFFLSNASRARSEQFINLR#EVSTRFRLPPGEYVVVPSTFEPNKEGDFVL 513 |
| Pongo pygmaeus | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 | || +|| ++||| +| ||+ |||||#|| ||+|||||||+|||||||+| ||| + Sbjct 452 LAGQPAVHLKRDFFLANASRARSEQFINLR#EVSTRFRLPPGEYVVVPSTFEPNKEGDFVL 511 |
| synthetic construct | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 | || +|| ++||| +| ||+ |||||#|| ||+|||||||+|||||||+| ||| + Sbjct 452 LVGQPAVHLKRDFFLANASRARSEQFINLR#EVSTRFRLPPGEYVVVPSTFEPNKEGDFVL 511 |
| Ovis aries | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 | || +|| ++||| +| ||+ |||||#|| ||+|||||||+|||||||+| ||| + Sbjct 66 LVGQPAVHLKRDFFLANASRARSEQFINLR#EVSTRFRLPPGEYVVVPSTFEPNKEGDFVL 125 |
| Sus scrofa | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 | || +|| ++||| +| ||+ |||||#|| ||+|||||||+|||||||+| ||| + Sbjct 453 LVGQP-VHLKRDFFLANASRARSEQFINLR#EVSTRFRLPPGEYVVVPSTFEPNKEGDFVL 511 |
| Xenopus laevis | Query 442 GQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 | |+|| |+||| ++ ||+||||||#|| + +||||||++||||||||| || | Sbjct 445 GCQNVHLKKDFFLRHQSCARSETFINLR#EVSTQIRLPPGEYIIVPSTFEPHKEADFII 502 |
| Canis familiaris | Query 447 HLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 ||+|+|| ++ || ||||||#|| +|||||||||+|+|||||||+ |||+ Sbjct 259 HLNKDFFRYHASQARSKTFINLR#EVSDRFKLPPGEYILIPSTFEPHQEADFCL 311 |
| Gallus gallus | Query 442 GQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 | + +|| ++|||+ +| ||+ |||||#|| | +||||||++|||||||++ ||| + Sbjct 455 GHSGVHLQRDFFLSNASRARSEQFINLR#EVSTRLRLPPGEYIVVPSTFEPNREGDFVL 512 |
| Xenopus laevis | Query 442 GQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDF 497 | |+|| |+||| ++ ||+||||||#|| + ||||||++||||||||| || Sbjct 445 GCQNVHLKKDFFLRHQSCARSETFINLR#EVSTKIHLPPGEYIIVPSTFEPHKEADF 500 |
| Danio rerio | Query 442 GQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 ||+ +|| ++|||| + ||+ |||||#|| +||+|| |||++|||||||+| || + Sbjct 444 GQSGVHLKRDFFLTHASSARSELFINLR#EVSSRFRLPAGEYIIVPSTFEPNKEADFVL 501 |
| Rattus norvegicus | Query 440 LTGQTNIHLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 | || +|| ++||| +| +|+ |||||#|| || +||||||++|||||||+| ||| + Sbjct 452 LAGQP-VHLKRDFFLANASRAQSEHFINLR#EVSNRIRLPPGEYIVVPSTFEPNKEGDFLL 510 |
| Rattus sp. | Query 455 TTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPH 492 |||||||||||||||#||||||||||||||||||||||| Query 455 TTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPH 492 |
| Macaca fascicularis | Query 447 HLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 || |+||| +| || |+||+|#|| ||+||| |||+||||+|||+ |+| + Sbjct 519 HLQKDFFLYNASRARSKTYINMR#EVSQRFRLPPSEYVIVPSTYEPHQEGEFIL 571 |
| Oryctolagus cuniculus | Query 447 HLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 || | ||| ++ || |+||+|#|| ||+||| |||+||||+|||+ |+| + Sbjct 451 HLQKEFFLYNASKARSKTYINMR#EVSQRFRLPPSEYVIVPSTYEPHQEGEFIL 503 |
| Oncorhynchus mykiss | Query 447 HLSKNFFLTTRARERSDTFINLR#EVLNRFKLPPGEYVLVPSTFEPHKNGDFCI 499 || |+|| ++ || |+||+|#|+ || |||| |+|||+||+|| || | Sbjct 417 HLGKDFFRYNGSKARSKTYINVR#EISERFTLPPGNYLLVPTTFQPHDEADFII 469 |
[Site 38] KNGDFCIRVF502-SEKKADYQTV
Phe502  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys493 | Asn494 | Gly495 | Asp496 | Phe497 | Cys498 | Ile499 | Arg500 | Val501 | Phe502 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser503 | Glu504 | Lys505 | Lys506 | Ala507 | Asp508 | Tyr509 | Gln510 | Thr511 | Val512 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| NRFKLPPGEYVLVPSTFEPHKNGDFCIRVFSEKKADYQTVDDEIEANIEEIEANEEDIGD |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 127.00 | 15 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 127.00 | 10 | calpain 2 |
| 3 | Mus musculus | 118.00 | 19 | calpain 2 |
| 4 | Homo sapiens | 109.00 | 14 | unnamed protein product |
| 5 | Homo sapiens | 107.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 107.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Gallus gallus | 94.40 | 3 | calpain 2, (m/II) large subunit |
| 8 | Danio rerio | 92.00 | 5 | calpain 2, (m/II) large subunit a |
| 9 | Tetraodon nigroviridis | 89.40 | 7 | unnamed protein product |
| 10 | Stizostedion vitreum vitreum | 87.80 | 1 | putative calpain-like protein |
| 11 | Xenopus tropicalis | 84.00 | 4 | calpain 2, (m/II) large subunit |
| 12 | Oncorhynchus mykiss | 80.10 | 1 | m-calpain |
| 13 | Rattus norvegicus | 79.70 | 1 | calpain 1, large subunit |
| 14 | Bos taurus | 77.40 | 5 | calpain 1, (mu/I) large subunit |
| 15 | Gallus gallus | 76.60 | 1 | mu-calpain large subunit |
| 16 | Canis familiaris | 75.90 | 21 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 17 | Xenopus laevis | 75.90 | 3 | hypothetical protein LOC398772 |
| 18 | Sus scrofa | 75.10 | 5 | calpain 1, large subunit |
| 19 | Canis familiaris | 75.10 | 2 | PREDICTED: similar to Calpain-9 (Digestive tract- |
| 20 | Pongo pygmaeus | 75.10 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Coturnix coturnix | 75.10 | 1 | quail calpain |
| 22 | Ovis aries | 74.70 | 3 | calpain 1 |
| 23 | Xenopus laevis | 74.70 | 1 | mu/m-calpain large subunit |
| 24 | synthetic construct | 74.30 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 25 | Danio rerio | 69.30 | 1 | calpain 1, large subunit |
| 26 | Oryctolagus cuniculus | 61.60 | 1 | calpain 3, (p94) |
| 27 | Drosophila melanogaster | 60.50 | 1 | calpain |
| 28 | Drosophila melanogaster | 60.50 | 1 | Calpain-B CG8107-PA |
| 29 | Drosophila pseudoobscura | 59.30 | 1 | GA20829-PA |
| 30 | Macaca fascicularis | 59.30 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 31 | Oncorhynchus mykiss | 59.30 | 1 | gill-specific calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDIGD 532 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDIGD 532 |
| Rattus norvegicus | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDIGD 532 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDIGD 532 |
| Mus musculus | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDIGD 532 |||||||||||||||||||||+||||||||#|||||||| ||||||||||||+|||||| | Sbjct 473 NRFKLPPGEYVLVPSTFEPHKDGDFCIRVF#SEKKADYQAVDDEIEANIEEIDANEEDIDD 532 |
| Homo sapiens | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDIGD 532 ||||||||||+||||||||+|+||||||||#|||||||| ||||||||+|| + +|+|| | Sbjct 56 NRFKLPPGEYILVPSTFEPNKDGDFCIRVF#SEKKADYQAVDDEIEANLEEFDISEDDIDD 115 |
| Homo sapiens | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDIGD 532 ||||||||||+||||||||+|+||||||||#|||||||| ||||||||+|| + +|+|| | Sbjct 473 NRFKLPPGEYILVPSTFEPNKDGDFCIRVF#SEKKADYQAVDDEIEANLEEFDISEDDIDD 532 |
| Macaca mulatta | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDIGD 532 ||||||||||+||||||||+|+||||||||#|||||||| ||||||||+|| + +|+|| | Sbjct 473 NRFKLPPGEYILVPSTFEPNKDGDFCIRVF#SEKKADYQAVDDEIEANLEEFDISEDDIDD 532 |
| Gallus gallus | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDI 530 |||||| |||++|||||||+ |||||+|||#||| |+ +||||||| || | +|+|| Sbjct 473 NRFKLPAGEYIIVPSTFEPNLNGDFCLRVF#SEKNANSTVIDDEIEANFEETEIDEDDI 530 |
| Danio rerio | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDI 530 |||||||||++|||||+||||||||+|||#|||+++ | || ||||+++ +||++ Sbjct 472 RFKLPPGEYLIVPSTFDPHKNGDFCVRVF#SEKQSEMQQCDDPIEANVDDEMVSEEEV 528 |
| Tetraodon nigroviridis | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDI 530 +|||||||||+++||||||| |||||||||#|||+|| | || + ||+|+ ++|+ Sbjct 470 SRFKLPPGEYLIIPSTFEPHLNGDFCIRVF#SEKQADTQPCDDPVSANLEDETVADQDV 527 |
| Stizostedion vitreum vitreum | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIE 521 +|| ||||||++||||||||||||||+|||#|||+||+| +|| +|+ +| Sbjct 474 SRFCLPPGEYLIVPSTFEPHKNGDFCVRVF#SEKQADFQEMDDPVESRVE 522 |
| Xenopus tropicalis | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDI 530 |||+|||||||++|||||||| ||||+|||#||| |+ + ||||+|+|+| + ||+ | Sbjct 473 NRFRLPPGEYVILPSTFEPHKVGDFCLRVF#SEKNAESKVPDDEIKADIDE-DVNEDAI 529 |
| Oncorhynchus mykiss | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEEDI 530 |||+|||||++|||||||| |||||||||#|||+ + + || ++|++++ ++ ++ Sbjct 472 RFKMPPGEYLIVPSTFEPHLNGDFCIRVF#SEKQTETRPCDDPVKADLDDETVSDAEV 528 |
| Rattus norvegicus | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANI-EEIEANEEDIG 531 || +||||||++|||||||+| ||| +| |#||||| | +||+|+||+ +| +||+| Sbjct 484 NRIRLPPGEYIVVPSTFEPNKEGDFLLRFF#SEKKAGTQELDDQIQANLPDEKVLSEEEID 543 Query 532 D 532 | Query 532 D 532 |
| Bos taurus | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANI--EEIEANEE 528 ||+|||||||+|||||||+| ||| +| |#||| | | +||+++||+ |++ + || Sbjct 488 RFRLPPGEYVVVPSTFEPNKEGDFVLRFF#SEKSAGTQELDDQVQANLPDEQVLSEEE 544 |
| Gallus gallus | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEE 522 | +||||||++|||||||++ ||| +|||#|||+|| + +||+||| + + Sbjct 487 RLRLPPGEYIVVPSTFEPNREGDFVLRVF#SEKRADTEEIDDKIEAKLPD 535 |
| Canis familiaris | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANI--EEIEANEE 528 ||+|||||||+|||||||+| ||| +| |#||| | + +||+|+||+ |++ + || Sbjct 484 RFRLPPGEYVVVPSTFEPNKEGDFVLRFF#SEKSAGTEELDDQIQANLPDEQVLSEEE 540 |
| Xenopus laevis | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANI-EEIEANEEDIGD 532 + +||||||++||||||||| || ||+|#+|| +| | +|+|+ |++ +| | |+|+ | Sbjct 477 QIRLPPGEYIIVPSTFEPHKEADFIIRIF#TEKHSDTQELDEEVSADLPDEEELTEDDVDD 536 |
| Sus scrofa | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANI--EEIEANEE 528 ||+|||||||+|||||||+| ||| +| |#||||| | +||+++| + |++ + || Sbjct 486 RFRLPPGEYVVVPSTFEPNKEGDFVLRFF#SEKKAGTQELDDQVQAILPDEQVLSEEE 542 |
| Canis familiaris | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEI 523 +|||||||||+|+|||||||+ |||+|+|#||||| + +| +++ ++ |+ Sbjct 285 DRFKLPPGEYILIPSTFEPHQEADFCLRIF#SEKKAITRDMDGDVDIDLPEL 335 |
| Pongo pygmaeus | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANI--EEIEANEE 528 ||+|||||||+|||||||+| ||| +| |#||| | +||+|+||+ |++ + || Sbjct 486 RFRLPPGEYVVVPSTFEPNKEGDFVLRFF#SEKSAGTAELDDQIQANLPDEQVLSEEE 542 |
| Coturnix coturnix | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANI-EEIEANEEDIG 531 |+ +||||||++||||||||| || +|||#+||++| +|+|| |++ +| | ++|| Sbjct 476 NQIRLPPGEYIVVPSTFEPHKEADFILRVF#TEKQSDTAELDEEISADLADEEEITDDDIE 535 Query 532 D 532 | Query 532 D 532 |
| Ovis aries | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANI--EEIEANEE 528 ||+|||||||+|||||||+| ||| +| |#||| | | +||+++||+ |++ + || Sbjct 99 TRFRLPPGEYVVVPSTFEPNKEGDFVLRFF#SEKSAGTQELDDQVQANLPDEQVLSEEE 156 |
| Xenopus laevis | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANI-EEIEANEEDIGD 532 + ||||||++||||||||| || +|+|#+|| +| | +|+|+ |++ || | |+|+ | Sbjct 477 KIHLPPGEYIIVPSTFEPHKEADFIMRIF#TEKHSDIQELDEEVTADLPEEEELTEDDVDD 536 |
| synthetic construct | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANI--EEIEANEE 528 ||+|||||||+|||||||+| ||| +| |#||| | +||+|+||+ |++ + || Sbjct 486 RFRLPPGEYVVVPSTFEPNKEGDFVLRFF#SEKSAGTVELDDQIQANLPDEQVLSEEE 542 |
| Danio rerio | Query 473 NRFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEEIEANEE 528 +||+|| |||++|||||||+| || +|||#||| |+ + +||++ | | | + +| Sbjct 475 SRFRLPAGEYIIVPSTFEPNKEADFVLRVF#SEKPANSEEMDDKVMAEIPEEQRLDE 530 |
| Oryctolagus cuniculus | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEAN 519 ||+||| |||+||||+|||+ |+| +|||#|||+ + |++ | + Sbjct 478 RFRLPPSEYVIVPSTYEPHQEGEFILRVF#SEKRNLSEEVENTISVD 523 |
| Drosophila melanogaster | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEI 516 ||||||| |++|||||+|++ |+| ||||#|| + + + |||+ Sbjct 688 RFKLPPGHYLIVPSTFDPNEEGEFIIRVF#SETRNNMEENDDEV 730 |
| Drosophila melanogaster | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEI 516 ||||||| |++|||||+|++ |+| ||||#|| + + + |||+ Sbjct 688 RFKLPPGHYLIVPSTFDPNEEGEFIIRVF#SETRNNMEENDDEV 730 |
| Drosophila pseudoobscura | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEI 516 ||||||| |++|||||+|++ |+| ||||#|| + + |||+ Sbjct 694 RFKLPPGHYLIVPSTFDPNEEGEFIIRVF#SETMNNMEENDDEV 736 |
| Macaca fascicularis | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEAN 519 ||+||| |||+||||+|||+ |+| +|||#|||+ + |++ | + Sbjct 546 RFRLPPSEYVIVPSTYEPHQEGEFILRVF#SEKRNLSEEVENTISVD 591 |
| Oncorhynchus mykiss | Query 474 RFKLPPGEYVLVPSTFEPHKNGDFCIRVF#SEKKADYQTVDDEIEANIEE 522 || |||| |+|||+||+|| || ||+|#||| | + + ++|++ + Sbjct 444 RFTLPPGNYLLVPTTFQPHDEADFIIRIF#SEKAAGTMEMGNTVDADLPD 492 |
[Site 39] RVFSEKKADY509-QTVDDEIEAN
Tyr509  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg500 | Val501 | Phe502 | Ser503 | Glu504 | Lys505 | Lys506 | Ala507 | Asp508 | Tyr509 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln510 | Thr511 | Val512 | Asp513 | Asp514 | Glu515 | Ile516 | Glu517 | Ala518 | Asn519 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GEYVLVPSTFEPHKNGDFCIRVFSEKKADYQTVDDEIEANIEEIEANEEDIGDGFRRLFA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 125.00 | 15 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 125.00 | 10 | calpain 2 |
| 3 | Mus musculus | 115.00 | 18 | m-calpain |
| 4 | Homo sapiens | 107.00 | 14 | unnamed protein product |
| 5 | Macaca mulatta | 105.00 | 2 | PREDICTED: calpain 2, large subunit |
| 6 | Homo sapiens | 105.00 | 2 | calpain 2, large subunit |
| 7 | Danio rerio | 86.30 | 5 | calpain 2, (m/II) large subunit a |
| 8 | Tetraodon nigroviridis | 84.70 | 7 | unnamed protein product |
| 9 | Gallus gallus | 84.00 | 3 | calpain 2, (m/II) large subunit |
| 10 | Stizostedion vitreum vitreum | 79.70 | 1 | putative calpain-like protein |
| 11 | Xenopus tropicalis | 76.60 | 4 | calpain 2, (m/II) large subunit |
| 12 | Oncorhynchus mykiss | 75.10 | 1 | m-calpain |
| 13 | Rattus norvegicus | 73.60 | 1 | calpain 1, large subunit |
| 14 | Xenopus laevis | 72.80 | 3 | hypothetical protein LOC398772 |
| 15 | Xenopus laevis | 72.40 | 1 | mu/m-calpain large subunit |
| 16 | Coturnix coturnix | 72.00 | 1 | quail calpain |
| 17 | Gallus gallus | 71.60 | 1 | mu-calpain large subunit |
| 18 | Sus scrofa | 71.20 | 5 | calpain II |
| 19 | Bos taurus | 70.50 | 5 | calpain 1, (mu/I) large subunit |
| 20 | Canis familiaris | 68.90 | 22 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 21 | Ovis aries | 67.80 | 3 | calpain 1 |
| 22 | Pongo pygmaeus | 67.80 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 23 | synthetic construct | 67.40 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 24 | Danio rerio | 66.20 | 1 | calpain 1, large subunit |
| 25 | Canis familiaris | 61.20 | 2 | PREDICTED: similar to Calpain-9 (Digestive tract- |
| 26 | Oncorhynchus mykiss | 52.40 | 1 | gill-specific calpain |
| 27 | Drosophila melanogaster | 48.50 | 1 | calpain |
| 28 | Oryctolagus cuniculus | 48.10 | 1 | calpain 3, (p94) |
| 29 | Macaca fascicularis | 48.10 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 30 | Drosophila melanogaster | 48.10 | 1 | Calpain-B CG8107-PA |
| 31 | Drosophila pseudoobscura | 47.00 | 1 | GA20829-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLFA 539 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLFA 539 |
| Rattus norvegicus | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLFA 539 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLFA 539 |
| Mus musculus | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLF 538 ||||||||||||||+|||||||||||||||#| ||||||||||||+|||||| ||||||| Sbjct 480 GEYVLVPSTFEPHKDGDFCIRVFSEKKADY#QAVDDEIEANIEEIDANEEDIDDGFRRLF 538 |
| Homo sapiens | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLFA 539 |||+||||||||+|+|||||||||||||||#| ||||||||+|| + +|+|| |||||||| Sbjct 63 GEYILVPSTFEPNKDGDFCIRVFSEKKADY#QAVDDEIEANLEEFDISEDDIDDGFRRLFA 122 |
| Macaca mulatta | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLFA 539 |||+||||||||+|+|||||||||||||||#| ||||||||+|| + +|+|| |||||||| Sbjct 480 GEYILVPSTFEPNKDGDFCIRVFSEKKADY#QAVDDEIEANLEEFDISEDDIDDGFRRLFA 539 |
| Homo sapiens | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLFA 539 |||+||||||||+|+|||||||||||||||#| ||||||||+|| + +|+|| |||||||| Sbjct 480 GEYILVPSTFEPNKDGDFCIRVFSEKKADY#QAVDDEIEANLEEFDISEDDIDDGFRRLFA 539 |
| Danio rerio | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLF 538 |||++|||||+||||||||+||||||+++ #| || ||||+++ +||++ ||| || Sbjct 478 GEYLIVPSTFDPHKNGDFCVRVFSEKQSEM#QQCDDPIEANVDDEMVSEEEVDSGFRGLF 536 |
| Tetraodon nigroviridis | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLFA 539 |||+++||||||| ||||||||||||+|| #| || + ||+|+ ++|+ ||| ||+ Sbjct 477 GEYLIIPSTFEPHLNGDFCIRVFSEKQADT#QPCDDPVSANLEDETVADQDVDPGFRGLFS 536 |
| Gallus gallus | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLFA 539 |||++|||||||+ |||||+|||||| |+ # +||||||| || | +|+|| |++|| Sbjct 480 GEYIIVPSTFEPNLNGDFCLRVFSEKNANS#TVIDDEIEANFEETEIDEDDIEPSFKKLFG 539 |
| Stizostedion vitreum vitreum | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLF 538 |||++||||||||||||||+||||||+||+#| +|| +|+ +| || +|+|+ | ||+|| Sbjct 481 GEYLIVPSTFEPHKNGDFCVRVFSEKQADF#QEMDDPVESRVETIEIDEDDVDDQFRKLF 539 |
| Xenopus tropicalis | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLF 538 ||||++|||||||| ||||+|||||| |+ #+ ||||+|+|+| + ||+ | |+|+| Sbjct 480 GEYVILPSTFEPHKVGDFCLRVFSEKNAES#KVPDDEIKADIDE-DVNEDAIDPNFKRMF 537 |
| Oncorhynchus mykiss | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLF 538 |||++|||||||| ||||||||||||+ + #+ || ++|++++ ++ ++ ||| || Sbjct 478 GEYLIVPSTFEPHLNGDFCIRVFSEKQTET#RPCDDPVKADLDDETVSDAEVDAGFRGLF 536 |
| Rattus norvegicus | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANI-EEIEANEEDIGDGFRRLF 538 |||++|||||||+| ||| +| |||||| #| +||+|+||+ +| +||+| | |+ || Sbjct 491 GEYIVVPSTFEPNKEGDFLLRFFSEKKAGT#QELDDQIQANLPDEKVLSEEEIDDNFKTLF 550 Query 539 A 539 + Sbjct 551 S 551 |
| Xenopus laevis | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANI-EEIEANEEDIGDGFRRLF 538 |||++||||||||| || ||+|+|| +| #| +|+|+ |++ +| | |+|+ | |+ +| Sbjct 483 GEYIIVPSTFEPHKEADFIIRIFTEKHSDT#QELDEEVSADLPDEEELTEDDVDDSFKAMF 542 |
| Xenopus laevis | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANI-EEIEANEEDIGDGFRRLF 538 |||++||||||||| || +|+|+|| +| #| +|+|+ |++ || | |+|+ | |+ +| Sbjct 483 GEYIIVPSTFEPHKEADFIMRIFTEKHSDI#QELDEEVTADLPEEEELTEDDVDDSFKAMF 542 |
| Coturnix coturnix | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANI-EEIEANEEDIGDGFRRLF 538 |||++||||||||| || +|||+||++| # +|+|| |++ +| | ++|| |||+ +| Sbjct 483 GEYIVVPSTFEPHKEADFILRVFTEKQSDT#AELDEEISADLADEEEITDDDIEDGFKNMF 542 |
| Gallus gallus | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANI-EEIEANEEDIGDGFRRLF 538 |||++|||||||++ ||| +||||||+|| #+ +||+||| + +| +| +| + |++|| Sbjct 493 GEYIVVPSTFEPNREGDFVLRVFSEKRADT#EEIDDKIEAKLPDEKVLSEGEIDENFKQLF 552 |
| Sus scrofa | Query 498 CIRVFSEKKADY#QTVDDEIEANIEEIEANEEDIGDGFRRLFA 539 ||||||||||||#| |||||||++|| +|+|+|| |||||||| Sbjct 1 CIRVFSEKKADY#QVVDDEIEADLEENDASEDDIDDGFRRLFA 42 |
| Bos taurus | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANI-EEIEANEEDIGDGFRRLF 538 ||||+|||||||+| ||| +| |||| | #| +||+++||+ +| +||+| + |+ || Sbjct 494 GEYVVVPSTFEPNKEGDFVLRFFSEKSAGT#QELDDQVQANLPDEQVLSEEEIDENFKSLF 553 |
| Canis familiaris | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANI-EEIEANEEDIGDGFRRLF 538 ||||+|||||||+| ||| +| |||| | #+ +||+|+||+ +| +||+| + |+ || Sbjct 490 GEYVVVPSTFEPNKEGDFVLRFFSEKSAGT#EELDDQIQANLPDEQVLSEEEIDENFKALF 549 |
| Ovis aries | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANI-EEIEANEEDIGDGFRRLF 538 ||||+|||||||+| ||| +| |||| | #| +||+++||+ +| +||+| + |+ || Sbjct 106 GEYVVVPSTFEPNKEGDFVLRFFSEKSAGT#QELDDQVQANLPDEQVLSEEEIDENFKSLF 165 |
| Pongo pygmaeus | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANI-EEIEANEEDIGDGFRRLF 538 ||||+|||||||+| ||| +| |||| | # +||+|+||+ +| +||+| + |+ || Sbjct 492 GEYVVVPSTFEPNKEGDFVLRFFSEKSAGT#AELDDQIQANLPDEQVLSEEEIDENFKALF 551 |
| synthetic construct | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANI-EEIEANEEDIGDGFRRLF 538 ||||+|||||||+| ||| +| |||| | # +||+|+||+ +| +||+| + |+ || Sbjct 492 GEYVVVPSTFEPNKEGDFVLRFFSEKSAGT#VELDDQIQANLPDEQVLSEEEIDENFKALF 551 |
| Danio rerio | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANI-EEIEANEEDIGDGFRRLF 538 |||++|||||||+| || +|||||| |+ #+ +||++ | | || +| | ||+ || Sbjct 482 GEYIIVPSTFEPNKEADFVLRVFSEKPANS#EEMDDKVMAEIPEEQRLDESQIDAGFKSLF 541 |
| Canis familiaris | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEEI-------EANEEDIGD 532 |||+|+|||||||+ |||+|+|||||| #+ +| +++ ++ |+ + ||++ Sbjct 292 GEYILIPSTFEPHQEADFCLRIFSEKKAIT#RDMDGDVDIDLPELPKPTPAGQETEEEL-- 349 Query 533 GFRRLF 538 || || Sbjct 350 QFRALF 355 |
| Oncorhynchus mykiss | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEANIEE-----IEANEEDIGDGF 534 | |+|||+||+|| || ||+|||| | # + + ++|++ + + | | | Sbjct 450 GNYLLVPTTFQPHDEADFIIRIFSEKAAGT#MEMGNTVDADLPDPPMPSLPEEETDEEKGL 509 Query 535 RRLF 538 ||+| Sbjct 510 RRMF 513 |
| Drosophila melanogaster | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEI 516 | |++|||||+|++ |+| |||||| + + #+ |||+ Sbjct 694 GHYLIVPSTFDPNEEGEFIIRVFSETRNNM#EENDDEV 730 |
| Oryctolagus cuniculus | Query 481 EYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEAN 519 |||+||||+|||+ |+| +||||||+ #+ |++ | + Sbjct 485 EYVIVPSTYEPHQEGEFILRVFSEKRNLS#EEVENTISVD 523 |
| Macaca fascicularis | Query 481 EYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEIEAN 519 |||+||||+|||+ |+| +||||||+ #+ |++ | + Sbjct 553 EYVIVPSTYEPHQEGEFILRVFSEKRNLS#EEVENTISVD 591 |
| Drosophila melanogaster | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEI 516 | |++|||||+|++ |+| |||||| + + #+ |||+ Sbjct 694 GHYLIVPSTFDPNEEGEFIIRVFSETRNNM#EENDDEV 730 |
| Drosophila pseudoobscura | Query 480 GEYVLVPSTFEPHKNGDFCIRVFSEKKADY#QTVDDEI 516 | |++|||||+|++ |+| |||||| + #+ |||+ Sbjct 700 GHYLIVPSTFDPNEEGEFIIRVFSETMNNM#EENDDEV 736 |
[Site 40] EKKADYQTVD513-DEIEANIEEI
Asp513  Asp
Asp
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu504 | Lys505 | Lys506 | Ala507 | Asp508 | Tyr509 | Gln510 | Thr511 | Val512 | Asp513 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asp514 | Glu515 | Ile516 | Glu517 | Ala518 | Asn519 | Ile520 | Glu521 | Glu522 | Ile523 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LVPSTFEPHKNGDFCIRVFSEKKADYQTVDDEIEANIEEIEANEEDIGDGFRRLFAQLAG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 125.00 | 14 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 125.00 | 9 | calpain 2 |
| 3 | Mus musculus | 115.00 | 20 | m-calpain large subunit |
| 4 | Homo sapiens | 106.00 | 15 | CAPN2 protein |
| 5 | Homo sapiens | 106.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 106.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Danio rerio | 85.10 | 5 | calpain 2, (m/II) large subunit a |
| 8 | Gallus gallus | 84.70 | 3 | calpain 2, (m/II) large subunit |
| 9 | Tetraodon nigroviridis | 83.20 | 7 | unnamed protein product |
| 10 | Stizostedion vitreum vitreum | 79.70 | 1 | putative calpain-like protein |
| 11 | Sus scrofa | 79.30 | 5 | calpain II |
| 12 | Xenopus tropicalis | 73.60 | 4 | calpain 2, (m/II) large subunit |
| 13 | Oncorhynchus mykiss | 73.60 | 1 | m-calpain |
| 14 | Gallus gallus | 70.50 | 1 | mu-calpain large subunit |
| 15 | Xenopus laevis | 70.10 | 3 | hypothetical protein LOC398772 |
| 16 | Rattus norvegicus | 70.10 | 1 | calpain 1, large subunit |
| 17 | Xenopus laevis | 69.30 | 1 | mu/m-calpain large subunit |
| 18 | Bos taurus | 68.90 | 5 | calpain 1, (mu/I) large subunit |
| 19 | Coturnix coturnix | 68.90 | 1 | quail calpain |
| 20 | Canis familiaris | 67.40 | 21 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 21 | Ovis aries | 66.60 | 3 | calpain 1 |
| 22 | Pongo pygmaeus | 66.60 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 23 | synthetic construct | 66.20 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 24 | Danio rerio | 65.10 | 1 | calpain 1, large subunit |
| 25 | Canis familiaris | 58.20 | 2 | PREDICTED: similar to Calpain-9 (Digestive tract- |
| 26 | Oncorhynchus mykiss | 52.00 | 1 | gill-specific calpain |
| 27 | Oryctolagus cuniculus | 47.40 | 1 | calpain 3, (p94) |
| 28 | Drosophila melanogaster | 44.70 | 1 | calpain |
| 29 | Drosophila melanogaster | 44.70 | 1 | Calpain-B CG8107-PA |
| 30 | Drosophila pseudoobscura | 42.40 | 1 | GA20829-PA |
| 31 | Macaca fascicularis | 42.40 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 |
| Rattus norvegicus | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 |
| Mus musculus | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 ||||||||||+|||||||||||||||| ||#||||||||||+|||||| ||||||| |||| Sbjct 484 LVPSTFEPHKDGDFCIRVFSEKKADYQAVD#DEIEANIEEIDANEEDIDDGFRRLFVQLAG 543 |
| Homo sapiens | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 ||||||||+|+|||||||||||||||| ||#||||||+|| + +|+|| |||||||||||| Sbjct 484 LVPSTFEPNKDGDFCIRVFSEKKADYQAVD#DEIEANLEEFDISEDDIDDGFRRLFAQLAG 543 |
| Homo sapiens | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 ||||||||+|+|||||||||||||||| ||#||||||+|| + +|+|| |||||||||||| Sbjct 484 LVPSTFEPNKDGDFCIRVFSEKKADYQAVD#DEIEANLEEFDISEDDIDDGFRRLFAQLAG 543 |
| Macaca mulatta | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 ||||||||+|+|||||||||||||||| ||#||||||+|| + +|+|| |||||||||||| Sbjct 484 LVPSTFEPNKDGDFCIRVFSEKKADYQAVD#DEIEANLEEFDISEDDIDDGFRRLFAQLAG 543 |
| Danio rerio | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 +|||||+||||||||+||||||+++ | |#| ||||+++ +||++ ||| || +||| Sbjct 482 IVPSTFDPHKNGDFCVRVFSEKQSEMQQCD#DPIEANVDDEMVSEEEVDSGFRGLFTKLAG 541 |
| Gallus gallus | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 +|||||||+ |||||+|||||| |+ +|#|||||| || | +|+|| |++|| |||| Sbjct 484 IVPSTFEPNLNGDFCLRVFSEKNANSTVID#DEIEANFEETEIDEDDIEPSFKKLFGQLAG 543 |
| Tetraodon nigroviridis | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 ++||||||| ||||||||||||+|| | |#| + ||+|+ ++|+ ||| ||++||| Sbjct 481 IIPSTFEPHLNGDFCIRVFSEKQADTQPCD#DPVSANLEDETVADQDVDPGFRGLFSKLAG 540 |
| Stizostedion vitreum vitreum | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 +||||||||||||||+||||||+||+| +|#| +|+ +| || +|+|+ | ||+|| |||| Sbjct 485 IVPSTFEPHKNGDFCVRVFSEKQADFQEMD#DPVESRVETIEIDEDDVDDQFRKLFEQLAG 544 |
| Sus scrofa | Query 498 CIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 ||||||||||||| ||#|||||++|| +|+|+|| |||||||||||| Sbjct 1 CIRVFSEKKADYQVVD#DEIEADLEENDASEDDIDDGFRRLFAQLAG 46 |
| Xenopus tropicalis | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 ++|||||||| ||||+|||||| |+ + |#|||+|+|+| + ||+ | |+|+| ||| Sbjct 484 ILPSTFEPHKVGDFCLRVFSEKNAESKVPD#DEIKADIDE-DVNEDAIDPNFKRMFLALAG 542 |
| Oncorhynchus mykiss | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEIEANEEDIGDGFRRLFAQLAG 543 +|||||||| ||||||||||||+ + + |#| ++|++++ ++ ++ ||| || +||| Sbjct 482 IVPSTFEPHLNGDFCIRVFSEKQTETRPCD#DPVKADLDDETVSDAEVDAGFRGLFTKLAG 541 |
| Gallus gallus | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI-EEIEANEEDIGDGFRRLFAQLA 542 +|||||||++ ||| +||||||+|| + +|#|+||| + +| +| +| + |++|| ||| Sbjct 497 VVPSTFEPNREGDFVLRVFSEKRADTEEID#DKIEAKLPDEKVLSEGEIDENFKQLFRQLA 556 Query 543 G 543 | Query 543 G 543 |
| Xenopus laevis | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI-EEIEANEEDIGDGFRRLFAQLA 542 +||||||||| || ||+|+|| +| | +|#+|+ |++ +| | |+|+ | |+ +| || Sbjct 487 IVPSTFEPHKEADFIIRIFTEKHSDTQELD#EEVSADLPDEEELTEDDVDDSFKAMFQHLA 546 Query 543 G 543 | Query 543 G 543 |
| Rattus norvegicus | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI-EEIEANEEDIGDGFRRLFAQLA 542 +|||||||+| ||| +| |||||| | +|#|+|+||+ +| +||+| | |+ ||++|| Sbjct 495 VVPSTFEPNKEGDFLLRFFSEKKAGTQELD#DQIQANLPDEKVLSEEEIDDNFKTLFSKLA 554 Query 543 G 543 | Query 543 G 543 |
| Xenopus laevis | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI-EEIEANEEDIGDGFRRLFAQLA 542 +||||||||| || +|+|+|| +| | +|#+|+ |++ || | |+|+ | |+ +| ||| Sbjct 487 IVPSTFEPHKEADFIMRIFTEKHSDIQELD#EEVTADLPEEEELTEDDVDDSFKAMFQQLA 546 Query 543 G 543 | Query 543 G 543 |
| Bos taurus | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI-EEIEANEEDIGDGFRRLFAQLA 542 +|||||||+| ||| +| |||| | | +|#|+++||+ +| +||+| + |+ || ||| Sbjct 498 VVPSTFEPNKEGDFVLRFFSEKSAGTQELD#DQVQANLPDEQVLSEEEIDENFKSLFRQLA 557 Query 543 G 543 | Query 543 G 543 |
| Coturnix coturnix | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI-EEIEANEEDIGDGFRRLFAQLA 542 +||||||||| || +|||+||++| +|#+|| |++ +| | ++|| |||+ +| ||| Sbjct 487 VVPSTFEPHKEADFILRVFTEKQSDTAELD#EEISADLADEEEITDDDIEDGFKNMFQQLA 546 Query 543 G 543 | Query 543 G 543 |
| Canis familiaris | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI-EEIEANEEDIGDGFRRLFAQLA 542 +|||||||+| ||| +| |||| | + +|#|+|+||+ +| +||+| + |+ || ||| Sbjct 494 VVPSTFEPNKEGDFVLRFFSEKSAGTEELD#DQIQANLPDEQVLSEEEIDENFKALFRQLA 553 Query 543 G 543 | Query 543 G 543 |
| Ovis aries | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI-EEIEANEEDIGDGFRRLFAQLA 542 +|||||||+| ||| +| |||| | | +|#|+++||+ +| +||+| + |+ || ||| Sbjct 110 VVPSTFEPNKEGDFVLRFFSEKSAGTQELD#DQVQANLPDEQVLSEEEIDENFKSLFRQLA 169 Query 543 G 543 | Query 543 G 543 |
| Pongo pygmaeus | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI-EEIEANEEDIGDGFRRLFAQLA 542 +|||||||+| ||| +| |||| | +|#|+|+||+ +| +||+| + |+ || ||| Sbjct 496 VVPSTFEPNKEGDFVLRFFSEKSAGTAELD#DQIQANLPDEQVLSEEEIDENFKALFRQLA 555 Query 543 G 543 | Query 543 G 543 |
| synthetic construct | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI-EEIEANEEDIGDGFRRLFAQLA 542 +|||||||+| ||| +| |||| | +|#|+|+||+ +| +||+| + |+ || ||| Sbjct 496 VVPSTFEPNKEGDFVLRFFSEKSAGTVELD#DQIQANLPDEQVLSEEEIDENFKALFRQLA 555 Query 543 G 543 | Query 543 G 543 |
| Danio rerio | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI-EEIEANEEDIGDGFRRLFAQLA 542 +|||||||+| || +|||||| |+ + +|#|++ | | || +| | ||+ || ||| Sbjct 486 IVPSTFEPNKEADFVLRVFSEKPANSEEMD#DKVMAEIPEEQRLDESQIDAGFKSLFRQLA 545 Query 543 G 543 | Query 543 G 543 |
| Canis familiaris | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEEI-------EANEEDIGDGFRR 536 |+|||||||+ |||+|+|||||| + +|# +++ ++ |+ + ||++ || Sbjct 296 LIPSTFEPHQEADFCLRIFSEKKAITRDMD#GDVDIDLPELPKPTPAGQETEEEL--QFRA 353 Query 537 LFAQLAG 543 || |+|| Sbjct 354 LFDQIAG 360 |
| Oncorhynchus mykiss | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANIEE-----IEANEEDIGDGFRRLF 538 |||+||+|| || ||+|||| | + #+ ++|++ + + | | | ||+| Sbjct 454 LVPTTFQPHDEADFIIRIFSEKAAGTMEMG#NTVDADLPDPPMPSLPEEETDEEKGLRRMF 513 Query 539 AQLAG 543 ++|| Sbjct 514 EKIAG 518 |
| Oryctolagus cuniculus | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEANI------EEIEANEEDIGDGFRRL 537 +||||+|||+ |+| +||||||+ + |+#+ | + + ||| || + Sbjct 488 IVPSTYEPHQEGEFILRVFSEKRNLSEEVE#NTISVDRPVPLPGNTDQENEEQ--QQFRNI 545 Query 538 FAQLAG 543 | |+|| Sbjct 546 FKQIAG 551 |
| Drosophila melanogaster | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DE---------IEANIEEIEANEEDIGD-- 532 +|||||+|++ |+| |||||| + + + |#|| | ++ ||| Sbjct 698 IVPSTFDPNEEGEFIIRVFSETRNNMEEND#DEVGFGETDDRIAPSLPPPTPKEEDDPQRI 757 Query 533 GFRRLFAQLAG 543 |||| +|| Sbjct 758 ALRRLFDSVAG 768 |
| Drosophila melanogaster | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DE---------IEANIEEIEANEEDIGD-- 532 +|||||+|++ |+| |||||| + + + |#|| | ++ ||| Sbjct 698 IVPSTFDPNEEGEFIIRVFSETRNNMEEND#DEVGFGETDDRIAPSLPPPTPKEEDDPQRI 757 Query 533 GFRRLFAQLAG 543 |||| +|| Sbjct 758 ALRRLFDSVAG 768 |
| Drosophila pseudoobscura | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DE---------IEANIEEIEANEEDIGD-- 532 +|||||+|++ |+| |||||| + + |#|| | + ||| Sbjct 704 IVPSTFDPNEEGEFIIRVFSETMNNMEEND#DEVGFGDTDDRIAPALPPPTPKEEDDPQRI 763 Query 533 GFRRLFAQLAG 543 |||| +|| Sbjct 764 ALRRLFDSVAG 774 |
| Macaca fascicularis | Query 484 LVPSTFEPHKNGDFCIRVFSEKKADYQTVD#DEIEAN 519 +||||+|||+ |+| +||||||+ + |+#+ | + Sbjct 556 IVPSTYEPHQEGEFILRVFSEKRNLSEEVE#NTISVD 591 |
[Site 41] DYQTVDDEIE517-ANIEEIEANE
Glu517  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asp508 | Tyr509 | Gln510 | Thr511 | Val512 | Asp513 | Asp514 | Glu515 | Ile516 | Glu517 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala518 | Asn519 | Ile520 | Glu521 | Glu522 | Ile523 | Glu524 | Ala525 | Asn526 | Glu527 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TFEPHKNGDFCIRVFSEKKADYQTVDDEIEANIEEIEANEEDIGDGFRRLFAQLAGEDAE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 124.00 | 16 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 2 | Rattus norvegicus | 124.00 | 10 | calpain 2 |
| 3 | Mus musculus | 114.00 | 21 | unnamed protein product |
| 4 | Homo sapiens | 104.00 | 15 | unnamed protein product |
| 5 | Homo sapiens | 104.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 104.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Sus scrofa | 85.10 | 5 | calpain II |
| 8 | Gallus gallus | 82.40 | 3 | calpain 2, (m/II) large subunit |
| 9 | Danio rerio | 81.60 | 5 | calpain 2, (m/II) large subunit a |
| 10 | Tetraodon nigroviridis | 79.30 | 7 | unnamed protein product |
| 11 | Stizostedion vitreum vitreum | 74.70 | 1 | putative calpain-like protein |
| 12 | Xenopus tropicalis | 71.20 | 4 | calpain 2, (m/II) large subunit |
| 13 | Oncorhynchus mykiss | 70.50 | 1 | m-calpain |
| 14 | Bos taurus | 67.80 | 5 | calpain 1, (mu/I) large subunit |
| 15 | Rattus norvegicus | 66.60 | 2 | calpain 1, large subunit |
| 16 | Gallus gallus | 66.60 | 1 | mu-calpain large subunit |
| 17 | Canis familiaris | 65.90 | 8 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 18 | Coturnix coturnix | 65.90 | 1 | quail calpain |
| 19 | Pongo pygmaeus | 65.10 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 20 | Xenopus laevis | 65.10 | 1 | mu/m-calpain large subunit |
| 21 | synthetic construct | 64.70 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 22 | Ovis aries | 64.30 | 2 | calpain 1 |
| 23 | Xenopus laevis | 62.80 | 3 | hypothetical protein LOC398772 |
| 24 | Danio rerio | 60.50 | 1 | calpain 1, large subunit |
| 25 | Canis familiaris | 51.60 | 2 | PREDICTED: similar to Calpain-9 (Digestive tract- |
| 26 | Oncorhynchus mykiss | 46.60 | 1 | gill-specific calpain |
| 27 | Drosophila melanogaster | 42.40 | 4 | calpain |
| 28 | Drosophila melanogaster | 42.40 | 2 | Calpain-B CG8107-PA |
| 29 | Oryctolagus cuniculus | 41.20 | 2 | calpain 3, (p94) |
| 30 | Drosophila pseudoobscura | 39.70 | 1 | GA20829-PA |
| 31 | Schistosoma japonicum | 35.40 | 1 | unknown |
| 32 | Ovis aries | 33.10 | 1 | calpain IV regulatory subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 |
| Rattus norvegicus | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 |
| Mus musculus | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 ||||||+|||||||||||||||| ||||||#||||||+|||||| ||||||| |||||||| Sbjct 488 TFEPHKDGDFCIRVFSEKKADYQAVDDEIE#ANIEEIDANEEDIDDGFRRLFVQLAGEDAE 547 |
| Homo sapiens | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 ||||+|+|||||||||||||||| ||||||#||+|| + +|+|| |||||||||||||||| Sbjct 71 TFEPNKDGDFCIRVFSEKKADYQAVDDEIE#ANLEEFDISEDDIDDGFRRLFAQLAGEDAE 130 |
| Homo sapiens | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 ||||+|+|||||||||||||||| ||||||#||+|| + +|+|| |||||||||||||||| Sbjct 488 TFEPNKDGDFCIRVFSEKKADYQAVDDEIE#ANLEEFDISEDDIDDGFRRLFAQLAGEDAE 547 |
| Macaca mulatta | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 ||||+|+|||||||||||||||| ||||||#||+|| + +|+|| |||||||||||||||| Sbjct 488 TFEPNKDGDFCIRVFSEKKADYQAVDDEIE#ANLEEFDISEDDIDDGFRRLFAQLAGEDAE 547 |
| Sus scrofa | Query 498 CIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 ||||||||||||| ||||||#|++|| +|+|+|| |||||||||||||||| Sbjct 1 CIRVFSEKKADYQVVDDEIE#ADLEENDASEDDIDDGFRRLFAQLAGEDAE 50 |
| Gallus gallus | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 ||||+ |||||+|||||| |+ +|||||#|| || | +|+|| |++|| |||| ||| Sbjct 488 TFEPNLNGDFCLRVFSEKNANSTVIDDEIE#ANFEETEIDEDDIEPSFKKLFGQLAGSDAE 547 |
| Danio rerio | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 ||+||||||||+||||||+++ | || ||#||+++ +||++ ||| || +|||+| | Sbjct 486 TFDPHKNGDFCVRVFSEKQSEMQQCDDPIE#ANVDDEMVSEEEVDSGFRGLFTKLAGDDME 545 |
| Tetraodon nigroviridis | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 ||||| ||||||||||||+|| | || + #||+|+ ++|+ ||| ||++||| | | Sbjct 485 TFEPHLNGDFCIRVFSEKQADTQPCDDPVS#ANLEDETVADQDVDPGFRGLFSKLAGPDME 544 |
| Stizostedion vitreum vitreum | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 |||||||||||+||||||+||+| +|| +|#+ +| || +|+|+ | ||+|| |||| | | Sbjct 489 TFEPHKNGDFCVRVFSEKQADFQEMDDPVE#SRVETIEIDEDDVDDQFRKLFEQLAGGDCE 548 |
| Xenopus tropicalis | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 |||||| ||||+|||||| |+ + ||||+#|+|+| + ||+ | |+|+| |||+| | Sbjct 488 TFEPHKVGDFCLRVFSEKNAESKVPDDEIK#ADIDE-DVNEDAIDPNFKRMFLALAGDDKE 546 |
| Oncorhynchus mykiss | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQLAGEDAE 547 ||||| ||||||||||||+ + + || ++#|++++ ++ ++ ||| || +|||+| | Sbjct 486 TFEPHLNGDFCIRVFSEKQTETRPCDDPVK#ADLDDETVSDAEVDAGFRGLFTKLAGDDME 545 |
| Bos taurus | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANI-EEIEANEEDIGDGFRRLFAQLAGEDA 546 ||||+| ||| +| |||| | | +||+++#||+ +| +||+| + |+ || |||||| Sbjct 502 TFEPNKEGDFVLRFFSEKSAGTQELDDQVQ#ANLPDEQVLSEEEIDENFKSLFRQLAGEDM 561 Query 547 E 547 | Query 547 E 547 |
| Rattus norvegicus | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANI-EEIEANEEDIGDGFRRLFAQLAGEDA 546 ||||+| ||| +| |||||| | +||+|+#||+ +| +||+| | |+ ||++|||+| Sbjct 499 TFEPNKEGDFLLRFFSEKKAGTQELDDQIQ#ANLPDEKVLSEEEIDDNFKTLFSKLAGDDM 558 Query 547 E 547 | Query 547 E 547 |
| Gallus gallus | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANI-EEIEANEEDIGDGFRRLFAQLAGEDA 546 ||||++ ||| +||||||+|| + +||+||#| + +| +| +| + |++|| |||| | Sbjct 501 TFEPNREGDFVLRVFSEKRADTEEIDDKIE#AKLPDEKVLSEGEIDENFKQLFRQLAGPDM 560 Query 547 E 547 | Query 547 E 547 |
| Canis familiaris | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANI-EEIEANEEDIGDGFRRLFAQLAGEDA 546 ||||+| ||| +| |||| | + +||+|+#||+ +| +||+| + |+ || |||||| Sbjct 498 TFEPNKEGDFVLRFFSEKSAGTEELDDQIQ#ANLPDEQVLSEEEIDENFKALFRQLAGEDM 557 Query 547 E 547 | Query 547 E 547 |
| Coturnix coturnix | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANI-EEIEANEEDIGDGFRRLFAQLAGEDA 546 |||||| || +|||+||++| +|+|| #|++ +| | ++|| |||+ +| |||||| Sbjct 491 TFEPHKEADFILRVFTEKQSDTAELDEEIS#ADLADEEEITDDDIEDGFKNMFQQLAGEDM 550 Query 547 E 547 | Query 547 E 547 |
| Pongo pygmaeus | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANI-EEIEANEEDIGDGFRRLFAQLAGEDA 546 ||||+| ||| +| |||| | +||+|+#||+ +| +||+| + |+ || |||||| Sbjct 500 TFEPNKEGDFVLRFFSEKSAGTAELDDQIQ#ANLPDEQVLSEEEIDENFKALFRQLAGEDM 559 Query 547 E 547 | Query 547 E 547 |
| Xenopus laevis | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANI-EEIEANEEDIGDGFRRLFAQLAGEDA 546 |||||| || +|+|+|| +| | +|+|+ #|++ || | |+|+ | |+ +| ||||+| Sbjct 491 TFEPHKEADFIMRIFTEKHSDIQELDEEVT#ADLPEEEELTEDDVDDSFKAMFQQLAGDDM 550 Query 547 E 547 | Query 547 E 547 |
| synthetic construct | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANI-EEIEANEEDIGDGFRRLFAQLAGEDA 546 ||||+| ||| +| |||| | +||+|+#||+ +| +||+| + |+ || |||||| Sbjct 500 TFEPNKEGDFVLRFFSEKSAGTVELDDQIQ#ANLPDEQVLSEEEIDENFKALFRQLAGEDM 559 Query 547 E 547 | Query 547 E 547 |
| Ovis aries | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANI-EEIEANEEDIGDGFRRLFAQLAGEDA 546 ||||+| ||| +| |||| | | +||+++#||+ +| +||+| + |+ || |||||| Sbjct 114 TFEPNKEGDFVLRFFSEKSAGTQELDDQVQ#ANLPDEQVLSEEEIDENFKSLFRQLAGEDM 173 Query 547 E 547 | Query 547 E 547 |
| Xenopus laevis | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANI-EEIEANEEDIGDGFRRLFAQLAGEDA 546 |||||| || ||+|+|| +| | +|+|+ #|++ +| | |+|+ | |+ +| |||+| Sbjct 491 TFEPHKEADFIIRIFTEKHSDTQELDEEVS#ADLPDEEELTEDDVDDSFKAMFQHLAGDDM 550 Query 547 E 547 | Query 547 E 547 |
| Danio rerio | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANI-EEIEANEEDIGDGFRRLFAQLAGEDA 546 ||||+| || +|||||| |+ + +||++ #| | || +| | ||+ || |||| | Sbjct 490 TFEPNKEADFVLRVFSEKPANSEEMDDKVM#AEIPEEQRLDESQIDAGFKSLFRQLAGADM 549 Query 547 E 547 | Query 547 E 547 |
| Canis familiaris | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEI-------EANEEDIGDGFRRLFAQ 540 |||||+ |||+|+|||||| + +| +++# ++ |+ + ||++ || || | Sbjct 300 TFEPHQEADFCLRIFSEKKAITRDMDGDVD#IDLPELPKPTPAGQETEEEL--QFRALFDQ 357 Query 541 LAGEDA 546 +||| + Sbjct 358 IAGETS 363 |
| Oncorhynchus mykiss | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEE-----IEANEEDIGDGFRRLFAQLA 542 ||+|| || ||+|||| | + + ++#|++ + + | | | ||+| ++| Sbjct 458 TFQPHDEADFIIRIFSEKAAGTMEMGNTVD#ADLPDPPMPSLPEEETDEEKGLRRMFEKIA 517 Query 543 GED 545 | | Sbjct 518 GSD 520 |
| Drosophila melanogaster | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDE---------IE#ANIEEIEANEEDIGD--GFRR 536 ||+|++ |+| |||||| + + + ||| | # ++ ||| || Sbjct 702 TFDPNEEGEFIIRVFSETRNNMEENDDEVGFGETDDRIA#PSLPPPTPKEEDDPQRIALRR 761 Query 537 LFAQLAGEDAE 547 || +|| | | Sbjct 762 LFDSVAGSDEE 772 |
| Drosophila melanogaster | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDE---------IE#ANIEEIEANEEDIGD--GFRR 536 ||+|++ |+| |||||| + + + ||| | # ++ ||| || Sbjct 702 TFDPNEEGEFIIRVFSETRNNMEENDDEVGFGETDDRIA#PSLPPPTPKEEDDPQRIALRR 761 Query 537 LFAQLAGEDAE 547 || +|| | | Sbjct 762 LFDSVAGSDEE 772 |
| Oryctolagus cuniculus | Query 488 TFEPHKNGDFCIRVFSEKKA------DYQTVDDEIE#ANIEEIEANEEDIGDGFRRLFAQL 541 |+|||+ |+| +||||||+ + +|| + # + ||| || +| |+ Sbjct 492 TYEPHQEGEFILRVFSEKRNLSEEVENTISVDRPVP#LPGNTDQENEEQ--QQFRNIFKQI 549 Query 542 AGEDAE 547 ||+| | Sbjct 550 AGDDME 555 |
| Drosophila pseudoobscura | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDE---------IE#ANIEEIEANEEDIGD--GFRR 536 ||+|++ |+| |||||| + + ||| | # + ||| || Sbjct 708 TFDPNEEGEFIIRVFSETMNNMEENDDEVGFGDTDDRIA#PALPPPTPKEEDDPQRIALRR 767 Query 537 LFAQLAGEDAE 547 || +|| | | Sbjct 768 LFDSVAGSDEE 778 |
| Schistosoma japonicum | Query 488 TFEPHKNGDFCIRVFSEKKADYQTVDDEIE#ANIEEIEANEEDI--------GDGFRRLFA 539 ||+|++ +| +|+||||| +|||| # + ++ + + ++ | Sbjct 136 TFQPNEEAEFMLRIFSEKKRSTHELDDEIG#ETQPALNVDDNTVVKPLTDVQVEALQKAFN 195 Query 540 QLAGEDAE 547 ++||+| | Sbjct 196 KVAGDDGE 203 |
| Ovis aries | Query 523 IEANEEDIGDGFRRLFAQLAGEDAE 547 +|||| + ||||||||||+| | Sbjct 8 MEANESEEVRQFRRLFAQLAGDDME 32 |
[Site 42] EIEANIEEIE524-ANEEDIGDGF
Glu524  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu515 | Ile516 | Glu517 | Ala518 | Asn519 | Ile520 | Glu521 | Glu522 | Ile523 | Glu524 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala525 | Asn526 | Glu527 | Glu528 | Asp529 | Ile530 | Gly531 | Asp532 | Gly533 | Phe534 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GDFCIRVFSEKKADYQTVDDEIEANIEEIEANEEDIGDGFRRLFAQLAGEDAEISAFELQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 120.00 | 23 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 120.00 | 8 | calpain 2 |
| 3 | Mus musculus | 112.00 | 24 | m-calpain large subunit |
| 4 | Homo sapiens | 105.00 | 15 | CAPN2 protein |
| 5 | Homo sapiens | 105.00 | 2 | calpain 2, large subunit |
| 6 | Macaca mulatta | 105.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Sus scrofa | 97.40 | 5 | calpain II |
| 8 | Gallus gallus | 82.80 | 3 | calpain 2, (m/II) large subunit |
| 9 | Tetraodon nigroviridis | 75.10 | 6 | unnamed protein product |
| 10 | Danio rerio | 74.30 | 5 | calpain 2, (m/II) large subunit a |
| 11 | Stizostedion vitreum vitreum | 69.70 | 1 | putative calpain-like protein |
| 12 | Xenopus tropicalis | 65.10 | 4 | calpain 2, (m/II) large subunit |
| 13 | Oncorhynchus mykiss | 64.70 | 1 | m-calpain |
| 14 | Gallus gallus | 63.90 | 1 | mu-calpain large subunit |
| 15 | Rattus norvegicus | 63.20 | 2 | calpain 1, large subunit |
| 16 | Bos taurus | 62.00 | 4 | calpain 1, (mu/I) large subunit |
| 17 | Coturnix coturnix | 61.60 | 1 | quail calpain |
| 18 | Xenopus laevis | 60.80 | 1 | mu/m-calpain large subunit |
| 19 | Ovis aries | 59.30 | 1 | calpain 1 |
| 20 | Canis familiaris | 58.90 | 7 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 21 | Xenopus laevis | 58.90 | 3 | hypothetical protein LOC398772 |
| 22 | synthetic construct | 58.90 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 23 | Pongo pygmaeus | 57.80 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 24 | Danio rerio | 56.20 | 1 | calpain 1, large subunit |
| 25 | Oryctolagus cuniculus | 46.20 | 2 | calpain, small subunit 1 |
| 26 | Oncorhynchus mykiss | 43.50 | 1 | gill-specific calpain |
| 27 | Xenopus tropicalis | 42.40 | 1 | calpain, small subunit 2 |
| 28 | Canis familiaris | 40.80 | 2 | PREDICTED: similar to Calpain-9 (Digestive tract- |
| 29 | Ovis aries | 40.40 | 1 | calpain IV regulatory subunit |
| 30 | Drosophila melanogaster | 37.40 | 3 | calpain |
| 31 | Drosophila melanogaster | 37.40 | 2 | Calpain-B CG8107-PA |
| 32 | Drosophila pseudoobscura | 35.00 | 1 | GA20829-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 |
| Rattus norvegicus | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 |
| Mus musculus | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 |||||||||||||||| ||||||||||||+#|||||| ||||||| ||||||||||||||| Sbjct 495 GDFCIRVFSEKKADYQAVDDEIEANIEEID#ANEEDIDDGFRRLFVQLAGEDAEISAFELQ 554 |
| Homo sapiens | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 |||||||||||||||| ||||||||+|| +# +|+|| ||||||||||||||||||||||| Sbjct 495 GDFCIRVFSEKKADYQAVDDEIEANLEEFD#ISEDDIDDGFRRLFAQLAGEDAEISAFELQ 554 |
| Homo sapiens | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 |||||||||||||||| ||||||||+|| +# +|+|| ||||||||||||||||||||||| Sbjct 495 GDFCIRVFSEKKADYQAVDDEIEANLEEFD#ISEDDIDDGFRRLFAQLAGEDAEISAFELQ 554 |
| Macaca mulatta | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 |||||||||||||||| ||||||||+|| +# +|+|| ||||||||||||||||||||||| Sbjct 495 GDFCIRVFSEKKADYQAVDDEIEANLEEFD#ISEDDIDDGFRRLFAQLAGEDAEISAFELQ 554 |
| Sus scrofa | Query 498 CIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 ||||||||||||| |||||||++|| +#|+|+|| ||||||||||||||||||||||| Sbjct 1 CIRVFSEKKADYQVVDDEIEADLEEND#ASEDDIDDGFRRLFAQLAGEDAEISAFELQ 57 |
| Gallus gallus | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 ||||+|||||| |+ +||||||| || |# +|+|| |++|| |||| |||||||||+ Sbjct 495 GDFCLRVFSEKNANSTVIDDEIEANFEETE#IDEDDIEPSFKKLFGQLAGSDAEISAFELR 554 |
| Tetraodon nigroviridis | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 |||||||||||+|| | || + ||+|+ # ++|+ ||| ||++||| | |||| ||| Sbjct 492 GDFCIRVFSEKQADTQPCDDPVSANLEDET#VADQDVDPGFRGLFSKLAGPDMEISAPELQ 551 |
| Danio rerio | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 ||||+||||||+++ | || ||||+++ # +||++ ||| || +|||+| |||| ||+ Sbjct 493 GDFCVRVFSEKQSEMQQCDDPIEANVDDEM#VSEEEVDSGFRGLFTKLAGDDMEISASELR 552 |
| Stizostedion vitreum vitreum | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 ||||+||||||+||+| +|| +|+ +| ||# +|+|+ | ||+|| |||| | |||++||| Sbjct 496 GDFCVRVFSEKQADFQEMDDPVESRVETIE#IDEDDVDDQFRKLFEQLAGGDCEISSYELQ 555 |
| Xenopus tropicalis | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFEL 553 ||||+|||||| |+ + ||||+|+|+| +# ||+ | |+|+| |||+| ||| || Sbjct 495 GDFCLRVFSEKNAESKVPDDEIKADIDE-D#VNEDAIDPNFKRMFLALAGDDKEISPHEL 552 |
| Oncorhynchus mykiss | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 |||||||||||+ + + || ++|++++ # ++ ++ ||| || +|||+| |||| ||+ Sbjct 493 GDFCIRVFSEKQTETRPCDDPVKADLDDET#VSDAEVDAGFRGLFTKLAGDDMEISAAELK 552 |
| Gallus gallus | Query 495 GDFCIRVFSEKKADYQTVDDEIEANI-EEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFEL 553 ||| +||||||+|| + +||+||| + +| # +| +| + |++|| |||| | ||| || Sbjct 508 GDFVLRVFSEKRADTEEIDDKIEAKLPDEKV#LSEGEIDENFKQLFRQLAGPDMEISVTEL 567 Query 554 Q 554 Query 554 Q 554 |
| Rattus norvegicus | Query 495 GDFCIRVFSEKKADYQTVDDEIEANI-EEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFEL 553 ||| +| |||||| | +||+|+||+ +| # +||+| | |+ ||++|||+| ||| || Sbjct 506 GDFLLRFFSEKKAGTQELDDQIQANLPDEKV#LSEEEIDDNFKTLFSKLAGDDMEISVKEL 565 Query 554 Q 554 Query 554 Q 554 |
| Bos taurus | Query 495 GDFCIRVFSEKKADYQTVDDEIEANI-EEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFEL 553 ||| +| |||| | | +||+++||+ +| # +||+| + |+ || |||||| ||| || Sbjct 509 GDFVLRFFSEKSAGTQELDDQVQANLPDEQV#LSEEEIDENFKSLFRQLAGEDMEISVKEL 568 Query 554 Q 554 Sbjct 569 R 569 |
| Coturnix coturnix | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +|||+||++| +|+|| |++ +| |# ++|| |||+ +| |||||| ||| |||+ Sbjct 499 DFILRVFTEKQSDTAELDEEISADLADEEE#ITDDDIEDGFKNMFQQLAGEDMEISVFELK 558 |
| Xenopus laevis | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +|+|+|| +| | +|+|+ |++ || |# |+|+ | |+ +| ||||+| ||| |||+ Sbjct 499 DFIMRIFTEKHSDIQELDEEVTADLPEEEE#LTEDDVDDSFKAMFQQLAGDDMEISVFELK 558 |
| Ovis aries | Query 495 GDFCIRVFSEKKADYQTVDDEIEANI-EEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFEL 553 ||| +| |||| | | +||+++||+ +| # +||+| + |+ || |||||| ||| || Sbjct 121 GDFVLRFFSEKSAGTQELDDQVQANLPDEQV#LSEEEIDENFKSLFRQLAGEDMEISVKEL 180 Query 554 Q 554 Sbjct 181 R 181 |
| Canis familiaris | Query 495 GDFCIRVFSEKKADYQTVDDEIEANI-EEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFEL 553 ||| +| |||| | + +||+|+||+ +| # +||+| + |+ || |||||| ||| || Sbjct 505 GDFVLRFFSEKSAGTEELDDQIQANLPDEQV#LSEEEIDENFKALFRQLAGEDMEISVKEL 564 Query 554 Q 554 Query 554 Q 554 |
| Xenopus laevis | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || ||+|+|| +| | +|+|+ |++ +| |# |+|+ | |+ +| |||+| ||| |||+ Sbjct 499 DFIIRIFTEKHSDTQELDEEVSADLPDEEE#LTEDDVDDSFKAMFQHLAGDDMEISVFELK 558 |
| synthetic construct | Query 495 GDFCIRVFSEKKADYQTVDDEIEANI-EEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFEL 553 ||| +| |||| | +||+|+||+ +| # +||+| + |+ || |||||| ||| || Sbjct 507 GDFVLRFFSEKSAGTVELDDQIQANLPDEQV#LSEEEIDENFKALFRQLAGEDMEISVKEL 566 Query 554 Q 554 Sbjct 567 R 567 |
| Pongo pygmaeus | Query 495 GDFCIRVFSEKKADYQTVDDEIEANI-EEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFEL 553 ||| +| |||| | +||+|+||+ +| # +||+| + |+ || |||||| | | || Sbjct 507 GDFVLRFFSEKSAGTAELDDQIQANLPDEQV#LSEEEIDENFKALFRQLAGEDMETSVKEL 566 Query 554 Q 554 Sbjct 567 R 567 |
| Danio rerio | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +|||||| |+ + +||++ | | || # +| | ||+ || |||| | ||| ||| Sbjct 498 DFVLRVFSEKPANSEEMDDKVMAEIPEEQR#LDESQIDAGFKSLFRQLAGADMEISVTELQ 557 |
| Oryctolagus cuniculus | Query 499 IRVFSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 | || | | + ||#||| + ||||||||||+| |+|| || Sbjct 64 ISAISEAAAQYNPEPPPPRTHYSNIE#ANESEEVRQFRRLFAQLAGDDMEVSATELM 119 |
| Oncorhynchus mykiss | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEE-----IE#ANEEDIGDGFRRLFAQLAGEDAEIS 549 || ||+|||| | + + ++|++ + + # | | | ||+| ++|| | || Sbjct 465 ADFIIRIFSEKAAGTMEMGNTVDADLPDPPMPSLP#EEETDEEKGLRRMFEKIAGSDQAIS 524 Query 550 AFELQ 554 | ||| Sbjct 525 ARELQ 529 |
| Xenopus tropicalis | Query 502 FSEKKADYQTVDDEIEANIEEIE#ANEEDIGDGFRRLFAQLAGEDAEISAFEL 553 || |+| ++ |+#+|| + |||||+||||+| |+|| || Sbjct 35 LSEAAANYNPQPPPPRSHPSVIQ#SNESEETRQFRRLFSQLAGDDMEVSATEL 86 |
| Canis familiaris | Query 495 GDFCIRVFSEKKADYQTVDDEIEANIEEI-------E#ANEEDIGDGFRRLFAQLAGEDAE 547 |||+|+|||||| + +| +++ ++ |+ +# ||++ || || |+||| + Sbjct 307 ADFCLRIFSEKKAITRDMDGDVDIDLPELPKPTPAGQ#ETEEEL--QFRALFDQIAGETSG 364 Query 548 ISAFELQ 554 | + Sbjct 365 NGKLEFK 371 |
| Ovis aries | Query 523 IE#ANEEDIGDGFRRLFAQLAGEDAEISAFEL 553 +|#||| + ||||||||||+| |+|| || Sbjct 8 ME#ANESEEVRQFRRLFAQLAGDDMEVSATEL 38 |
| Drosophila melanogaster | Query 495 GDFCIRVFSEKKADYQTVDDE---------IEANIEEIE#ANEEDIGD--GFRRLFAQLAG 543 |+| |||||| + + + ||| | ++ # ||| |||| +|| Sbjct 709 GEFIIRVFSETRNNMEENDDEVGFGETDDRIAPSLPPPT#PKEEDDPQRIALRRLFDSVAG 768 Query 544 EDAEISAFELQ 554 | |+ ||+ Sbjct 769 SDEEVDWQELK 779 |
| Drosophila melanogaster | Query 495 GDFCIRVFSEKKADYQTVDDE---------IEANIEEIE#ANEEDIGD--GFRRLFAQLAG 543 |+| |||||| + + + ||| | ++ # ||| |||| +|| Sbjct 709 GEFIIRVFSETRNNMEENDDEVGFGETDDRIAPSLPPPT#PKEEDDPQRIALRRLFDSVAG 768 Query 544 EDAEISAFELQ 554 | |+ ||+ Sbjct 769 SDEEVDWQELK 779 |
| Drosophila pseudoobscura | Query 495 GDFCIRVFSEKKADYQTVDDE---------IEANIEEIE#ANEEDIGD--GFRRLFAQLAG 543 |+| |||||| + + ||| | + # ||| |||| +|| Sbjct 715 GEFIIRVFSETMNNMEENDDEVGFGDTDDRIAPALPPPT#PKEEDDPQRIALRRLFDSVAG 774 Query 544 EDAEISAFELQ 554 | |+ ||+ Sbjct 775 SDEEVDWQELK 785 |
[Site 43] IEANIEEIEA525-NEEDIGDGFR
Ala525  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile516 | Glu517 | Ala518 | Asn519 | Ile520 | Glu521 | Glu522 | Ile523 | Glu524 | Ala525 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn526 | Glu527 | Glu528 | Asp529 | Ile530 | Gly531 | Asp532 | Gly533 | Phe534 | Arg535 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DFCIRVFSEKKADYQTVDDEIEANIEEIEANEEDIGDGFRRLFAQLAGEDAEISAFELQT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 120.00 | 24 | A Chain A, Crystal Structure Of M-Calpain |
| 2 | Rattus norvegicus | 120.00 | 8 | calpain 2 |
| 3 | Mus musculus | 112.00 | 23 | unnamed protein product |
| 4 | Homo sapiens | 105.00 | 15 | CAPN2 protein |
| 5 | Macaca mulatta | 105.00 | 2 | PREDICTED: calpain 2, large subunit |
| 6 | Homo sapiens | 105.00 | 2 | calpain 2, large subunit |
| 7 | Sus scrofa | 99.80 | 5 | calpain II |
| 8 | Gallus gallus | 81.30 | 3 | calpain 2, (m/II) large subunit |
| 9 | Tetraodon nigroviridis | 74.30 | 6 | unnamed protein product |
| 10 | Danio rerio | 73.90 | 5 | calpain 2, (m/II) large subunit a |
| 11 | Stizostedion vitreum vitreum | 67.40 | 1 | putative calpain-like protein |
| 12 | Xenopus tropicalis | 64.30 | 4 | CAPN2 protein |
| 13 | Oncorhynchus mykiss | 64.30 | 1 | m-calpain |
| 14 | Gallus gallus | 63.50 | 1 | mu-calpain large subunit |
| 15 | Coturnix coturnix | 63.50 | 1 | quail calpain |
| 16 | Rattus norvegicus | 62.80 | 2 | calpain 1, large subunit |
| 17 | Xenopus laevis | 62.40 | 1 | mu/m-calpain large subunit |
| 18 | Bos taurus | 61.60 | 4 | calpain 1, (mu/I) large subunit |
| 19 | Xenopus laevis | 60.80 | 3 | hypothetical protein LOC398772 |
| 20 | Ovis aries | 58.90 | 1 | calpain 1 |
| 21 | Canis familiaris | 58.50 | 8 | PREDICTED: similar to Calpain-1 catalytic subunit |
| 22 | synthetic construct | 58.50 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 23 | Danio rerio | 58.20 | 1 | calpain 1, large subunit |
| 24 | Pongo pygmaeus | 57.00 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 25 | Oryctolagus cuniculus | 45.80 | 2 | calpain, small subunit 1 |
| 26 | Oncorhynchus mykiss | 43.50 | 1 | gill-specific calpain |
| 27 | Xenopus tropicalis | 42.00 | 1 | calpain, small subunit 2 |
| 28 | Ovis aries | 40.00 | 1 | calpain IV regulatory subunit |
| 29 | Canis familiaris | 39.30 | 2 | PREDICTED: similar to Calpain-9 (Digestive tract- |
| 30 | Drosophila melanogaster | 35.00 | 3 | calpain |
| 31 | Drosophila melanogaster | 35.00 | 2 | Calpain-B CG8107-PA |
| 32 | Bos taurus x Bos indicus | 31.20 | 1 | micromolar calcium activated neutral protease 1 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 |
| Rattus norvegicus | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 |
| Mus musculus | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 ||||||||||||||| ||||||||||||+|#||||| ||||||| |||||||||||||||| Sbjct 496 DFCIRVFSEKKADYQAVDDEIEANIEEIDA#NEEDIDDGFRRLFVQLAGEDAEISAFELQT 555 |
| Homo sapiens | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 ||||||||||||||| ||||||||+|| + #+|+|| |||||||||||||||||||||||| Sbjct 496 DFCIRVFSEKKADYQAVDDEIEANLEEFDI#SEDDIDDGFRRLFAQLAGEDAEISAFELQT 555 |
| Macaca mulatta | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 ||||||||||||||| ||||||||+|| + #+|+|| |||||||||||||||||||||||| Sbjct 496 DFCIRVFSEKKADYQAVDDEIEANLEEFDI#SEDDIDDGFRRLFAQLAGEDAEISAFELQT 555 |
| Homo sapiens | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 ||||||||||||||| ||||||||+|| + #+|+|| |||||||||||||||||||||||| Sbjct 496 DFCIRVFSEKKADYQAVDDEIEANLEEFDI#SEDDIDDGFRRLFAQLAGEDAEISAFELQT 555 |
| Sus scrofa | Query 498 CIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 ||||||||||||| |||||||++|| +|#+|+|| |||||||||||||||||||||||| Sbjct 1 CIRVFSEKKADYQVVDDEIEADLEENDA#SEDDIDDGFRRLFAQLAGEDAEISAFELQT 58 |
| Gallus gallus | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 |||+|||||| |+ +||||||| || | #+|+|| |++|| |||| |||||||||++ Sbjct 496 DFCLRVFSEKNANSTVIDDEIEANFEETEI#DEDDIEPSFKKLFGQLAGSDAEISAFELRS 555 |
| Tetraodon nigroviridis | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +|||||| ||+| +|| ++ ++|+|+ #+|+|| + |+ || +|||+| |||||||| Sbjct 502 DFYLRVFSEKNADFQEIDDSVKCHVEQIDI#DEDDISNHFKSLFGKLAGQDVEISAFELQ 560 |
| Danio rerio | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 |||+||||||+++ | || ||||+++ #+||++ ||| || +|||+| |||| ||+| Sbjct 494 DFCVRVFSEKQSEMQQCDDPIEANVDDEMV#SEEEVDSGFRGLFTKLAGDDMEISASELRT 553 |
| Stizostedion vitreum vitreum | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 |||+||||||+||+| +|| +|+ +| || #+|+|+ | ||+|| |||| | |||++||| Sbjct 497 DFCVRVFSEKQADFQEMDDPVESRVETIEI#DEDDVDDQFRKLFEQLAGGDCEISSYELQ 555 |
| Xenopus tropicalis | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +|+|+|| +| | +|+|+ |++ || | # |+|+ | |+ +| |||||| ||| |||+ Sbjct 499 DFVLRIFTEKHSDIQELDEEVSADLPEEEEL#TEDDVDDSFKAMFRQLAGEDMEISVFELK 558 Query 555 T 555 | Query 555 T 555 |
| Oncorhynchus mykiss | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQT 555 ||||||||||+ + + || ++|++++ #++ ++ ||| || +|||+| |||| ||+| Sbjct 494 DFCIRVFSEKQTETRPCDDPVKADLDDETV#SDAEVDAGFRGLFTKLAGDDMEISAAELKT 553 |
| Gallus gallus | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +||||||+|| + +||+||| + +| #+| +| + |++|| |||| | ||| ||| Sbjct 509 DFVLRVFSEKRADTEEIDDKIEAKLPDEKVL#SEGEIDENFKQLFRQLAGPDMEISVTELQ 568 Query 555 T 555 | Query 555 T 555 |
| Coturnix coturnix | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +|||+||++| +|+|| |++ +| | # ++|| |||+ +| |||||| ||| |||+ Sbjct 499 DFILRVFTEKQSDTAELDEEISADLADEEEI#TDDDIEDGFKNMFQQLAGEDMEISVFELK 558 Query 555 T 555 | Query 555 T 555 |
| Rattus norvegicus | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +| |||||| | +||+|+||+ +| #+||+| | |+ ||++|||+| ||| ||| Sbjct 507 DFLLRFFSEKKAGTQELDDQIQANLPDEKVL#SEEEIDDNFKTLFSKLAGDDMEISVKELQ 566 Query 555 T 555 | Query 555 T 555 |
| Xenopus laevis | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +|+|+|| +| | +|+|+ |++ || | # |+|+ | |+ +| ||||+| ||| |||+ Sbjct 499 DFIMRIFTEKHSDIQELDEEVTADLPEEEEL#TEDDVDDSFKAMFQQLAGDDMEISVFELK 558 Query 555 T 555 | Query 555 T 555 |
| Bos taurus | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +| |||| | | +||+++||+ +| #+||+| + |+ || |||||| ||| ||+ Sbjct 510 DFVLRFFSEKSAGTQELDDQVQANLPDEQVL#SEEEIDENFKSLFRQLAGEDMEISVKELR 569 Query 555 T 555 | Query 555 T 555 |
| Xenopus laevis | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || ||+|+|| +| | +|+|+ |++ +| | # |+|+ | |+ +| |||+| ||| |||+ Sbjct 499 DFIIRIFTEKHSDTQELDEEVSADLPDEEEL#TEDDVDDSFKAMFQHLAGDDMEISVFELK 558 Query 555 T 555 | Query 555 T 555 |
| Ovis aries | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA-#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +| |||| | | +||+++||+ + + #+||+| + |+ || |||||| ||| ||+ Sbjct 122 DFVLRFFSEKSAGTQELDDQVQANLPDEQVL#SEEEIDENFKSLFRQLAGEDMEISVKELR 181 Query 555 T 555 | Query 555 T 555 |
| Canis familiaris | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +| |||| | + +||+|+||+ +| #+||+| + |+ || |||||| ||| ||| Sbjct 506 DFVLRFFSEKSAGTEELDDQIQANLPDEQVL#SEEEIDENFKALFRQLAGEDMEISVKELQ 565 Query 555 T 555 | Query 555 T 555 |
| synthetic construct | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +| |||| | +||+|+||+ +| #+||+| + |+ || |||||| ||| ||+ Sbjct 508 DFVLRFFSEKSAGTVELDDQIQANLPDEQVL#SEEEIDENFKALFRQLAGEDMEISVKELR 567 Query 555 T 555 | Query 555 T 555 |
| Danio rerio | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +|||||| |+ + +||++ | | || #+| | ||+ || |||| | ||| ||| Sbjct 498 DFVLRVFSEKPANSEEMDDKVMAEIPEEQRL#DESQIDAGFKSLFRQLAGADMEISVTELQ 557 Query 555 T 555 | Query 555 T 555 |
| Pongo pygmaeus | Query 496 DFCIRVFSEKKADYQTVDDEIEANI-EEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 || +| |||| | +||+|+||+ +| #+||+| + |+ || |||||| | | ||+ Sbjct 508 DFVLRFFSEKSAGTAELDDQIQANLPDEQVL#SEEEIDENFKALFRQLAGEDMETSVKELR 567 Query 555 T 555 | Query 555 T 555 |
| Oryctolagus cuniculus | Query 499 IRVFSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFELQ 554 | || | | + |||#|| + ||||||||||+| |+|| || Sbjct 64 ISAISEAAAQYNPEPPPPRTHYSNIEA#NESEEVRQFRRLFAQLAGDDMEVSATELM 119 |
| Oncorhynchus mykiss | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEE-----IEA#NEEDIGDGFRRLFAQLAGEDAEISA 550 || ||+|||| | + + ++|++ + + # | | | ||+| ++|| | ||| Sbjct 466 DFIIRIFSEKAAGTMEMGNTVDADLPDPPMPSLPE#EETDEEKGLRRMFEKIAGSDQAISA 525 Query 551 FELQ 554 ||| Sbjct 526 RELQ 529 |
| Xenopus tropicalis | Query 502 FSEKKADYQTVDDEIEANIEEIEA#NEEDIGDGFRRLFAQLAGEDAEISAFEL 553 || |+| ++ |++#|| + |||||+||||+| |+|| || Sbjct 35 LSEAAANYNPQPPPPRSHPSVIQS#NESEETRQFRRLFSQLAGDDMEVSATEL 86 |
| Ovis aries | Query 523 IEA#NEEDIGDGFRRLFAQLAGEDAEISAFEL 553 +||#|| + ||||||||||+| |+|| || Sbjct 8 MEA#NESEEVRQFRRLFAQLAGDDMEVSATEL 38 |
| Canis familiaris | Query 496 DFCIRVFSEKKADYQTVDDEIEANIEEIEA-------#NEEDIGDGFRRLFAQLAGEDA 546 |||+|+|||||| + +| +++ ++ |+ # ||++ || || |+||| + Sbjct 308 DFCLRIFSEKKAITRDMDGDVDIDLPELPKPTPAGQE#TEEEL--QFRALFDQIAGETS 363 |
| Drosophila melanogaster | Query 496 DFCIRVFSEKKADYQTVDDE---------IEANIEEIEA#NEEDIGD--GFRRLFAQLAGE 544 +| |||||| + + + ||| | ++ # ||| |||| +|| Sbjct 710 EFIIRVFSETRNNMEENDDEVGFGETDDRIAPSLPPPTP#KEEDDPQRIALRRLFDSVAGS 769 Query 545 DAEISAFELQ 554 | |+ ||+ Sbjct 770 DEEVDWQELK 779 |
| Drosophila melanogaster | Query 496 DFCIRVFSEKKADYQTVDDE---------IEANIEEIEA#NEEDIGD--GFRRLFAQLAGE 544 +| |||||| + + + ||| | ++ # ||| |||| +|| Sbjct 710 EFIIRVFSETRNNMEENDDEVGFGETDDRIAPSLPPPTP#KEEDDPQRIALRRLFDSVAGS 769 Query 545 DAEISAFELQ 554 | |+ ||+ Sbjct 770 DEEVDWQELK 779 |
| Bos taurus x Bos indicus | Query 507 ADYQTVDDEIEANIEEIEA-#NEEDIGDGFRRLFAQLAG 543 | | +||+++||+ + + #+||+| + |+ || |||| Sbjct 2 AGTQELDDQVQANLPDEQVL#SEEEIDENFKSLFRQLAG 39 |
[Site 44] TKIQKYQKIY611-REIDVDRSGT
Tyr611  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Thr602 | Lys603 | Ile604 | Gln605 | Lys606 | Tyr607 | Gln608 | Lys609 | Ile610 | Tyr611 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg612 | Glu613 | Ile614 | Asp615 | Val616 | Asp617 | Arg618 | Ser619 | Gly620 | Thr621 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DMLDEDGSGKLGLKEFYILWTKIQKYQKIYREIDVDRSGTMNSYEMRKALEEAGFKLPCQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 126.00 | 25 | calpain 2 |
| 2 | N/A | 126.00 | 22 | A Chain A, Crystal Structure Analysis Of Rat M-Ca |
| 3 | Rattus norvegicus | 126.00 | 7 | calpain 2 |
| 4 | Sus scrofa | 124.00 | 6 | calpain II |
| 5 | Homo sapiens | 123.00 | 15 | AF261089_1 calpain large polypeptide L2 |
| 6 | Macaca mulatta | 123.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Homo sapiens | 123.00 | 2 | calpain 2, large subunit |
| 8 | Gallus gallus | 114.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Xenopus tropicalis | 88.60 | 4 | calpain 2, (m/II) large subunit |
| 10 | Danio rerio | 87.80 | 5 | AF282675_1 calpain 1 |
| 11 | Danio rerio | 87.40 | 1 | calpain 1, large subunit |
| 12 | Tetraodon nigroviridis | 84.70 | 5 | unnamed protein product |
| 13 | Stizostedion vitreum vitreum | 83.60 | 1 | putative calpain-like protein |
| 14 | Canis familiaris | 82.00 | 2 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 15 | Xenopus laevis | 79.70 | 3 | calpain 1, (mu/I) large subunit |
| 16 | Coturnix coturnix | 79.30 | 1 | quail calpain |
| 17 | Macaca fascicularis | 78.60 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 18 | Canis familiaris | 78.20 | 21 | PREDICTED: similar to Calpain-3 (Calpain L3) (Cal |
| 19 | synthetic construct | 78.20 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 20 | Pongo pygmaeus | 78.20 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Rattus norvegicus | 77.80 | 2 | calpain 1, large subunit |
| 22 | Sus scrofa | 77.40 | 1 | skeletal muscle specific calpain |
| 23 | Bos taurus | 77.00 | 5 | calpain 1, (mu/I) large subunit |
| 24 | Ovis aries | 76.60 | 1 | skeletal muscle-specific calpain |
| 25 | Oryctolagus cuniculus | 76.30 | 2 | calpain 3, (p94) |
| 26 | Gallus gallus | 75.90 | 1 | mu-calpain large subunit |
| 27 | Oncorhynchus mykiss | 73.90 | 1 | m-calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 |
| N/A | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 |
| Rattus norvegicus | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 |
| Sus scrofa | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 |||| ||| |||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 85 DMLDSDGSAKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 144 |
| Homo sapiens | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||||+||| Sbjct 582 DMLDSDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKMPCQ 641 |
| Macaca mulatta | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||||+||| Sbjct 582 DMLDSDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKMPCQ 641 |
| Homo sapiens | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 |||| |||||||||||||||||||||||||#||||||||||||||||||||||||||+||| Sbjct 582 DMLDSDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKMPCQ 641 |
| Gallus gallus | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 |+|| |||||||||||+ ||||||||||||#|||||||||||||||||+||| ||||| || Sbjct 582 DLLDNDGSGKLGLKEFHTLWTKIQKYQKIY#REIDVDRSGTMNSYEMRRALEAAGFKLSCQ 641 |
| Xenopus tropicalis | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 |+|| ||+|||||||| |||||| ||||||# +| | |||+|||||| ||| || |+ Sbjct 581 DLLDSDGTGKLGLKEFKILWTKILKYQKIY#SSVDRDHSGTINSYEMRGALEAAGIKV 637 |
| Danio rerio | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++| |||||||| ||++|| ||++| +|+#|+ |||+||||+||||||||| ||||| Sbjct 585 NLMDTDGSGKLGLVEFHVLWEKIKRYLQIF#RDHDVDKSGTMSSYEMRKALETAGFKL 641 |
| Danio rerio | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++| |||||||| ||++|| ||++| +|+#|+ |||+||||+||||||||| ||||| Sbjct 585 NLVDTDGSGKLGLVEFHVLWEKIKRYLQIF#RDHDVDKSGTMSSYEMRKALETAGFKL 641 |
| Tetraodon nigroviridis | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++| |||||||| ||++|| ||++| |+#| |+|+||||+||||| ||| ||||| Sbjct 611 NLMDLDGSGKLGLTEFHVLWEKIKRYLTIF#RTFDMDKSGTMSSYEMRMALESAGFKL 667 |
| Stizostedion vitreum vitreum | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +|||+|||||||| || +|||||+ | +|#|+ ||| ||||+| ||| |+||||| | Sbjct 583 NMLDKDGSGKLGLLEFKVLWTKIESYLSVY#RQKDVDNSGTMSSTEMRMAVEEAGFSL 639 |
| Canis familiaris | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++|+|||||||| || ||| ||+|+ |+#|| | | ||++|||||| |+|+|| || Sbjct 553 NLMDKDGSGKLGLPEFQILWKKIKKWTDIF#RECDEDHSGSLNSYEMRLAIEKAGIKL 609 |
| Xenopus laevis | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++|+||+||||| || ||| ||+ | |+#|+ |+|+||+|++|||| ||| ||||| Sbjct 594 NLMDKDGNGKLGLVEFNILWNKIKNYLTIF#RKFDMDKSGSMSAYEMRLALESAGFKL 650 |
| Coturnix coturnix | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++|+||| +||| || ||| ||+ + |+#|+ |+|+||||+||||| ||| ||||| Sbjct 586 NLMDKDGSARLGLVEFQILWNKIRSWLTIF#RQYDLDKSGTMSSYEMRMALESAGFKL 642 |
| Macaca fascicularis | Query 583 MLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 ++| |||||| |+||+ || ||+ +|||+#+ | |+|||+|||||| |+ +||| | | Sbjct 697 LMDTDGSGKLNLQEFHHLWNKIKAWQKIF#KHYDTDQSGTINSYEMRNAVNDAGFHLNNQ 755 |
| Canis familiaris | Query 583 MLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 +|| ||||+| |+||+ || ||+ +|||+#+ | |+|||+|||||| |+ +||| | | Sbjct 582 LLDTDGSGRLNLQEFHHLWKKIKAWQKIF#KHYDTDQSGTINSYEMRNAVNDAGFHLNSQ 640 |
| synthetic construct | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++| ||+||||| || ||| +|+ | |+#|+ |+|+||+|++|||| |+| ||||| Sbjct 595 NLMDRDGNGKLGLVEFNILWNRIRNYLSIF#RKFDLDKSGSMSAYEMRMAIESAGFKL 651 |
| Pongo pygmaeus | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++| ||+||||| || ||| +|+ | |+#|+ |+|+||+|++|||| |+| ||||| Sbjct 595 NLMDRDGNGKLGLVEFNILWNRIRNYLSIF#RKFDLDKSGSMSAYEMRMAIESAGFKL 651 |
| Rattus norvegicus | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++| ||+||||| || ||| +|+ | |+#|+ |+|+||+|++|||| |+| ||||| Sbjct 594 NLMDRDGNGKLGLVEFNILWNRIRNYLTIF#RKFDLDKSGSMSAYEMRMAIEAAGFKL 650 |
| Sus scrofa | Query 583 MLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 ++| ||||+| |+||+ || ||+ +|||+#+ | |+|||+|||||| |+ +||| | | Sbjct 103 LMDTDGSGRLNLQEFHHLWKKIKSWQKIF#KHYDTDQSGTINSYEMRNAVNDAGFHLNNQ 161 |
| Bos taurus | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++| ||+||||| || ||| +|+ | |+#|+ |+|+||+|++|||| |+| ||||| Sbjct 597 NLMDRDGNGKLGLVEFNILWNRIRNYLSIF#RKFDLDKSGSMSAYEMRMAIEFAGFKL 653 |
| Ovis aries | Query 583 MLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 ++| ||||+| |+||+ || ||+ +|||+#+ | |+|||+|||||| |+ +||| | | Sbjct 704 LMDTDGSGRLNLQEFHHLWKKIKTWQKIF#KHYDTDQSGTINSYEMRNAVNDAGFHLNNQ 762 |
| Oryctolagus cuniculus | Query 583 MLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKLPCQ 641 +| ||||+| |+||+ || ||+ +|||+#+ | | |||+|||||| |+++||| | | Sbjct 591 FMDTDGSGRLNLQEFHHLWKKIKAWQKIF#KRYDTDHSGTINSYEMRNAVKDAGFHLNSQ 649 |
| Gallus gallus | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++|+||+||||| || +|| +|+ | ++#|+ |+|+||+|++|||| ||| +|+|| Sbjct 596 NLMDKDGNGKLGLVEFNVLWNRIRNYLSVF#RKFDLDKSGSMSAYEMRMALEASGYKL 652 |
| Oncorhynchus mykiss | Query 582 DMLDEDGSGKLGLKEFYILWTKIQKYQKIY#REIDVDRSGTMNSYEMRKALEEAGFKL 638 +++|+ |+||||| || || |+||| ||#++ |+| ||||++ ||| ||+|||| | Sbjct 580 NLMDDSGNGKLGLGEFATLWKKVQKYLGIY#KKNDMDNSGTMSTPEMRMALKEAGFTL 636 |
[Site 45] KIYREIDVDR618-SGTMNSYEMR
Arg618  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys609 | Ile610 | Tyr611 | Arg612 | Glu613 | Ile614 | Asp615 | Val616 | Asp617 | Arg618 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser619 | Gly620 | Thr621 | Met622 | Asn623 | Ser624 | Tyr625 | Glu626 | Met627 | Arg628 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SGKLGLKEFYILWTKIQKYQKIYREIDVDRSGTMNSYEMRKALEEAGFKLPCQLHQVIVA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Sus scrofa | 127.00 | 5 | calpain II |
| 2 | Mus musculus | 126.00 | 25 | calpain 2 |
| 3 | N/A | 126.00 | 22 | A Chain A, Crystal Structure Of M-Calpain |
| 4 | Rattus norvegicus | 126.00 | 8 | calpain 2 |
| 5 | Homo sapiens | 125.00 | 17 | AF261089_1 calpain large polypeptide L2 |
| 6 | Macaca mulatta | 125.00 | 2 | PREDICTED: calpain 2, large subunit |
| 7 | Homo sapiens | 125.00 | 2 | calpain 2, large subunit |
| 8 | Gallus gallus | 118.00 | 4 | calpain 2, (m/II) large subunit |
| 9 | Danio rerio | 85.90 | 5 | AF282675_1 calpain 1 |
| 10 | Danio rerio | 85.90 | 1 | calpain 1, large subunit |
| 11 | Stizostedion vitreum vitreum | 83.60 | 1 | putative calpain-like protein |
| 12 | Tetraodon nigroviridis | 83.20 | 4 | unnamed protein product |
| 13 | Xenopus tropicalis | 82.80 | 4 | calpain 2, (m/II) large subunit |
| 14 | Xenopus laevis | 80.50 | 3 | calpain 1, (mu/I) large subunit |
| 15 | Coturnix coturnix | 80.50 | 1 | quail calpain |
| 16 | Canis familiaris | 79.70 | 2 | PREDICTED: similar to Calpain-11 (Calcium-activat |
| 17 | Rattus norvegicus | 77.00 | 2 | calpain 1, large subunit |
| 18 | synthetic construct | 75.50 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 19 | Macaca fascicularis | 75.50 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) ( |
| 20 | Pongo pygmaeus | 75.50 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 21 | Gallus gallus | 75.50 | 1 | mu-calpain large subunit |
| 22 | Canis familiaris | 75.10 | 21 | PREDICTED: similar to Calpain-3 (Calpain L3) (Cal |
| 23 | Bos taurus | 73.90 | 5 | calpain 3 |
| 24 | Oryctolagus cuniculus | 73.90 | 2 | calpain 3, (p94) |
| 25 | Oncorhynchus mykiss | 73.90 | 1 | m-calpain |
| 26 | Ovis aries | 73.90 | 1 | skeletal muscle-specific calpain |
| 27 | Sus scrofa | 73.20 | 1 | skeletal muscle specific calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| Sus scrofa | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 | ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 92 SAKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 151 |
| Mus musculus | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 |
| N/A | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 |
| Rattus norvegicus | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 |
| Homo sapiens | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 ||||||||||||||||||||||||||||||#|||||||||||||||||||+|||||||||| Sbjct 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKMPCQLHQVIVA 648 |
| Macaca mulatta | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 ||||||||||||||||||||||||||||||#|||||||||||||||||||+|||||||||| Sbjct 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKMPCQLHQVIVA 648 |
| Homo sapiens | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 ||||||||||||||||||||||||||||||#|||||||||||||||||||+|||||||||| Sbjct 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKMPCQLHQVIVA 648 |
| Gallus gallus | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 |||||||||+ |||||||||||||||||||#||||||||||+||| ||||| |||||+||| Sbjct 589 SGKLGLKEFHTLWTKIQKYQKIYREIDVDR#SGTMNSYEMRRALEAAGFKLSCQLHQIIVA 648 |
| Danio rerio | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIV 647 |||||| ||++|| ||++| +|+|+ |||+#||||+||||||||| ||||| | |+|+ Sbjct 592 SGKLGLVEFHVLWEKIKRYLQIFRDHDVDK#SGTMSSYEMRKALETAGFKLNNHLFQLII 650 |
| Danio rerio | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIV 647 |||||| ||++|| ||++| +|+|+ |||+#||||+||||||||| ||||| | |+|+ Sbjct 592 SGKLGLVEFHVLWEKIKRYLQIFRDHDVDK#SGTMSSYEMRKALETAGFKLNNHLFQLII 650 |
| Stizostedion vitreum vitreum | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 |||||| || +|||||+ | +||+ ||| #||||+| ||| |+||||| | |||++|| Sbjct 590 SGKLGLLEFKVLWTKIESYLSVYRQKDVDN#SGTMSSTEMRMAVEEAGFSLNNALHQILVA 649 |
| Tetraodon nigroviridis | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIV 647 |||||| ||++|| ||++| |+| |+|+#||||+||||| ||| ||||| | |+|+ Sbjct 618 SGKLGLTEFHVLWEKIKRYLTIFRTFDMDK#SGTMSSYEMRMALESAGFKLTNNLFQLII 676 |
| Xenopus tropicalis | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 +|||||||| |||||| |||||| +| | #|||+|||||| ||| || |+ +++++|| Sbjct 588 TGKLGLKEFKILWTKILKYQKIYSSVDRDH#SGTINSYEMRGALEAAGIKVNSTINELLVA 647 |
| Xenopus laevis | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIV 647 +||||| || ||| ||+ | |+|+ |+|+#||+|++|||| ||| ||||| |||+|+ Sbjct 601 NGKLGLVEFNILWNKIKNYLTIFRKFDMDK#SGSMSAYEMRLALESAGFKLTNSLHQLII 659 |
| Coturnix coturnix | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 | +||| || ||| ||+ + |+|+ |+|+#||||+||||| ||| ||||| + |||+|| Sbjct 593 SARLGLVEFQILWNKIRSWLTIFRQYDLDK#SGTMSSYEMRMALESAGFKLNNKPHQVVVA 652 |
| Canis familiaris | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 |||||| || ||| ||+|+ |+|| | | #||++|||||| |+|+|| || ++ ||+|| Sbjct 560 SGKLGLPEFQILWKKIKKWTDIFRECDEDH#SGSLNSYEMRLAIEKAGIKLSNKVTQVLVA 619 |
| Rattus norvegicus | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIV 647 +||||| || ||| +|+ | |+|+ |+|+#||+|++|||| |+| ||||| +||++|+ Sbjct 601 NGKLGLVEFNILWNRIRNYLTIFRKFDLDK#SGSMSAYEMRMAIEAAGFKLNKKLHELII 659 |
| synthetic construct | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIV 647 +||||| || ||| +|+ | |+|+ |+|+#||+|++|||| |+| ||||| +|+++|+ Sbjct 602 NGKLGLVEFNILWNRIRNYLSIFRKFDLDK#SGSMSAYEMRMAIESAGFKLNKKLYELII 660 |
| Macaca fascicularis | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVI 646 |||| |+||+ || ||+ +|||++ | |+#|||+|||||| |+ +||| | ||+ +| Sbjct 703 SGKLNLQEFHHLWNKIKAWQKIFKHYDTDQ#SGTINSYEMRNAVNDAGFHLNNQLYDII 760 |
| Pongo pygmaeus | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIV 647 +||||| || ||| +|+ | |+|+ |+|+#||+|++|||| |+| ||||| +|+++|+ Sbjct 602 NGKLGLVEFNILWNRIRNYLSIFRKFDLDK#SGSMSAYEMRMAIESAGFKLNKKLYELII 660 |
| Gallus gallus | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIV 647 +||||| || +|| +|+ | ++|+ |+|+#||+|++|||| ||| +|+|| +|||+++ Sbjct 603 NGKLGLVEFNVLWNRIRNYLSVFRKFDLDK#SGSMSAYEMRMALEASGYKLTQKLHQLLI 661 |
| Canis familiaris | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVI 646 ||+| |+||+ || ||+ +|||++ | |+#|||+|||||| |+ +||| | ||+ +| Sbjct 588 SGRLNLQEFHHLWKKIKAWQKIFKHYDTDQ#SGTINSYEMRNAVNDAGFHLNSQLYDII 645 |
| Bos taurus | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIV 647 ||+| |+||+ || ||+ +|||++ | |+#|||+|||||| |+++||| | ||+ +| Sbjct 710 SGRLNLQEFHHLWKKIKTWQKIFKHYDTDQ#SGTINSYEMRNAVKDAGFHLNNQLYDIIT 768 |
| Oryctolagus cuniculus | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIV 647 ||+| |+||+ || ||+ +|||++ | | #|||+|||||| |+++||| | ||+ +| Sbjct 597 SGRLNLQEFHHLWKKIKAWQKIFKRYDTDH#SGTINSYEMRNAVKDAGFHLNSQLYDIIT 655 |
| Oncorhynchus mykiss | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIVA 648 +||||| || || |+||| ||++ |+| #||||++ ||| ||+|||| | ++|++|| Sbjct 587 NGKLGLGEFATLWKKVQKYLGIYKKNDMDN#SGTMSTPEMRMALKEAGFTLNNGIYQILVA 646 |
| Ovis aries | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVIV 647 ||+| |+||+ || ||+ +|||++ | |+#|||+|||||| |+ +||| | ||+ +| Sbjct 710 SGRLNLQEFHHLWKKIKTWQKIFKHYDTDQ#SGTINSYEMRNAVNDAGFHLNNQLYDIIT 768 |
| Sus scrofa | Query 589 SGKLGLKEFYILWTKIQKYQKIYREIDVDR#SGTMNSYEMRKALEEAGFKLPCQLHQVI 646 ||+| |+||+ || ||+ +|||++ | |+#|||+|||||| |+ +||| | ||+ +| Sbjct 109 SGRLNLQEFHHLWKKIKSWQKIFKHYDTDQ#SGTINSYEMRNAVNDAGFHLNNQLYDII 166 |
* References
[PubMed ID: 23495712] Weber H, Muller L, Jonas L, Schult C, Sparmann G, Schuff-Werner P, Calpain mediates caspase-dependent apoptosis initiated by hydrogen peroxide in pancreatic acinar AR42J cells. Free Radic Res. 2013 May;47(5):432-46. doi: 10.3109/10715762.2013.785633. Epub
[PubMed ID: 22711986] ... Wang CF, Huang YS, Calpain 2 activated through N-methyl-D-aspartic acid receptor signaling cleaves CPEB3 and abrogates CPEB3-repressed translation in neurons. Mol Cell Biol. 2012 Aug;32(16):3321-32. doi: 10.1128/MCB.00296-12. Epub 2012 Jun
[PubMed ID: 22547201] ... Xu PT, Li Q, Sheng JJ, Chang H, Song Z, Yu ZB, Passive stretch reduces calpain activity through nitric oxide pathway in unloaded soleus muscles. Mol Cell Biochem. 2012 Aug;367(1-2):113-24. doi: 10.1007/s11010-012-1325-8. Epub
[PubMed ID: 22870316] ... Miao Y, Dong LD, Chen J, Hu XC, Yang XL, Wang Z, Involvement of calpain/p35-p25/Cdk5/NMDAR signaling pathway in glutamate-induced neurotoxicity in cultured rat retinal neurons. PLoS One. 2012;7(8):e42318. doi: 10.1371/journal.pone.0042318. Epub 2012 Aug 1.
[PubMed ID: 21549862] ... Chou JS, Impens F, Gevaert K, Davies PL, m-Calpain activation in vitro does not require autolysis or subunit dissociation. Biochim Biophys Acta. 2011 Jul;1814(7):864-72. doi: 10.1016/j.bbapap.2011.04.007.
[PubMed ID: 11102442] ... Hosfield CM, Moldoveanu T, Davies PL, Elce JS, Jia Z, Calpain mutants with increased Ca2+ sensitivity and implications for the role of the C(2)-like domain. J Biol Chem. 2001 Mar 9;276(10):7404-7. Epub 2000 Dec 1.
[PubMed ID: 10601010] ... Hosfield CM, Elce JS, Davies PL, Jia Z, Crystal structure of calpain reveals the structural basis for Ca(2+)-dependent protease activity and a novel mode of enzyme activation. EMBO J. 1999 Dec 15;18(24):6880-9.
[PubMed ID: 7635186] ... Arthur JS, Gauthier S, Elce JS, Active site residues in m-calpain: identification by site-directed mutagenesis. FEBS Lett. 1995 Jul 24;368(3):397-400.
[PubMed ID: 7982961] ... Graham-Siegenthaler K, Gauthier S, Davies PL, Elce JS, Active recombinant rat calpain II. Bacterially produced large and small subunits associate both in vivo and in vitro. J Biol Chem. 1994 Dec 2;269(48):30457-60.
[PubMed ID: 8218419] ... DeLuca CI, Davies PL, Samis JA, Elce JS, Molecular cloning and bacterial expression of cDNA for rat calpain II 80 kDa subunit. Biochim Biophys Acta. 1993 Oct 19;1216(1):81-93.