SB0132 : dUTPase
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial isoform 2 [Homo sapiens]; deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial isoform 2; dUTP pyrophosphatase; dUTP nucleotidohydrolase; deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial; dUTP diphosphatase |
| Gene Names | DUT; deoxyuridine triphosphatase |
| Gene Locus | 15q21.1; chromosome 15 |
| GO Function | Not available |
* Information From OMIM
Description: Deoxyuridine triphosphate nucleotidohydrolase (dUTPase; EC 3.6.1.23) hydrolyzes dUTP to dUMP and pyrophosphate (Ladner and Caradonna, 1997).
Function: Mutations in dUTPase of E. coli lead to increased dUTP levels and unusually high levels of dUMP incorporation into DNA during replication and repair which, in turn, causes DNA fragmentation and cell death (Lindahl, 1982; el-Hajj et al., 1988). Presumably for this reason, null mutants of dUTPase in yeast are lethal. A second function for dUTPase is to provide dUMP for synthesis of thymidylate. {2,1:Canman et al. (1992, 1994)} showed that increased dUTPase levels in some cancer cell lines accounted for their resistance to the thymidine synthase inhibitor fluorodeoxyuridine (FUdR).
* Structure Information
1. Primary Information
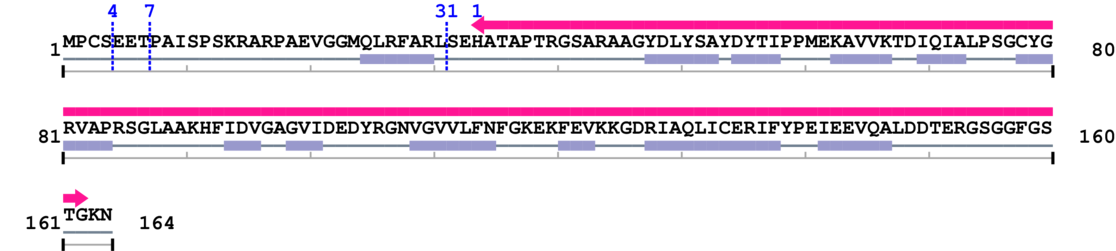
Length: 164 aa
Average Mass: 17.748 kDa
Monoisotopic Mass: 17.737 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| --- cleavage 4 --- | ||||
| --- cleavage 7 --- | ||||
| --- cleavage 31 --- | ||||
| dUTPase 1. | 34 | 162 | 2.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0132.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
3 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 21625588] Bozoky Z, Rona G, Klement E, Medzihradszky KF, Merenyi G, Vertessy BG, Friedrich P, Calpain-catalyzed proteolysis of human dUTPase specifically removes the nuclear localization signal peptide. PLoS One. 2011;6(5):e19546. doi: 10.1371/journal.pone.0019546. Epub 2011 May 19.
Cleavage sites (±10aa)
[Site 1] MPCS4-EETPAISPSK
Ser4  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | Met1 | Pro2 | Cys3 | Ser4 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu5 | Glu6 | Thr7 | Pro8 | Ala9 | Ile10 | Ser11 | Pro12 | Ser13 | Lys14 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MPCSEETPAISPSKRARPAEVGGMQLRFARLSEH |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 73.20 | 5 | deoxyuridine triphosphatase isoform 2 |
| 2 | synthetic construct | 73.20 | 2 | dUTP pyrophosphatase |
| 3 | Canis familiaris | 48.90 | 1 | PREDICTED: similar to Deoxyuridine 5-triphosphate |
| 4 | Mus musculus | 43.90 | 1 | AF091101_1 dUTPase |
| 5 | Mus musculus | 43.90 | 1 | deoxyuridine triphosphatase |
| 6 | Bos taurus | 43.90 | 1 | deoxyuridine triphosphatase |
| 7 | Danio rerio | 31.60 | 1 | deoxyuridine triphosphatase |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 1 MPCS#EETPAISPSKRARPAEVGGMQLRFARLSEH 34 ||||#|||||||||||||||||||||||||||||| Query 1 MPCS#EETPAISPSKRARPAEVGGMQLRFARLSEH 34 |
| synthetic construct | Query 1 MPCS#EETPAISPSKRARPAEVGGMQLRFARLSEH 34 ||||#|||||||||||||||||||||||||||||| Query 1 MPCS#EETPAISPSKRARPAEVGGMQLRFARLSEH 34 |
| Canis familiaris | Query 3 CS#EETPAISPSKRARPAEVGGMQLRFARLSEH 34 | # |||||||||||||| ||+||| ||||| Sbjct 54 CR#ATTPAISPSKRARPAE-DGMRLRFVRLSEH 84 |
| Mus musculus | Query 1 MPCS#EETPAISPSKRARPAEVGGMQLRFARLSEH 34 ||||#|+ |+| ||||| + | ||| ||||| Sbjct 1 MPCS#EDAAAVSASKRARAED--GASLRFVRLSEH 32 |
| Mus musculus | Query 1 MPCS#EETPAISPSKRARPAEVGGMQLRFARLSEH 34 ||||#|+ |+| ||||| + | ||| ||||| Sbjct 1 MPCS#EDAAAVSASKRARAED--GASLRFVRLSEH 32 |
| Bos taurus | Query 5 EETPAISPSKRARPAEVGGMQLRFARLSEH 34 +| +||||||| | | |+||||||||| Sbjct 26 KEAQVVSPSKRARATEAGDMRLRFARLSEH 55 |
| Danio rerio | Query 4 S#EETPAISPSKRARPAEVGGMQ----LRFARLSEH 34 +#| | |+|| |||+ | | + |+||+|+|| Sbjct 23 A#EVTEAVSPHKRAKSDAVNGAEERAVLKFAKLTEH 57 |
[Site 2] MPCSEET7-PAISPSKRAR
Thr7  Pro
Pro
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | - | Met1 | Pro2 | Cys3 | Ser4 | Glu5 | Glu6 | Thr7 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Pro8 | Ala9 | Ile10 | Ser11 | Pro12 | Ser13 | Lys14 | Arg15 | Ala16 | Arg17 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MPCSEETPAISPSKRARPAEVGGMQLRFARLSEHATA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | synthetic construct | 78.60 | 2 | dUTP pyrophosphatase |
| 2 | Homo sapiens | 78.20 | 6 | deoxyuridine triphosphatase isoform 2 |
| 3 | Canis familiaris | 53.50 | 1 | PREDICTED: similar to Deoxyuridine 5-triphosphate |
| 4 | Bos taurus | 48.50 | 1 | deoxyuridine triphosphatase |
| 5 | Mus musculus | 48.10 | 1 | deoxyuridine triphosphatase |
| 6 | Mus musculus | 47.00 | 2 | AF091101_1 dUTPase |
| 7 | Rattus norvegicus | 35.00 | 1 | deoxyuridine triphosphatase isoform 1 |
| 8 | Danio rerio | 34.30 | 1 | deoxyuridine triphosphatase |
| 9 | N/A | 31.20 | 1 | A Chain A, Human Dutp Pyrophosphatase Complex Wit |
Top-ranked sequences
| organism | matching |
|---|---|
| synthetic construct | Query 1 MPCSEET#PAISPSKRARPAEVGGMQLRFARLSEHATA 37 |||||||#|||||||||||||||||||||||||||||| Query 1 MPCSEET#PAISPSKRARPAEVGGMQLRFARLSEHATA 37 |
| Homo sapiens | Query 1 MPCSEET#PAISPSKRARPAEVGGMQLRFARLSEHATA 37 |||||||#|||||||||||||||||||||||||||||| Query 1 MPCSEET#PAISPSKRARPAEVGGMQLRFARLSEHATA 37 |
| Canis familiaris | Query 3 CSEET#PAISPSKRARPAEVGGMQLRFARLSEHATA 37 | |#||||||||||||| ||+||| |||||||| Sbjct 54 CRATT#PAISPSKRARPAE-DGMRLRFVRLSEHATA 87 |
| Bos taurus | Query 5 EET#PAISPSKRARPAEVGGMQLRFARLSEHATA 37 +| # +||||||| | | |+|||||||||||| Sbjct 26 KEA#QVVSPSKRARATEAGDMRLRFARLSEHATA 58 |
| Mus musculus | Query 1 MPCSEET#PAISPSKRARPAEVGGMQLRFARLSEHATA 37 |||||+ # |+| ||||| + | ||| |||||||| Sbjct 1 MPCSEDA#AAVSASKRARAED--GASLRFVRLSEHATA 35 |
| Mus musculus | Query 1 MPCSEET#PAISPSKRARPAEVGGMQLRFARLSEHAT 36 |||||+ # |+| ||||| + | ||| ||||||| Sbjct 1 MPCSEDA#AAVSASKRARAED--GASLRFVRLSEHAT 34 |
| Rattus norvegicus | Query 8 PAISPSKRARPAEVGGMQLRFARLSEHATA 37 ||+| ||||| + ||| |||||||| Sbjct 51 PAVSVSKRARAED--DASLRFVRLSEHATA 78 |
| Danio rerio | Query 4 SEET#PAISPSKRARPAEVGGMQ----LRFARLSEHAT 36 +| |# |+|| |||+ | | + |+||+|+|||| Sbjct 23 AEVT#EAVSPHKRAKSDAVNGAEERAVLKFAKLTEHAT 59 |
| N/A | Query 24 MQLRFARLSEHATA 37 |||||||||||||| Query 24 MQLRFARLSEHATA 37 |
[Site 3] GGMQLRFARL31-SEHATAPTRG
Leu31  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly22 | Gly23 | Met24 | Gln25 | Leu26 | Arg27 | Phe28 | Ala29 | Arg30 | Leu31 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser32 | Glu33 | His34 | Ala35 | Thr36 | Ala37 | Pro38 | Thr39 | Arg40 | Gly41 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PCSEETPAISPSKRARPAEVGGMQLRFARLSEHATAPTRGSARAAGYDLYSAYDYTIPPM |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 124.00 | 7 | deoxyuridine triphosphatase isoform 2 |
| 2 | synthetic construct | 124.00 | 2 | dUTP pyrophosphatase |
| 3 | Bos taurus | 96.70 | 1 | deoxyuridine triphosphatase |
| 4 | Canis familiaris | 95.90 | 1 | PREDICTED: similar to Deoxyuridine 5-triphosphate |
| 5 | Mus musculus | 91.70 | 1 | deoxyuridine triphosphatase |
| 6 | Mus musculus | 90.10 | 2 | AF091101_1 dUTPase |
| 7 | N/A | 82.00 | 4 | A Chain A, Human Dutp Pyrophosphatase Complex Wit |
| 8 | Rattus norvegicus | 81.60 | 1 | deoxyuridine triphosphatase isoform 1 |
| 9 | Danio rerio | 77.40 | 1 | deoxyuridine triphosphatase |
| 10 | Tetraodon nigroviridis | 68.90 | 1 | unnamed protein product |
| 11 | Drosophila pseudoobscura | 53.90 | 1 | GA18276-PA |
| 12 | Ectromelia virus | 52.00 | 1 | EVM026 |
| 13 | Drosophila melanogaster | 50.80 | 2 | Deoxyuridine triphosphatase CG4584-PB, isoform B |
| 14 | Apis mellifera | 50.80 | 1 | PREDICTED: similar to Deoxyuridine 5-triphosphate |
| 15 | Cowpox virus | 50.40 | 2 | CPXV049 protein |
| 16 | Vaccinia virus | 50.40 | 1 | dUTPase |
| 17 | Vaccinia virus (strain Tian Tan) | 50.40 | 1 | TF2L |
| 18 | Variola virus | 50.10 | 1 | hypothetical protein VARVgp026 |
| 19 | Rabbitpox virus | 50.10 | 1 | DUT_RABPU Deoxyuridine 5'-triphosphate nucleotido |
| 20 | Monkeypox virus | 50.10 | 1 | C8L |
| 21 | Myxoma virus | 49.30 | 1 | deoxyuridine 5'triphosphatenucleotidohydrolase |
| 22 | Yarrowia lipolytica | 48.90 | 1 | hypothetical protein |
| 23 | Grouper iridovirus | 47.40 | 1 | dUTPase |
| 24 | Candida glabrata | 47.40 | 1 | unnamed protein product |
| 25 | Invertebrate iridescent virus 6 | 46.60 | 1 | 438L |
| 26 | Mule deer poxvirus | 46.60 | 1 | DUTPase |
| 27 | Camelpox virus | 46.20 | 1 | dUTPase; CMLV037 |
| 28 | Yaba monkey tumor virus | 45.80 | 1 | 17L |
| 29 | Gibberella zeae PH-1 | 45.40 | 1 | hypothetical protein FG00904.1 |
| 30 | Aspergillus fumigatus Af293 | 45.10 | 1 | dUTPase (Dut), putaive |
| 31 | Saccharomyces cerevisiae | 45.10 | 1 | dUTP pyrophosphatase |
| 32 | Strongylocentrotus purpuratus | 45.10 | 1 | PREDICTED: similar to ENSANGP00000012604, partial |
| 33 | Saccharomyces cerevisiae | 44.70 | 1 | dUTPase, catalyzes hydrolysis of dUTP to dUMP and |
| 34 | Fowl adenovirus A | 44.70 | 1 | dUTP pyrophosphatase |
| 35 | fowl adenovirus 1 | 44.70 | 1 | 19.3 kda polypeptide |
| 36 | Yaba-like disease virus | 44.70 | 1 | 17L protein |
| 37 | Spodoptera litura nucleopolyhedrovirus | 44.30 | 1 | dUTP pyrophosphatase |
| 38 | Rabbit fibroma virus | 44.30 | 1 | gp012L |
| 39 | Ashbya gossypii ATCC 10895 | 43.50 | 1 | AGR249Cp |
| 40 | Swinepox virus | 43.10 | 1 | SPV013 dUTPase |
| 41 | orf virus strain D1701 | 43.10 | 1 | dUTPase homolog |
| 42 | Emiliania huxleyi virus 86 | 43.10 | 1 | putative deoxyuridine 5'-triphosphate nucleotidoh |
| 43 | Orf virus | 43.10 | 1 | DUT_ORFN2 Deoxyuridine 5'-triphosphate nucleotidoh |
| 44 | Schizosaccharomyces pombe 972h- | 42.40 | 1 | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| 45 | Oryza sativa (japonica cultivar-group) | 42.00 | 1 | putative deoxyuridine triphosphatase |
| 46 | Fowlpox virus | 42.00 | 1 | ORF FPV038 dUTP pyrophosphatase vaccinia F2L homo |
| 47 | Tiger frog virus | 41.60 | 1 | AF389451_13 dUTPase-like protein |
| 48 | Rana grylio virus 9506 | 41.60 | 1 | deoxyuridine triphosphatase |
| 49 | Aspergillus nidulans FGSC A4 | 41.20 | 1 | hypothetical protein AN0271.2 |
| 50 | Cryptococcus neoformans var. neoformans JEC21 | 40.80 | 2 | microtubule binding protein |
| 51 | Neurospora crassa | 40.80 | 1 | DUT_NEUCR Deoxyuridine 5'-triphosphate nucleotido |
| 52 | Tree shrew adenovirus | 40.40 | 1 | E4 ORFA |
| 53 | Aspergillus oryzae RIB40 | 40.40 | 1 | hypothetical protein |
| 54 | Arabidopsis thaliana | 39.70 | 1 | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| 55 | lumpy skin disease virus | 39.30 | 2 | dUTPase |
| 56 | Staphylococcus phage EW | 39.30 | 1 | ORF024 |
| 57 | Lumpy skin disease virus NI-2490 | 39.30 | 1 | LSDV018 dUTPase |
| 58 | Debaryomyces hansenii CBS767 | 39.30 | 1 | hypothetical protein DEHA0D16357g |
| 59 | Sheeppox virus | 39.30 | 1 | dUTPase |
| 60 | Chaetomium globosum CBS 148.51 | 39.30 | 1 | dUTP pyrophosphatase |
| 61 | Ustilago maydis 521 | 38.90 | 1 | hypothetical protein UM02908.1 |
| 62 | Entamoeba histolytica HM-1:IMSS | 38.50 | 3 | deoxyuridine 5'-triphosphate nucleotidohydrolase, |
| 63 | Paramecium bursaria Chlorella virus 1 | 38.50 | 1 | hypothetical protein PBCV1_A551L |
| 64 | Bacillus clausii KSM-K16 | 38.10 | 1 | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| 65 | Dictyostelium discoideum AX4 | 37.40 | 1 | dUTP diphosphatase |
| 66 | Caenorhabditis elegans | 37.40 | 1 | DeoxyUTPase family member (dut-1) |
| 67 | Caenorhabditis briggsae AF16 | 37.40 | 1 | Hypothetical protein CBG03725 |
| 68 | Lycopersicon esculentum | 37.40 | 1 | DUT_LYCES Deoxyuridine 5'-triphosphate nucleotidoh |
| 69 | Lactococcus phage Tuc2009 | 36.20 | 1 | dUTPase |
| 70 | Fowl adenovirus 8 | 36.20 | 1 | ORF1 |
| 71 | Candida albicans SC5314 | 35.80 | 1 | dUTP pyrophosphatase |
| 72 | Lactococcus lactis subsp. cremoris SK11 | 35.00 | 3 | dUTPase |
| 73 | Agrotis segetum granulovirus | 35.00 | 1 | unknown |
| 74 | Staphylococcus haemolyticus JCSC1435 | 34.70 | 1 | putative dUTP pyrophosphatase |
| 75 | Entamoeba histolytica HM-1:IMSS | 34.70 | 1 | dUTP diphosphatase |
| 76 | Fowl adenovirus D | 34.70 | 1 | dUTPAse homolog |
| 77 | Cryptococcus neoformans var. neoformans B-3501A | 34.70 | 1 | hypothetical protein CNBM1290 |
| 78 | Lactococcus phage TP901-1 | 34.30 | 1 | DUT |
| 79 | Amsacta moorei entomopoxvirus | 34.30 | 1 | putative dUTPase |
| 80 | Chrysodeixis chalcites nucleopolyhedrovirus | 33.50 | 1 | dUTPase |
| 81 | Fowl adenovirus 4 | 33.50 | 1 | ORF1 |
| 82 | Fowl adenovirus 10 | 33.50 | 1 | ORF1 |
| 83 | Lactococcus phage ul36 | 33.50 | 1 | putative dUTPase |
| 84 | Lactococcus phage bIL309 | 33.50 | 1 | dUTPase |
| 85 | Lactococcus phage bIL285 | 33.50 | 1 | dUTPase |
| 86 | Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 | 33.50 | 1 | dUTPase |
| 87 | Bacillus thuringiensis serovar israelensis ATCC 35646 | 33.10 | 1 | Deoxyuridine 5'-triphosphate nucleotidohydrolase |
| 88 | Heliothis zea virus 1 | 33.10 | 1 | dUTP pyrophosphatase |
| 89 | Wolinella succinogenes DSM 1740 | 33.10 | 1 | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| 90 | Corynebacterium jeikeium K411 | 33.10 | 1 | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| 91 | Lactococcus phage bIL286 | 32.70 | 1 | dUTPase |
| 92 | Spodoptera exigua nucleopolyhedrovirus | 32.30 | 1 | ORF55 |
| 93 | Rhodobacter sphaeroides ATCC 17025 | 32.30 | 1 | hypothetical protein Rsph17025_4038 |
| 94 | Staphylococcus phage 71 | 32.00 | 1 | ORF026 |
| 95 | bacteriophage phi31.1 | 32.00 | 1 | AF208055_15 Orf139b |
| 96 | Kluyveromyces lactis | 31.60 | 1 | unnamed protein product |
| 97 | Lymantria dispar nucleopolyhedrovirus | 31.20 | 1 | dUTPase |
| 98 | Taro bacilliform virus | 31.20 | 1 | polyprotein |
| 99 | Helicobacter hepaticus ATCC 51449 | 31.20 | 1 | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| 100 | Encephalitozoon cuniculi GB-M1 | 31.20 | 1 | DEOXYURIDINE 5' TRIPHOSPHATE NUCLEOTIDOHYDROLASE |
| 101 | Lactococcus lactis | 30.80 | 1 | dUTPase |
| 102 | Staphylococcus aureus subsp. aureus MRSA252 | 30.80 | 1 | putative dUTP pyrophosphatase |
| 103 | Staphylococcus phage 187 | 30.80 | 1 | ORF025 |
| 104 | Staphylococcus phage 11 | 30.80 | 1 | dUTPase |
| 105 | Staphylococcus phage ROSA | 30.80 | 1 | ORF023 |
| 106 | Staphylococcus phage phiSLT | 30.80 | 1 | phi PVL ORF 53 analogue |
| 107 | Staphylococcus aureus subsp. aureus USA300 | 30.80 | 1 | dUTP diphosphatase |
| 108 | Staphylococcus aureus subsp. aureus NCTC 8325 | 30.80 | 1 | dUTP pyrophosphatase |
| 109 | Staphylococcus phage 29 | 30.40 | 1 | ORF025 |
| 110 | Staphylococcus phage PVL | 30.40 | 1 | dUTPase |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 2 PCSEETPAISPSKRARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 PCSEETPAISPSKRARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 |
| synthetic construct | Query 2 PCSEETPAISPSKRARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 2 PCSEETPAISPSKRARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 |
| Bos taurus | Query 5 EETPAISPSKRARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 +| +||||||| | | |+||||||#||||||||+|||||||||||||||||+||| Sbjct 26 KEAQVVSPSKRARATEAGDMRLRFARL#SEHATAPTKGSARAAGYDLYSAYDYTVPPM 82 |
| Canis familiaris | Query 3 CSEETPAISPSKRARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 | |||||||||||||| ||+||| ||#||||||||+|| ||||||||||||| +||| Sbjct 54 CRATTPAISPSKRARPAE-DGMRLRFVRL#SEHATAPTKGSPRAAGYDLYSAYDYILPPM 111 |
| Mus musculus | Query 2 PCSEETPAISPSKRARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 ||||+ |+| ||||| + | ||| ||#|||||||||||||||||||+||||||| || Sbjct 2 PCSEDAAAVSASKRARAED--GASLRFVRL#SEHATAPTRGSARAAGYDLFSAYDYTISPM 59 |
| Mus musculus | Query 2 PCSEETPAISPSKRARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 ||||+ |+| ||||| + | ||| ||#||||| |||||||||||||+||||||| || Sbjct 2 PCSEDAAAVSASKRARAED--GASLRFVRL#SEHATTPTRGSARAAGYDLFSAYDYTISPM 59 |
| N/A | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 ||||||||#|||||||||||||||||||||||||||||| Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 |
| Rattus norvegicus | Query 8 PAISPSKRARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 ||+| ||||| + ||| ||#|||||||||||||||||||||||||||| | Sbjct 51 PAVSVSKRARAED--DASLRFVRL#SEHATAPTRGSARAAGYDLYSAYDYTIPSM 102 |
| Danio rerio | Query 4 SEETPAISPSKRARPAEVGGMQ----LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 +| | |+|| |||+ | | + |+||+|#+|||| |+||| |||||||||||||+| Sbjct 23 AEVTEAVSPHKRAKSDAVNGAEERAVLKFAKL#TEHATTPSRGSNRAAGYDLYSAYDYSIG 82 Query 60 PM 61 || Query 60 PM 61 |
| Tetraodon nigroviridis | Query 10 ISPSKRARPAEVGGMQ--LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 +||||| | + ||||+|#||||| ||||| +|||+|||||||| | |+ Sbjct 17 VSPSKRTRTETAAEDRPVLRFAKL#SEHATTPTRGSVKAAGFDLYSAYDYAIDPL 70 |
| Drosophila pseudoobscura | Query 16 ARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 |+ ++ ||||+|#+||| | ||||+||| || |||| +| Sbjct 5 AKKMKIDKCVLRFAKL#TEHALEPVRGSAKAAGVDLRSAYDVVVP 48 |
| Ectromelia virus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|| + #+ | +||| | |||||||||||||||| Sbjct 10 VRFVKE#TNRAKSPTRQSNHAAGYDLYSAYDYTIPP 44 |
| Drosophila melanogaster | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 ||||+|#+|+| | ||||+||| || |||| +| Sbjct 8 LRFAKL#TENALEPVRGSAKAAGVDLRSAYDVVVP 41 |
| Apis mellifera | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 |+||+|#+| | ||+|| |||||| |||+| +|| Sbjct 4 LKFAKL#TEKAFIPTKGSKYAAGYDLRSAYEYIVPP 38 |
| Cowpox virus | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 ++|| + #+ | +||| | |||||||||||||||| Sbjct 9 RVRFVKE#TNRAKSPTRQSPGAAGYDLYSAYDYTIPP 44 |
| Vaccinia virus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|| + #+ | +||| | |||||||||||||||| Sbjct 10 VRFVKE#TNRAKSPTRQSPYAAGYDLYSAYDYTIPP 44 |
| Vaccinia virus (strain Tian Tan) | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|| + #+ | +||| | |||||||||||||||| Sbjct 10 VRFVKE#TNRAKSPTRQSPGAAGYDLYSAYDYTIPP 44 |
| Variola virus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|| + #+ | +||| | |||||||||||||||| Sbjct 10 VRFVKE#TNRAKSPTRQSPYAAGYDLYSAYDYTIPP 44 |
| Rabbitpox virus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|| + #+ | +||| | |||||||||||||||| Sbjct 10 VRFVKE#TNRAKSPTRQSPYAAGYDLYSAYDYTIPP 44 |
| Monkeypox virus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|| + #+ | +||| | |||||||||||||||| Sbjct 14 VRFVKE#TNRAKSPTRQSPGAAGYDLYSAYDYTIPP 48 |
| Myxoma virus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 ++ + #||||+ ||+ +| |||||||||| | ||| Sbjct 11 VKCVKF#SEHASLPTKATADAAGYDLYSAYAYIIPP 45 |
| Yarrowia lipolytica | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | |+ | |#+|+| |||||| |||||||+| + ||| Sbjct 1 MSLKVAFL#NENARAPTRGSVHAAGYDLYAAEEKTIP 36 |
| Grouper iridovirus | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|||+++#++ || ||+|| +|||||||| | | | Sbjct 11 KLRFSKI#TDRATTPTKGSKLSAGYDLYSAQDLAIEP 46 |
| Candida glabrata | Query 23 GMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 | |+ #||| |||+|| |||||+|++ || || | Sbjct 3 GNVLKVQLR#SEHGIAPTKGSVYAAGYDIYASADYVIPAM 41 |
| Invertebrate iridescent virus 6 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 +|++|| |#+|+| | ||| +|| || |||+| +| Sbjct 154 LQIKFAAL#TEYAQKPKRGSEFSAGIDLRSAYEYLVP 189 |
| Mule deer poxvirus | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 + ++ +|#|| || | + + +||||||||||||+ | Sbjct 4 ITVKCLKL#SEFATIPKKATYNSAGYDLYSAYDYTVNPF 41 |
| Camelpox virus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|| + #+ | +||| | |||||||||| ||||| Sbjct 10 VRFVKE#TNRAKSPTRQSPYAAGYDLYSAYYYTIPP 44 |
| Yaba monkey tumor virus | Query 32 SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 || || ||| | ++||||||||||| + | Sbjct 12 SEFATIPTRSSKKSAGYDLYSAYDYLVRP 40 |
| Gibberella zeae PH-1 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 |+ +|#|| ||||| |||||+|||+| ||| Sbjct 32 LQIKKL#SEKGRLPTRGSEFAAGYDIYSAHDTTIP 65 |
| Aspergillus fumigatus Af293 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | +|#+| | ||||||| ||||||||| + || Sbjct 113 LLVKKL#TESAQAPTRGSAFAAGYDLYSAKETVIP 146 |
| Saccharomyces cerevisiae | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 +||| #| || ||+||| |||||+|++ | ||| | Sbjct 11 IQLR----#SASATVPTKGSATAAGYDIYASQDITIPAM 44 |
| Strongylocentrotus purpuratus | Query 7 TPAI-SPSKRARPAEVGGM-------QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTI 58 || + ||+|| + || || |+| ||#++ | || ||| |||+||+| | + Sbjct 2 TPGVESPAKRQK-MEVNGMGDAVVEPVLQFKRL#ND-AFAPVRGSDHAAGFDLFSNKDMVV 59 Query 59 P 59 | Query 59 P 59 |
| Saccharomyces cerevisiae | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 +||| #| || ||+||| |||||+|++ | ||| | Sbjct 11 IQLR----#SASATVPTKGSATAAGYDIYASQDITIPAM 44 |
| Fowl adenovirus A | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 +| | ||#| || | | + ||||||+|||| +| Sbjct 21 KLYFVRL#SPHAVPPVRATHGAAGYDLFSAYDIKVP 55 |
| fowl adenovirus 1 | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 +| | ||#| || | | + ||||||+|||| +| Sbjct 21 KLYFVRL#SPHAVPPVRATHGAAGYDLFSAYDIKVP 55 |
| Yaba-like disease virus | Query 32 SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 || || ||+ | ++||||||||||| + | Sbjct 12 SESATIPTKSSKKSAGYDLYSAYDYLVCP 40 |
| Spodoptera litura nucleopolyhedrovirus | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 ||++ +|#|||| || || ||| || ||+|| | Sbjct 27 QLKYLKL#SEHAYEPTYGSDGAAGLDLKSAHDYIIK 61 |
| Rabbit fibroma virus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 ++ + #||||+ ||+ + |||||||||| | | | Sbjct 5 VKCVKF#SEHASLPTKATKDAAGYDLYSAYAYIIRP 39 |
| Ashbya gossypii ATCC 10895 | Query 12 PSKRARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 |+|+ | +||| #||+| |||+||| |||||+|++ | || Sbjct 5 PAKKVHSAPTLKVQLR----#SENAIAPTKGSAAAAGYDIYASQDCVIP 48 |
| Swinepox virus | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 + ++ +|#| +| | | ++ +||||||||| ||+ | Sbjct 3 LYVKCVKL#SNNAIIPNRSTSGSAGYDLYSAYSYTVKP 39 |
| orf virus strain D1701 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 |+ ||#|++|| | ||| ||| || |||| || Sbjct 9 LQVVRL#SQNATIPARGSPGAAGLDLCSAYDCVIP 42 |
| Emiliania huxleyi virus 86 | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | +||#++ | ||+|+ +|||||||+ | ||| Sbjct 7 NLYVSRL#TDDAKLPTKGTTLSAGYDLYSSEDVKIPP 42 |
| Orf virus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 |+ ||#|++|| | ||| ||| || |||| || Sbjct 9 LQVVRL#SQNATIPARGSPGAAGLDLCSAYDCVIP 42 |
| Schizosaccharomyces pombe 972h- | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | +|#|| || ||+||| +||||||+| + +| Sbjct 1 MSFFVQKL#SEKATIPTKGSANSAGYDLYAAAECIVP 36 |
| Oryza sativa (japonica cultivar-group) | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 |+ +|#||+| |+|||| |||||| || + +| Sbjct 88 LKVKKL#SENAVLPSRGSALAAGYDLSSAAEVVVP 121 |
| Fowlpox virus | Query 30 RL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 ++#|+ | | + ++ +||||||||||| | || Sbjct 8 KI#SDRAKLPYKQTSYSAGYDLYSAYDYVIEPM 39 |
| Tiger frog virus | Query 23 GMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | |++ +|#||||+ |||| |||||| +|+ +| Sbjct 3 GNSLQYVKL#SEHASGLIRGSAGAAGYDLAAAHPVVVP 39 |
| Rana grylio virus 9506 | Query 23 GMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | |++ +|#||||+ |||| |||||| +|+ +| Sbjct 3 GNSLQYVKL#SEHASGLIRGSAGAAGYDLAAAHPVVVP 39 |
| Aspergillus nidulans FGSC A4 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | +|#++ | ||||||| |||||||+| + || Sbjct 70 LLVKKL#TDTARAPTRGSAFAAGYDLYAAKETIIP 103 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 2 PCSEETPAISPSKRARPAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | +| ++|| | +|#| || |||||+ +|||||||| + +| Sbjct 530 PKTENNSVVAPSN----------TLLVKKL#SPFATLPTRGSSLSAGYDLYSAEETVVP 577 |
| Neurospora crassa | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | +|#|+ | |||||| |||||+|++ + ||| Sbjct 18 LLIKKL#SDKARLPTRGSAFAAGYDIYASKETTIP 51 |
| Tree shrew adenovirus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | | ||#| || || |||| |||+|| + + ||| Sbjct 4 LYFVRL#SPHAYAPERGSALAAGFDLRAPREVLIPP 38 |
| Aspergillus oryzae RIB40 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | +|#+| ||||||| |||||+|+| + || Sbjct 57 LLIKKL#NEGGRAPTRGSAFAAGYDVYAAKETVIP 90 |
| Arabidopsis thaliana | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 + +|#|| | ||||| +||||| || | +| Sbjct 29 FKVKKL#SEKAVIPTRGSPLSAGYDLSSAVDSKVP 62 |
| lumpy skin disease virus | Query 30 RL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 ||#| | ||+ + +||||||||||| + Sbjct 12 RL#SAFAKLPTKSTDFSAGYDLYSAYDYIVK 41 |
| Staphylococcus phage EW | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 ||+ |#||+|| ||| + ||+|+|+| |+ | Sbjct 3 QLQIKLL#SENATLPTRNHSTDAGFDIYAAETITLEP 38 |
| Lumpy skin disease virus NI-2490 | Query 30 RL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 ||#| | ||+ + +||||||||||| + Sbjct 12 RL#SAFAKLPTKSTDFSAGYDLYSAYDYIVK 41 |
| Debaryomyces hansenii CBS767 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 || #||+|| ||||| +||||+|++ + || Sbjct 20 LRVFLR#SENATLPTRGSVLSAGYDIYASEEAVIP 53 |
| Sheeppox virus | Query 30 RL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 ||#| | ||+ + +||||||||||| + Sbjct 12 RL#SAFAKLPTKSTDFSAGYDLYSAYDYIVK 41 |
| Chaetomium globosum CBS 148.51 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | +|#|+ | |||||| ||||||++| ||| Sbjct 38 LLIKKL#SDKARLPTRGSAFAAGYDLFAARPTTIP 71 |
| Ustilago maydis 521 | Query 12 PSKRARPAEVGGM------QLRFARL#SEH--ATAPTRGSARAAGYDLYSAYDYTIP 59 |+| |+ | | | +| | # | | |||||| ||||||||| +| Sbjct 17 PTKLAKLEEAGTMSVNSTVRLLVQRH#PSHPLAKLPTRGSALAAGYDLYSAEKVVLP 72 |
| Entamoeba histolytica HM-1:IMSS | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | +|# | | ||||| ||| |||| ++ | | Sbjct 5 LLVKKL#VEDAIVPTRGSKCAAGIDLYSNTNFIIQP 39 |
| Paramecium bursaria Chlorella virus 1 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 | +|# | || | ||| |||||+ | | +| | Sbjct 4 LLVKKL#VESATTPMRGSEGAAGYDISSVEDVVVPAM 39 |
| Bacillus clausii KSM-K16 | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +++ |#+|+| || || |||+|||+| | | | Sbjct 3 RVKVKLL#NENAVMPTYGSEGAAGFDLYAAEDVIIEP 38 |
| Dictyostelium discoideum AX4 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 + +|#|+ | | ||| |||||| ||++ +| Sbjct 38 FKVKKL#SDKAIIPQRGSKGAAGYDLSSAHELVVP 71 |
| Caenorhabditis elegans | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 + +|| +|#+|+| || || ||| ||||| | |+| Sbjct 185 ITVRFTQL#NENAQTPTYGSEEAAGADLYSAEDITVP 220 Query 5 EETPAISPSKRAR-PAEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | ||+ || + + ++ + #+++| || ||| ||| ||||| | |+| Sbjct 328 EAEPALKKSKTEQIKTSATSVTIQITKS#NDNAQMPTYGSAEAAGADLYSAEDVTVP 383 Query 2 PCSEETPAISPSKRARPAE-VGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 || + ||| || + +|| +# | || || +|| ||||| | +| Sbjct 8 PCVQ--TVTEPSKMEMQAESTSHITIRFTEM#VGDAQKPTYGSISSAGADLYSAEDVVVP 64 |
| Caenorhabditis briggsae AF16 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 + +|| +|#+|+| ||| || ||| ||||| | +| Sbjct 185 IAVRFTQL#NENAHAPTYGSEEAAGADLYSAEDVVVP 220 Query 5 EETPAISPSKRARPAEVG--GMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | | + | +| | + + | + #+|+| || || ||| ||||| | |+| Sbjct 328 ENEPVVKKVK-TNQSETGLISVSIEFTKS#TENAHMPTNGSETAAGADLYSAEDVTVP 383 Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 + ||+ |#+ | || || ||| ||||| | +| Sbjct 29 LTLRYTEL#NSDAKMPTYGSPSAAGADLYSAEDVVVP 64 |
| Lycopersicon esculentum | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | +|#||+| |+| |+ |||||| || + +| Sbjct 32 FRVKKL#SENAVLPSRASSLAAGYDLSSAAETKVP 65 |
| Lactococcus phage Tuc2009 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | +| +|#+|+|| | | + +||||+ ++ || | Sbjct 1 MTRKFKKL#NENATIPERATKHSAGYDISASETVTIQP 37 |
| Fowl adenovirus 8 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 ++| + | #| | || + ++ +|||||+|| | +|| Sbjct 13 VKLLYQRH#SPFAIAPAKATSGSAGYDLFSAADVVVPP 49 |
| Candida albicans SC5314 | Query 37 APTRGSARAAGYDLYSAYDYTIP 59 ||+||| ||||||||| ||| Sbjct 30 VPTKGSALAAGYDLYSAEAATIP 52 |
| Lactococcus lactis subsp. cremoris SK11 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | | +|#+|+|| | | + +||||+ ++ || | Sbjct 1 MTREFKKL#NENATIPERATKHSAGYDISASETVTIQP 37 |
| Agrotis segetum granulovirus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | + +|# + | || ||| ||| || | + ||| Sbjct 16 LFYTKL#RKDAHAPVRGSVGAAGLDLASVENIQIPP 50 |
| Staphylococcus haemolyticus JCSC1435 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | +|#|| | ||| + +| ||| | | ||| Sbjct 4 LPIKKL#SEKAILPTRANPTDSGLDLYVAEDTTIP 37 |
| Entamoeba histolytica HM-1:IMSS | Query 28 FARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|# + | ||||+ ++ |+|||| | | | Sbjct 1 IKKL#HKDAIIPTRGTEQSVGFDLYSIDDNIIYP 33 |
| Fowl adenovirus D | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 ++| | + #| | | | ++ |||||| |+ | +|| Sbjct 13 VKLLFKKH#SPFAVTPQRATSGAAGYDLCSSADVVVPP 49 |
| Cryptococcus neoformans var. neoformans B-3501A | Query 31 L#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 |#|+ || || || ||| |||+| || Sbjct 6 L#SDRATLPTHGSVFAAGMDLYAAEHKIIP 34 |
| Lactococcus phage TP901-1 | Query 28 FARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | +|#+|+|| | | + +||||+ ++ || | Sbjct 5 FKKL#NENATIPERATEHSAGYDISASETVTIQP 37 |
| Amsacta moorei entomopoxvirus | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTI 58 ++ +#+|+| | | ++++|| || ||||| + Sbjct 5 FKYMFV#TENAFEPIRQTSKSAGMDLKSAYDYIV 37 |
| Chrysodeixis chalcites nucleopolyhedrovirus | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 + | +|#++ | || | + +||||||+ | + | Sbjct 28 NMYFKKL#NKDAIAPQRATLGSAGYDLYTPTDVCLKP 63 |
| Fowl adenovirus 4 | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +| + ++# | || | | ||| ||+| | +|| Sbjct 12 KLLYKKV#KSDAFAPVRMSPDAAGLDLFSCEDVVVPP 47 |
| Fowl adenovirus 10 | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +| + ++# | || | | ||| ||+| | +|| Sbjct 30 KLLYKKV#KSDAFAPVRMSPDAAGLDLFSCEDVVVPP 65 |
| Lactococcus phage ul36 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 |+ +| +|#+ | | | + +||||+ |+ || | Sbjct 1 MKRKFLKL#NTQAIKPERATKHSAGYDISSSETVTIQP 37 |
| Lactococcus phage bIL309 | Query 28 FARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | +|# |+|| | | + +||||+ ++ || | Sbjct 5 FKKL#DENATIPERATEHSAGYDISASETVTIQP 37 |
| Lactococcus phage bIL285 | Query 28 FARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | +|# |+|| | | + +||||+ ++ || | Sbjct 5 FKKL#DENATIPERATEHSAGYDISASETVTIQP 37 |
| Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 | Query 32 SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 |+ | | | + +||||| +| | ||| + Sbjct 13 DENITVPKRSTQQAAGYDFEAAVDITIPSV 42 |
| Bacillus thuringiensis serovar israelensis ATCC 35646 | Query 10 ISPSKRARPAEV-GGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 + +|+ | || + | #| ||| + |||||+ + || ||| Sbjct 6 LFKNKKVRGFEVLNDFKEHFENF#S--VLKPTRKTLNAAGYDIAAVGDYVIPP 55 |
| Heliothis zea virus 1 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 |++ +|#|++| | + +||+|| ||++ || Sbjct 6 LKYRKL#SKNAKMPIHSTEESAGFDLCSAHEVVIP 39 |
| Wolinella succinogenes DSM 1740 | Query 19 AEVGGMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPPM 61 +|+ |++| +|# +| | || |||+|| |||+ Sbjct 2 SELKKMKIRLLKL#HPNAVIPAYQSAGAAGFDLCCVESLEIPPL 44 |
| Corynebacterium jeikeium K411 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 || ||# + | | || |||+| | || | Sbjct 12 LRIVRL#DKELPLPRRAHPTDAGIDLYTAQDVTIAP 46 |
| Lactococcus phage bIL286 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | +| +|#+ +|| | | + +||||+ ++ || | Sbjct 1 MTRKFKKL#NGNATIPERATEHSAGYDISASETVTIQP 37 |
| Spodoptera exigua nucleopolyhedrovirus | Query 23 GMQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTI 58 | ||| + #|+ | | | ||||||++ |+ | Sbjct 3 GQVLRFKKT#SKSAYTPRMASDGAAGYDLHTPVDFVI 38 |
| Rhodobacter sphaeroides ATCC 17025 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDL 50 | |+ ||#| | | ||| |||||| Sbjct 1 MALQVQRL#STTACLPVRGSDEAAGYDL 27 |
| Staphylococcus phage 71 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 |+ |#||+| | | ||||++|| + + | Sbjct 5 LQVKLL#SENARMPERNHKTDAGYDIFSAENVVLEP 39 |
| bacteriophage phi31.1 | Query 28 FARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | +|# +|| | | + +||||+ |+ || | Sbjct 5 FKKL#DGNATIPERATEHSAGYDISSSETVTIQP 37 |
| Kluyveromyces lactis | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 +||| #| | ||+|| |||||+|++ || Sbjct 11 VQLR----#SADAKVPTKGSTTAAGYDIYASQPGVIP 42 |
| Lymantria dispar nucleopolyhedrovirus | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +||+ + #|| | || ||| || || + ||| Sbjct 7 ELRYVKT#SERAGDLAHASAGAAGLDLRSAEEAVIPP 42 |
| Taro bacilliform virus | Query 38 PTRGSARAAGYDLYSAYDYTIPP 60 ||+ | ++||||| | | ||| Sbjct 949 PTKKSNKSAGYDLQSNIDIEIPP 971 |
| Helicobacter hepaticus ATCC 51449 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIP 59 | ++ +|# || |+ + +|||+||++ | | Sbjct 5 MHIKIKKL#HSHAVIPSYQTPQAAGFDLHAVEDSLIK 40 |
| Encephalitozoon cuniculi GB-M1 | Query 25 QLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 ++| ++#+ | | | | |||||++| ||| Sbjct 5 EIRVRKI#NPRAKIPKRQSEGAAGYDIHSVESGRIPP 40 |
| Lactococcus lactis | Query 28 FARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 | +|# +|| | | + +||||+ ++ || | Sbjct 5 FKKL#DGNATIPERATKHSAGYDISASETVTIQP 37 |
| Staphylococcus aureus subsp. aureus MRSA252 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|+| |#||+| | | ||||++|| + | Sbjct 5 LQVRL--L#SENARMPERNHKTDAGYDIFSAETVVLEP 39 |
| Staphylococcus phage 187 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 |+ |#||+| | | ||||++|| + | Sbjct 5 LQVKLL#SENARMPERNHKTDAGYDIFSAETVVLEP 39 |
| Staphylococcus phage 11 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|+| |#||+| | | ||||++|| + | Sbjct 5 LQVRL--L#SENARMPERNHKTDAGYDIFSAETVVLEP 39 |
| Staphylococcus phage ROSA | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 |+ |#||+| | | ||||++|| + | Sbjct 5 LQVKLL#SENARMPERNHKTDAGYDIFSAETVVLEP 39 |
| Staphylococcus phage phiSLT | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 |+ |#||+| | | ||||++|| + | Sbjct 5 LQVKLL#SENARMPERNHKTDAGYDIFSAETVVLEP 39 |
| Staphylococcus aureus subsp. aureus USA300 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 |+ |#||+| | | ||||++|| + | Sbjct 5 LQVKLL#SENARMPERNHKTDAGYDIFSAETVVLEP 39 |
| Staphylococcus aureus subsp. aureus NCTC 8325 | Query 24 MQLRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 +|+| |#||+| | | ||||++|| + | Sbjct 14 LQVRL--L#SENARMPERNHKTDAGYDIFSAETVVLEP 48 |
| Staphylococcus phage 29 | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 |+ |#||+| | | ||||++|| + | Sbjct 5 LQVKLL#SENARMPERNHKTDAGYDIFSAETVVLEP 39 |
| Staphylococcus phage PVL | Query 26 LRFARL#SEHATAPTRGSARAAGYDLYSAYDYTIPP 60 |+ |#||+| | | ||||++|| + | Sbjct 11 LQVKLL#SENARMPERNHKTDAGYDIFSAETVVLEP 45 |
* References
[PubMed ID: 24311590] Rona G, Marfori M, Borsos M, Scheer I, Takacs E, Toth J, Babos F, Magyar A, Erdei A, Bozoky Z, Buday L, Kobe B, Vertessy BG, Phosphorylation adjacent to the nuclear localization signal of human dUTPase abolishes nuclear import: structural and mechanistic insights. Acta Crystallogr D Biol Crystallogr. 2013 Dec;69(Pt 12):2495-505. doi:
[PubMed ID: 23894549] ... Ariza ME, Rivailler P, Glaser R, Chen M, Williams MV, Epstein-Barr virus encoded dUTPase containing exosomes modulate innate and adaptive immune responses in human dendritic cells and peripheral blood mononuclear cells. PLoS One. 2013 Jul 22;8(7):e69827. doi: 10.1371/journal.pone.0069827. Print 2013.
[PubMed ID: 22658674] ... Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW, Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012 Jun 8;149(6):1393-406. doi: 10.1016/j.cell.2012.04.031. Epub 2012 May
[PubMed ID: 22172489] ... Wilson PM, LaBonte MJ, Lenz HJ, Mack PC, Ladner RD, Inhibition of dUTPase induces synthetic lethality with thymidylate synthase-targeted therapies in non-small cell lung cancer. Mol Cancer Ther. 2012 Mar;11(3):616-28. doi: 10.1158/1535-7163.MCT-11-0781. Epub
[PubMed ID: 21780905] ... Merenyi G, Kovari J, Toth J, Takacs E, Zagyva I, Erdei A, Vertessy BG, Cellular response to efficient dUTPase RNAi silencing in stable HeLa cell lines perturbs expression levels of genes involved in thymidylate metabolism. Nucleosides Nucleotides Nucleic Acids. 2011 Jun;30(6):369-90. doi:
[PubMed ID: 8631817] ... Ladner RD, Carr SA, Huddleston MJ, McNulty DE, Caradonna SJ, Identification of a consensus cyclin-dependent kinase phosphorylation site unique to the nuclear form of human deoxyuridine triphosphate nucleotidohydrolase. J Biol Chem. 1996 Mar 29;271(13):7752-7.
[PubMed ID: 8631817] ... Ladner RD, Carr SA, Huddleston MJ, McNulty DE, Caradonna SJ, Identification of a consensus cyclin-dependent kinase phosphorylation site unique to the nuclear form of human deoxyuridine triphosphate nucleotidohydrolase. J Biol Chem. 1996 Mar 29;271(13):7752-7.
[PubMed ID: 8631816] ... Ladner RD, McNulty DE, Carr SA, Roberts GD, Caradonna SJ, Characterization of distinct nuclear and mitochondrial forms of human deoxyuridine triphosphate nucleotidohydrolase. J Biol Chem. 1996 Mar 29;271(13):7745-51.
[PubMed ID: 8389461] ... Strahler JR, Zhu XX, Hora N, Wang YK, Andrews PC, Roseman NA, Neel JV, Turka L, Hanash SM, Maturation stage and proliferation-dependent expression of dUTPase in human T cells. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):4991-5.
[PubMed ID: 1325640] ... McIntosh EM, Ager DD, Gadsden MH, Haynes RH, Human dUTP pyrophosphatase: cDNA sequence and potential biological importance of the enzyme. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8020-4.
[PubMed ID: 1091006] ... Babicev SI, Briskin BS, [Method for plastic surgery of the small intestine after the resection of the esophagus and proximal portion of the stomach for cancer]. Rozhl Chir. 1975 Jan;54(1):11-6.