SB0134 : Glycogen synthase kinase-3[beta] (GSK-3[beta])
[ CaMP Format ]
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | glycogen synthase kinase-3 beta [Rattus norvegicus]; glycogen synthase kinase-3 beta; FA; factor A; GSK-3 beta; serine/threonine-protein kinase GSK3B; Glycogen synthase kinase-3 beta; 2.7.11.26; Factor A; Serine/threonine-protein kinase GSK3B; 2.7.11.1 |
| Gene Names | Gsk3b; glycogen synthase kinase 3 beta |
| Gene Locus | 11q21; chromosome 11 |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 420 aa
Average Mass: 46.710 kDa
Monoisotopic Mass: 46.681 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
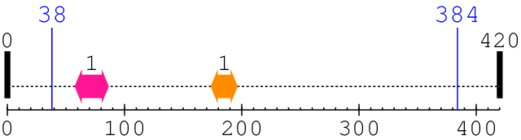
| --- cleavage 38 --- | ||||
| ABC1 family 1. | 58 | 86 | 15.0 | 0.0 |
| Phosphotransferase enzyme family 1. | 174 | 196 | 165.0 | 0.1 |
3. Sequence Information
Fasta Sequence: SB0134.fasta
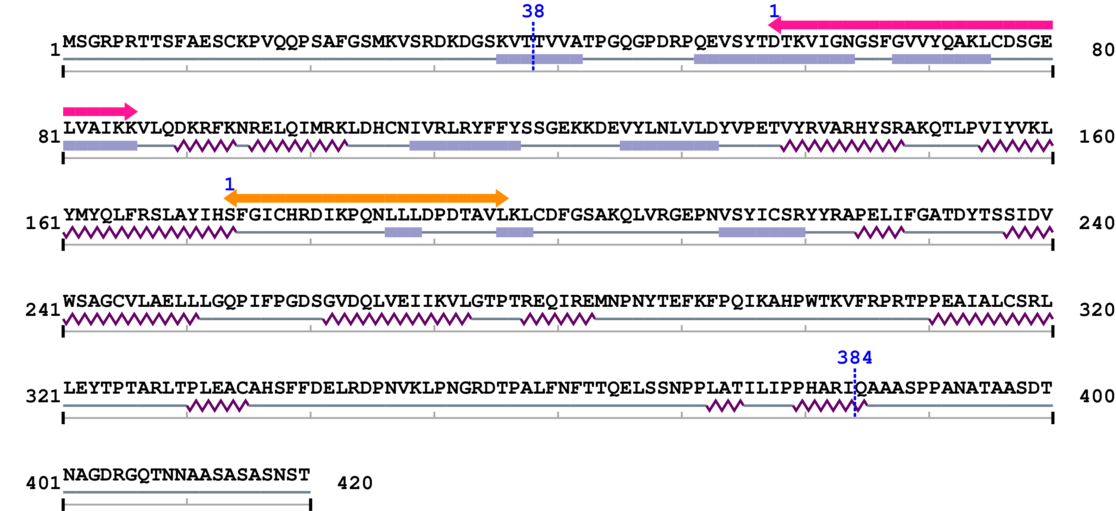
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 22496446] Ma S, Liu S, Huang Q, Xie B, Lai B, Wang C, Song B, Li M, Site-specific phosphorylation protects glycogen synthase kinase-3beta from calpain-mediated truncation of its N and C termini. J Biol Chem. 2012 Jun 29;287(27):22521-32. doi: 10.1074/jbc.M111.321349. Epub
Cleavage sites (±10aa)
[Site 1] SRDKDGSKVT38-TVVATPGQGP
Thr38  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser29 | Arg30 | Asp31 | Lys32 | Asp33 | Gly34 | Ser35 | Lys36 | Val37 | Thr38 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr39 | Val40 | Val41 | Ala42 | Thr43 | Pro44 | Gly45 | Gln46 | Gly47 | Pro48 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SFAESCKPVQQPSAFGSMKVSRDKDGSKVTTVVATPGQGPDRPQEVSYTDTKVIGNGSFG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 124.00 | 15 | protein kinase |
| 2 | Canis familiaris | 124.00 | 9 | PREDICTED: similar to Glycogen synthase kinase-3 |
| 3 | Homo sapiens | 124.00 | 3 | glycogen synthase kinase 3 beta |
| 4 | Rattus norvegicus | 124.00 | 2 | GSK3B_RAT Glycogen synthase kinase-3 beta (GSK-3 |
| 5 | synthetic construct | 124.00 | 1 | glycogen synthase kinase 3 beta |
| 6 | Mus musculus | 124.00 | 1 | glycogen synthase kinase 3 beta |
| 7 | Mus musculus | 122.00 | 2 | unnamed protein product |
| 8 | Danio rerio | 121.00 | 2 | glycogen synthase kinase 3 beta |
| 9 | Danio rerio | 121.00 | 2 | glycogen synthase kinase 3 beta |
| 10 | Xenopus laevis | 119.00 | 1 | intracellular kinase |
| 11 | Tetraodon nigroviridis | 110.00 | 2 | unnamed protein product |
| 12 | Apis mellifera | 78.60 | 1 | PREDICTED: similar to shaggy CG2621-PA, isoform A |
| 13 | Ciona intestinalis | 74.30 | 1 | glycogen synthase kinase alpha/beta |
| 14 | Drosophila melanogaster | 73.90 | 9 | protein kinase |
| 15 | Drosophila melanogaster | 73.60 | 5 | protein kinase; sgg protein kinase |
| 16 | Paracentrotus lividus | 72.40 | 1 | GSK3 beta |
| 17 | Glossina morsitans morsitans | 69.70 | 1 | shaggy |
| 18 | Hydra vulgaris | 68.60 | 1 | AF272672_1 serine/threonine kinase GSK3 |
| 19 | Suberites domuncula | 64.70 | 1 | glycogen synthase kinase 3 |
| 20 | Drosophila pseudoobscura | 49.70 | 1 | GA15928-PA |
| 21 | Homo sapiens | 42.00 | 1 | glycogen synthase kinase 3 beta |
| 22 | Caenorhabditis elegans | 40.80 | 3 | AF159950_1 GSK-3 |
| 23 | Caenorhabditis briggsae | 40.80 | 1 | Hypothetical protein CBG07972 |
| 24 | Ustilago maydis 521 | 36.60 | 1 | hypothetical protein UM00560.1 |
| 25 | Ustilago hordei | 35.00 | 1 | glycogen synthase kinase |
| 26 | Triticum aestivum | 31.20 | 1 | GSK-like kinase |
| 27 | Arabidopsis thaliana | 30.80 | 2 | shaggy-like kinase alpha |
| 28 | Cryptococcus neoformans var. neoformans JEC21 | 30.80 | 1 | glycogen synthase kinase 3 |
| 29 | Streptomyces avermitilis MA-4680 | 30.40 | 1 | hypothetical protein SAV_7456 |
| 30 | Neurospora crassa OR74A | 30.40 | 1 | protein kinase gsk3 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |
| Canis familiaris | Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |
| Homo sapiens | Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |
| Rattus norvegicus | Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |
| synthetic construct | Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |
| Mus musculus | Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |
| Mus musculus | Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |
| Danio rerio | Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 9 SFAESCKPVPQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |
| Danio rerio | Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 9 SFAESCKPVPQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |
| Xenopus laevis | Query 9 SFAESCKPVQQPSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |||||||||||||+||||||||||||||||#|||||||||||| |||+||||||||||||| Sbjct 9 SFAESCKPVQQPSSFGSMKVSRDKDGSKVT#TVVATPGQGPDRQQEVTYTDTKVIGNGSFG 68 |
| Tetraodon nigroviridis | Query 9 SFAESCKPVQQPSAFGSMKVSR---------DKDGSKVT#TVVATPGQGPDRPQEVSYTDT 59 ||||||||| |||||||||||+ ||||||||#||||||||||||||||||||| Sbjct 9 SFAESCKPVPQPSAFGSMKVSQTLNGPWRGGDKDGSKVT#TVVATPGQGPDRPQEVSYTDT 68 Query 60 KVIGNGSFG 68 ||||||||| Query 60 KVIGNGSFG 68 |
| Apis mellifera | Query 24 GSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 | + + +|+||+|||#||||||| |||||||+|||||||||||||| Sbjct 33 GGVTLKKDRDGNKVT#TVVATPGAGPDRPQEISYTDTKVIGNGSFG 77 |
| Ciona intestinalis | Query 21 SAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 + |+|| ||||+ ||+|#||||| | ||||||||+||||||||||||| Sbjct 7 TTLGNMKGSRDKE-SKIT#TVVATHGHGPDRPQEVAYTDTKVIGNGSFG 53 |
| Drosophila melanogaster | Query 9 SFAESCKPVQQPS-AFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSF 67 |||| | | || | +| +||||+|#||||||||| || |||||||||||||||| Sbjct 9 SFAEGNK--QSPSLVLGGVKTC-SRDGSKIT#TVVATPGQGTDRVQEVSYTDTKVIGNGSF 65 Query 68 G 68 | Query 68 G 68 |
| Drosophila melanogaster | Query 9 SFAESCKPVQQPS-AFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSF 67 |||| | | || | +| +||||+|#||||||||| || |||||||||||||||| Sbjct 9 SFAEGNK--QSPSLVLGGVKTC-SRDGSKIT#TVVATPGQGTDRVQEVSYTDTKVIGNGSF 65 Query 68 G 68 | Query 68 G 68 |
| Paracentrotus lividus | Query 21 SAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |++| +||||||| ||+|#+| || | ||| +|+|||||+|||||||| Sbjct 21 SSYGGLKVSRDKDSSKIT#SVTATEGPPPDRTKEISYTDTRVIGNGSFG 68 |
| Glossina morsitans morsitans | Query 32 KDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 +||+|+|#||||||||| || ||||||||||||||||| Sbjct 267 RDGAKIT#TVVATPGQGTDRVQEVSYTDTKVIGNGSFG 303 |
| Hydra vulgaris | Query 20 PSAFGSMKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 || | ++++|||||||||#+|+|| ||+ +|+|| ||||||||||| Sbjct 44 PSNNGGIRITRDKDGSKVT#SVLATTASYPDQTEEISYCDTKVIGNGSFG 92 |
| Suberites domuncula | Query 27 KVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 +++|||||+|||#||+|| |+ ||| ||++| + ||||||||| Sbjct 36 RITRDKDGNKVT#TVLATVGKPPDRQQEINYCEVKVIGNGSFG 77 |
| Drosophila pseudoobscura | Query 35 SKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 +|| #||||| | | | |+|||||||+|||||| Sbjct 7 NKVI#TVVATNGYGADTMSEISYTDTKVVGNGSFG 40 |
| Homo sapiens | Query 9 SFAESCKPVQQPSAFGSMK 27 ||||||||||||||||||| Query 9 SFAESCKPVQQPSAFGSMK 27 |
| Caenorhabditis elegans | Query 32 KDGSKVT#TVVATPG-QGPDRPQEVSYTDTKVIGNGSFG 68 | | +||# |||+ | |+ |+|| | ||||||||| Sbjct 11 KSGKQVT#MVVASVATDGVDQQVEISYYDQKVIGNGSFG 48 |
| Caenorhabditis briggsae | Query 32 KDGSKVT#TVVATPG-QGPDRPQEVSYTDTKVIGNGSFG 68 | | +||# |||+ | |+ |+|| | ||||||||| Sbjct 11 KSGKQVT#MVVASVATDGVDQQVEISYYDQKVIGNGSFG 48 |
| Ustilago maydis 521 | Query 26 MKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 +|++ | +|| # |+|+ |+ ++ +|++||+ ||||||||| Sbjct 9 VKLNPLDDPNKVI#KVLASDGKTGEQ-REIAYTNCKVIGNGSFG 50 |
| Ustilago hordei | Query 26 MKVSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 +|++ | +|| # |+|+ + ++ +|++||+ ||||||||| Sbjct 9 VKLNPLDDPNKVI#KVLASDSKTAEQ-REIAYTNCKVIGNGSFG 50 |
| Triticum aestivum | Query 24 GSMKVSRDKDGSKVT#T-----------VVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 |+| + |+| + # | + |+ | +|| +|+|||||| Sbjct 2 GNMSIRDDRDPEDIV#VNGNGTEPGHIIVTSIEGRNGQAKQTISYMAERVVGNGSFG 57 |
| Arabidopsis thaliana | Query 20 PSAFGSMKVSRDK-------DGSKVT#T---VVATPGQGPDRP-QEVSYTDTKVIGNGSFG 68 | ||+ || ||+ #| +| | | +| | +|| +|+|+|||| Sbjct 22 PEEMNDMKIRDDKEMEATVVDGNGTE#TGHIIVTTIGGRNGQPKQTISYMAERVVGHGSFG 81 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 26 MKVSRDKDGSKVT#TVVATPGQ-GPDRPQEVSYTDTKVIGNGSFG 68 ++|+ | | ++| #|| | |+ | | +|||+ | +|||||| Sbjct 8 VRVATD-DPNRVI#TVSAQWGKTGAD--TTISYTNCKAVGNGSFG 48 |
| Streptomyces avermitilis MA-4680 | Query 13 SCKPVQQPSAFGSMK---VSRDKDGSKVT#TVVATPGQGPD 49 || | + |++| | + |+|| +| #| | + ||+ Sbjct 172 SCDPAELPASFTDSKPYEIWTDRDGRRVA#TTVVSSNDGPE 211 |
| Neurospora crassa OR74A | Query 19 QPSAFGSMK---VSRDKDGSKVT#TVVATPGQGPDRPQEVSYTDTKVIGNGSFG 68 +|+|| +++ | |+| ||# +|+ || |++|||||| Sbjct 5 RPAAFNTLRMGEVIREKVQDGVT#----------GETREIQYTQCKIVGNGSFG 47 |
[Site 2] TILIPPHARI384-QAAASPPANA
Ile384  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Thr375 | Ile376 | Leu377 | Ile378 | Pro379 | Pro380 | His381 | Ala382 | Arg383 | Ile384 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln385 | Ala386 | Ala387 | Ala388 | Ser389 | Pro390 | Pro391 | Ala392 | Asn393 | Ala394 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DTPALFNFTTQELSSNPPLATILIPPHARIQAAASPPANATAASDTNAGDRGQTNNAASA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Rattus norvegicus | 119.00 | 2 | GSK3B_RAT Glycogen synthase kinase-3 beta (GSK-3 |
| 2 | Mus musculus | 119.00 | 1 | glycogen synthase kinase 3 beta |
| 3 | Canis familiaris | 103.00 | 9 | PREDICTED: similar to Glycogen synthase kinase-3 |
| 4 | N/A | 101.00 | 13 | GSK3B_HUMAN Glycogen synthase kinase-3 beta (GSK- |
| 5 | Homo sapiens | 101.00 | 3 | glycogen synthase kinase 3 beta |
| 6 | synthetic construct | 101.00 | 1 | glycogen synthase kinase 3 beta |
| 7 | Danio rerio | 73.90 | 2 | glycogen synthase kinase 3 beta |
| 8 | Danio rerio | 72.00 | 2 | glycogen synthase kinase 3 beta |
| 9 | Xenopus laevis | 58.50 | 1 | intracellular kinase |
| 10 | Ciona intestinalis | 38.50 | 1 | glycogen synthase kinase alpha/beta |
| 11 | Drosophila melanogaster | 37.00 | 13 | SD09379p |
| 12 | Drosophila melanogaster | 37.00 | 6 | shaggy CG2621-PG, isoform G |
| 13 | Mus musculus | 37.00 | 2 | Gsk3a protein |
| 14 | Aspergillus fumigatus Af293 | 37.00 | 1 | glycogen synthase kinase (Skp1) |
| 15 | Cryptococcus neoformans var. neoformans JEC21 | 36.60 | 1 | glycogen synthase kinase 3 |
| 16 | Aspergillus nidulans FGSC A4 | 35.80 | 1 | hypothetical protein AN6508.2 |
| 17 | Paracentrotus lividus | 35.80 | 1 | GSK3 beta |
| 18 | Magnaporthe grisea | 35.00 | 1 | glycogen synthase kinase |
| 19 | Aspergillus oryzae RIB40 | 35.00 | 1 | hypothetical protein |
| 20 | Yarrowia lipolytica | 35.00 | 1 | hypothetical protein |
| 21 | Hydra vulgaris | 34.70 | 1 | AF272672_1 serine/threonine kinase GSK3 |
| 22 | Arabidopsis thaliana | 34.30 | 4 | Identical to A. thaliana AtK-1 (gb |
| 23 | Medicago sativa | 34.30 | 2 | AF432225_1 GSK-3-like protein MsK4 |
| 24 | Chaetomium globosum CBS 148.51 | 34.30 | 1 | hypothetical protein CHGG_07166 |
| 25 | Gibberella zeae PH-1 | 34.30 | 1 | hypothetical protein FG07329.1 |
| 26 | Arabidopsis thaliana | 34.30 | 1 | shaggy-like kinase kappa |
| 27 | Physcomitrella patens | 33.90 | 3 | shaggy-related protein kinase 3 |
| 28 | Tetraodon nigroviridis | 33.90 | 1 | unnamed protein product |
| 29 | Petunia x hybrida | 33.10 | 1 | shaggy kinase 7 |
| 30 | Schizosaccharomyces pombe 972h- | 32.30 | 2 | serine/threonine protein kinase Gsk3 |
| 31 | Nicotiana tabacum | 32.00 | 2 | shaggy-like kinase 91 |
| 32 | Ustilago maydis 521 | 32.00 | 1 | hypothetical protein UM00560.1 |
| 33 | Neurospora crassa OR74A | 32.00 | 1 | protein kinase gsk3 |
| 34 | Nicotiana tabacum | 32.00 | 1 | NSK6; Shaggy-like kinase 6 |
| 35 | Schizosaccharomyces pombe | 31.60 | 1 | pi064 |
| 36 | Dictyostelium discoideum AX4 | 31.60 | 1 | glycogen synthase kinase 3 |
| 37 | Medicago sativa subsp. x varia | 31.60 | 1 | wound-induced GSK-3-like protein |
| 38 | Schizosaccharomyces pombe | 31.20 | 2 | unnamed protein product |
| 39 | Ustilago hordei | 30.80 | 1 | glycogen synthase kinase |
Top-ranked sequences
| organism | matching |
|---|---|
| Rattus norvegicus | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDRGQTNNAASA 414 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDRGQTNNAASA 414 |
| Mus musculus | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDRGQTNNAASA 414 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDRGQTNNAASA 414 |
| Canis familiaris | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDRGQT 408 ||||||||||||||||||||||||||||||#||||| | ||||||| |||||||| Sbjct 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASTPTNATAASDANAGDRGQT 408 |
| N/A | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDRGQT 408 ||||||||||||||||||||||||||||||#||||| | ||||||| | |||||| Sbjct 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASTPTNATAASDANTGDRGQT 408 |
| Homo sapiens | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDRGQT 408 ||||||||||||||||||||||||||||||#||||| | ||||||| | |||||| Sbjct 368 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASTPTNATAASDANTGDRGQT 421 |
| synthetic construct | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDRGQT 408 ||||||||||||||||||||||||||||||#||||| | ||||||| | |||||| Sbjct 368 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASTPTNATAASDANTGDRGQT 421 |
| Danio rerio | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDRGQ 407 + |+||||||||||||| ||+|||| ||| #|| || | | +| || |+||| | Sbjct 355 EKPSLFNFTTQELSSNPTLASILIPAHARN#QAGASTPTNPSATSDANSGDRSQ 407 |
| Danio rerio | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDR 405 + |+||||||||||||| ||+|||| ||| #|| || | | +| || |+||| Sbjct 355 EKPSLFNFTTQELSSNPTLASILIPAHARN#QAGASTPTNPSATSDANSGDR 405 |
| Xenopus laevis | Query 357 PALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDRGQT 408 ||||||||||||||| |++|||| ||| #||| | +| |+ ||+| |+|| | Sbjct 357 PALFNFTTQELSSNPSLSSILIPAHARN#QAAVSTTSNTTSTSDSNTGERGST 408 |
| Ciona intestinalis | Query 357 PALFNFTTQELSSNPPLATILIPPHAR 383 | ||||| +||| || |||| ||| Sbjct 342 PPLFNFTDKELSIKSPLNNILIPLHAR 368 |
| Drosophila melanogaster | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPAN 393 | | ||||| ||| | | |+| | +#| |+ | | Sbjct 336 DMPPLFNFTEHELSIQPSLVPQLLPKH--L#QNASGPGGN 372 |
| Drosophila melanogaster | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAASPPAN 393 | | ||||| ||| | | |+| | +#| |+ | | Sbjct 336 DMPPLFNFTEHELSIQPSLVPQLLPKH--L#QNASGPGGN 372 |
| Mus musculus | Query 357 PALFNFTTQELSSNPPLATILIPPHARI#QAAASPPANATAASDTNAGDRGQTNN 410 | ||||+ ||| | | |||||| | # | + | + + | || Sbjct 195 PPLFNFSPGELSIQPSLNAILIPPHLRS#PAGPASPLTTSYNPSSQALTEAQTGQ 248 |
| Aspergillus fumigatus Af293 | Query 355 DTPALFNFTTQELSSNPPLATILIPPHAR 383 | | ||+|+ ||| | | + |+||||| Sbjct 358 DLPNLFDFSRHELSIAPALNSRLVPPHAR 386 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 355 DTPALFNFTTQELSSNPPLATILIPPHA 382 | | ||||| +|||| | | |+| || Sbjct 344 DMPELFNFTHEELSSRPDLIRQLVPTHA 371 |
| Aspergillus nidulans FGSC A4 | Query 355 DTPALFNFTTQELSSNPPLATILIPPHAR 383 | | ||+|+ ||| | + + |+|||+| Sbjct 340 DLPNLFDFSRHELSIAPSMNSRLVPPHSR 368 |
| Paracentrotus lividus | Query 355 DTPALFNFTTQELSSNPPLATILIPPH-ARI#QAAASPPANAT 395 + | |||||+ ||+ | | ||||| ++ # ++++ | +|| Sbjct 355 ELPPLFNFTSSELAIKQSLRTALIPPHISQN#SSSSTAPHDAT 396 |
| Magnaporthe grisea | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#QAAA 388 | | ||+| ||| | | |+||| | # || Sbjct 340 DLPPLFDFNRHELSIAPQLNHQLVPPHVRP#TLAA 373 |
| Aspergillus oryzae RIB40 | Query 355 DTPALFNFTTQELSSNPPLATILIPPHAR 383 + | ||+|+ ||| | + + ||||||| Sbjct 340 ELPNLFDFSRHELSIAPAMNSRLIPPHAR 368 |
| Yarrowia lipolytica | Query 357 PALFNFTTQELSSNPPLATILIPPHAR 383 ||||||| ||| | | |+| ||| Sbjct 262 PALFNFTRHELSIAPELNQKLVPAHAR 288 |
| Hydra vulgaris | Query 355 DTPALFNFTTQELSSNP---PLATILIPPHARI#QAAASPPANATAASDTNAGDRGQTNNA 411 + | ||||+ ||||| | | + |+ | # + || |+ + +| ++ | + Sbjct 379 EMPKLFNFSAQELSSKPSFSPSYSPLVKKHQSS#TSLASTPSTISNQVNTTDSNKTSINIS 438 Query 412 ASA 414 | Sbjct 439 TEA 441 |
| Arabidopsis thaliana | Query 357 PALFNFTTQELSSNPP-LATILIPPHARI#Q 385 | |||| |||| || + |+| ||| #| Sbjct 410 PPLFNFKPQELSGIPPEIVNRLVPEHARK#Q 439 |
| Medicago sativa | Query 357 PALFNFTTQELSSNPP-LATILIPPHARI#Q 385 | |||| |||| || + ||| ||| #| Sbjct 395 PPLFNFKPQELSGIPPDVINRLIPEHARK#Q 424 |
| Chaetomium globosum CBS 148.51 | Query 355 DTPALFNFTTQELSSNPPLATILIPPH 381 | ||||+|+ ||| | | |+||| Sbjct 340 DLPALFDFSRHELSIAPHLNDQLVPPH 366 |
| Gibberella zeae PH-1 | Query 355 DTPALFNFTTQELSSNPPLATILIPPHAR 383 | ||||+|| ||| | | |+| | | Sbjct 340 DLPALFDFTRHELSIAPSLNPKLVPAHIR 368 |
| Arabidopsis thaliana | Query 357 PALFNFTTQELSSNPP-LATILIPPHARI#Q 385 | |||| |||| || + |+| ||| #| Sbjct 338 PPLFNFKPQELSGIPPEIVNRLVPEHARK#Q 367 |
| Physcomitrella patens | Query 357 PALFNFTTQELS-SNPPLATILIPPHARI#Q 385 |+|||| |||| + | + ||| ||| #| Sbjct 388 PSLFNFKTQELKGATPDILQRLIPEHARK#Q 417 |
| Tetraodon nigroviridis | Query 362 FTTQELSSNPPLATILIPPHARI#QAAAS 389 | ||| | | +|||||||| # || Sbjct 370 FCVTELSIQPQLNSILIPPHART#YTTAS 397 |
| Petunia x hybrida | Query 357 PALFNFTTQELSSNPP-LATILIPPHAR 383 | ||||| |||| | | ||| ||| Sbjct 431 PPLFNFTPQELSGAPTELRQRLIPEHAR 458 |
| Schizosaccharomyces pombe 972h- | Query 357 PALFNFTTQELSSNPPLATILIPPHAR 383 | ||||+ ||| | | ||| ||| Sbjct 340 PELFNFSPFELSIRPDLNQKLIPSHAR 366 |
| Nicotiana tabacum | Query 357 PALFNFTTQELSSNPP-LATILIPPHAR 383 | ||||| |||| | | ||| | | Sbjct 443 PPLFNFTAQELSGAPADLRKRLIPEHMR 470 |
| Ustilago maydis 521 | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#Q 385 + | |||+| +||| | | + |+| || #+ Sbjct 348 ELPPLFNWTKEELSVRPDLISRLVPQHAEA#E 378 |
| Neurospora crassa OR74A | Query 355 DTPALFNFTTQELSSNPPLATILIPPHAR 383 | | ||+|| ||| | | |+| | | Sbjct 340 DLPPLFDFTRHELSIAPQLNHKLVPSHMR 368 |
| Nicotiana tabacum | Query 357 PALFNFTTQELSSNPP-LATILIPPHAR 383 | ||||| |||| | | ||| | | Sbjct 443 PPLFNFTAQELSGAPADLRKRLIPEHMR 470 |
| Schizosaccharomyces pombe | Query 357 PALFNFTTQELSSNPPLATILIPPH 381 | |||| ||| | | ++||| Sbjct 302 PPLFNFNLAELSIRPNLNKAILPPH 326 |
| Dictyostelium discoideum AX4 | Query 357 PALFNFTTQELSS-NPPLATILIPPHARI#QAAASPP 391 | ||||| | +| | || ||| || #| | Sbjct 356 PPLFNFTIAEQTSIGPKLAKTLIPSHAMN#QIELPSP 391 |
| Medicago sativa subsp. x varia | Query 357 PALFNFTTQELSSNPP-LATILIPPHAR 383 | ||||| ||| + | | ||| ||| Sbjct 440 PPLFNFTPQELVNAPEDLRQRLIPEHAR 467 |
| Schizosaccharomyces pombe | Query 357 PALFNFTTQELSSNPPLATILIPPH 381 | |||| ||| | | ++||| Sbjct 342 PPLFNFNLAELSIRPNLNKAILPPH 366 |
| Ustilago hordei | Query 355 DTPALFNFTTQELSSNPPLATILIPPHARI#Q 385 + | ||++| +||| | | + |+| || #+ Sbjct 349 EVPPLFDWTKEELSVRPDLISRLVPQHAEA#E 379 |
* References
[PubMed ID: 24425879] Desai SS, Modali SD, Parekh VI, Kebebew E, Agarwal SK, GSK-3beta protein phosphorylates and stabilizes HLXB9 protein in insulinoma cells to form a targetable mechanism of controlling insulinoma cell proliferation. J Biol Chem. 2014 Feb 28;289(9):5386-98. doi: 10.1074/jbc.M113.533612. Epub 2014
[PubMed ID: 24275526] ... Weng HR, Gao M, Maixner DW, Glycogen synthase kinase 3 beta regulates glial glutamate transporter protein expression in the spinal dorsal horn in rats with neuropathic pain. Exp Neurol. 2014 Feb;252:18-27. doi: 10.1016/j.expneurol.2013.11.018. Epub 2013
[PubMed ID: 23860320] ... Song R, Wang X, Mao Y, Li H, Li Z, Xu W, Wang R, Guo T, Jin L, Zhang X, Zhang Y, Zhou N, Hu R, Jia J, Lei Z, Irwin DM, Niu G, Tan H, Resistin disrupts glycogen synthesis under high insulin and high glucose levels by down-regulating the hepatic levels of GSK3beta. Gene. 2013 Oct 15;529(1):50-6. doi: 10.1016/j.gene.2013.06.085. Epub 2013 Jul 13.
[PubMed ID: 23940672] ... Hernandez-Baltazar D, Mendoza-Garrido ME, Martinez-Fong D, Activation of GSK-3beta and caspase-3 occurs in Nigral dopamine neurons during the development of apoptosis activated by a striatal injection of 6-hydroxydopamine. PLoS One. 2013 Aug 5;8(8):e70951. doi: 10.1371/journal.pone.0070951. Print 2013.
[PubMed ID: 23864697] ... Nelson CD, Kim MJ, Hsin H, Chen Y, Sheng M, Phosphorylation of threonine-19 of PSD-95 by GSK-3beta is required for PSD-95 mobilization and long-term depression. J Neurosci. 2013 Jul 17;33(29):12122-35. doi: 10.1523/JNEUROSCI.0131-13.2013.
[PubMed ID: 8524413] ... Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA, Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995 Dec 21-28;378(6559):785-9.
[PubMed ID: 8253190] ... Kobayashi S, Ishiguro K, Omori A, Takamatsu M, Arioka M, Imahori K, Uchida T, A cdc2-related kinase PSSALRE/cdk5 is homologous with the 30 kDa subunit of tau protein kinase II, a proline-directed protein kinase associated with microtubule. FEBS Lett. 1993 Dec 6;335(2):171-5.
[PubMed ID: 7686508] ... Ishiguro K, Shiratsuchi A, Sato S, Omori A, Arioka M, Kobayashi S, Uchida T, Imahori K, Glycogen synthase kinase 3 beta is identical to tau protein kinase I generating several epitopes of paired helical filaments. FEBS Lett. 1993 Jul 5;325(3):167-72.
[PubMed ID: 8382613] ... Hughes K, Nikolakaki E, Plyte SE, Totty NF, Woodgett JR, Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. EMBO J. 1993 Feb;12(2):803-8.
[PubMed ID: 2164470] ... Woodgett JR, Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 1990 Aug;9(8):2431-8.