SB0135 : Retinoid X receptor [alpha]
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | retinoic acid receptor RXR-alpha isoform a [Homo sapiens]; retinoic acid receptor RXR-alpha isoform a; retinoic acid receptor RXR-alpha; retinoid X nuclear receptor alpha; nuclear receptor subfamily 2 group B member 1; Retinoic acid receptor RXR-alpha; Nuclear receptor subfamily 2 group B member 1; Retinoid X receptor alpha |
| Gene Names | RXRA; NR2B1; retinoid X receptor, alpha |
| Gene Locus | 9q34.3; chromosome 9 |
| GO Function | Not available |
* Information From OMIM
Function: Zhou et al. (1995) used an in vitro model system of cardiac muscle cell hypertrophy to identify a retinoic acid-mediated pathway that suppresses the acquisition of specific features of the hypertrophic phenotype after exposure to the alpha-adrenergic receptor agonist phenylephrine. They found that retinoic acid at physiologic concentrations suppressed the increase in cell size and induction of a genetic marker for hypertrophy, namely the atrial natriuretic factor gene (ANF; OMIM:108780). Retinoic acid also suppressed endothelin-1 (EDN1; OMIM:131240) pathways for cardiac muscle cell hypertrophy. These and results of further studies suggested the possibility that a pathway for suppression of hypertrophy may exist in vivo, which may have potential therapeutic value.
* Structure Information
1. Primary Information
Length: 462 aa
Average Mass: 50.811 kDa
Monoisotopic Mass: 50.778 kDa
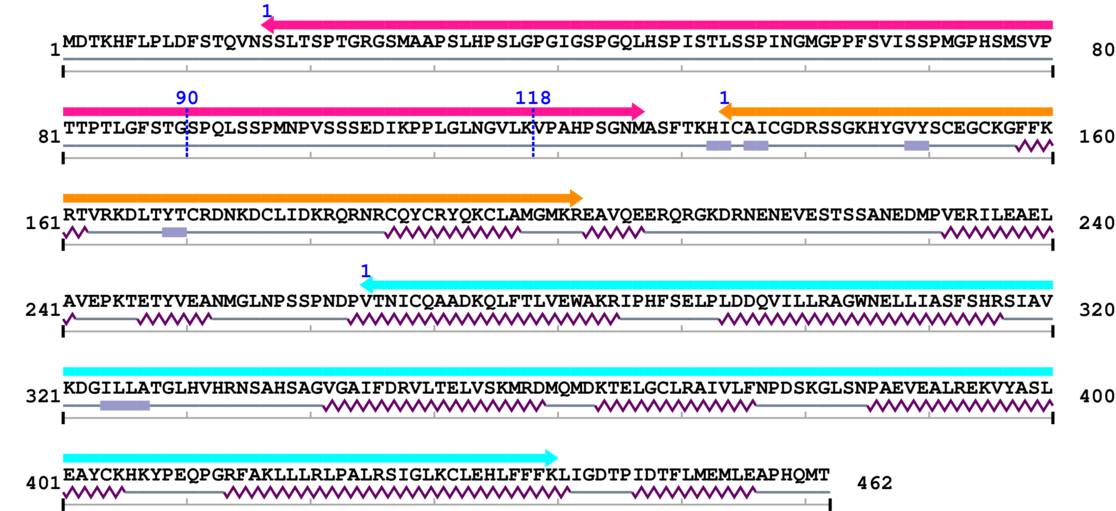
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Nuclear/hormone receptor activator site AF-1 1. | 17 | 127 | 1.0 | 21.5 |
| --- cleavage 90 (inside Nuclear/hormone receptor activator site AF-1 17..127) --- | ||||
| --- cleavage 118 (inside Nuclear/hormone receptor activator site AF-1 17..127) --- | ||||
| Zinc finger, C4 type (two domains) 1. | 134 | 202 | 2.0 | 16.3 |
| Ligand-binding domain of nuclear hormone receptor 1. | 265 | 440 | 13.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0135.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1BY4 (X-ray; 210 A; A/B/C/D=129-209), 1DSZ (X-ray; 170 A; B=129-212), 1FBY (X-ray; 225 A; A/B=224-462), 1FM6 (X-ray; 210 A; A/U=225-462), 1FM9 (X-ray; 210 A; A=225-462), 1G1U (X-ray; 250 A; A/B/C/D=225-462), 1G5Y (X-ray; 200 A; A/B/C/D=225-462), 1K74 (X-ray; 230 A; A=225-462), 1LBD (X-ray; 270 A; A=201-460), 1MV9 (X-ray; 190 A; A=223-462), 1MVC (X-ray; 190 A; A=223-462), 1MZN (X-ray; 190 A; A/C/E/G=223-462), 1R0N (X-ray; 260 A; A=130-206), 1RDT (X-ray; 240 A; A=225-462), 1RXR (NMR; -; A=130-212), 1XLS (X-ray; 296 A; A/B/C/D=227-458), 1XV9 (X-ray; 270 A; A/C=227-462), 1XVP (X-ray; 260 A; A/C=227-462), 1YNW (X-ray; 300 A; B=130-228), 2ACL (X-ray; 280 A; A/C/E/G=225-462), 2NLL (X-ray; 190 A; A=135-200), 2P1T (X-ray; 180 A; A=223-462), 2P1U (X-ray; 220 A; A=223-462), 2P1V (X-ray; 220 A; A=223-462), 2ZXZ (X-ray; 300 A; A=223-462), 2ZY0 (X-ray; 290 A; A/C=223-462), 3DZU (X-ray; 320 A; A=11-462), 3DZY (X-ray; 310 A; A=11-462), 3E00 (X-ray; 310 A; A=11-462), 3E94 (X-ray; 190 A; A=223-462), 3FAL (X-ray; 236 A; A/C=225-462), 3FC6 (X-ray; 206 A; A/C=225-462), 3FUG (X-ray; 200 A; A=223-462), 3H0A (X-ray; 210 A; A=228-455), 3KWY (X-ray; 230 A; A=223-462), 3NSP (X-ray; 290 A; A/B=223-462), 3NSQ (X-ray; 260 A; A/B=223-462), 3OAP (X-ray; 205 A; A=228-458), 3OZJ (X-ray; 210 A; A/C=225-462), 3PCU (X-ray; 200 A; A=229-458), 3R29 (X-ray; 290 A; A/B=223-462), 3R2A (X-ray; 300 A; A/B/C/D=223-462), 3R5M (X-ray; 280 A; A/C=223-462), 3UVV (X-ray; 295 A; B=225-462), 4CN2 (X-ray; 207 A; C/D=130-212), 4CN3 (X-ray; 235 A; A/B/C=130-212, D=130-173, D=175-212), 4CN5 (X-ray; 200 A; A/B=130-212), 4CN7 (X-ray; 234 A; A/B/E/F=130-212), 4J5W (X-ray; 280 A; C/D=227-462), 4J5X (X-ray; 280 A; C/D=227-462), 4K4J (X-ray; 200 A; A=228-458), 4K6I (X-ray; 210 A; A=228-458), 4M8E (X-ray; 240 A; A=228-458), 4M8H (X-ray; 220 A; A=228-458), 4N5G (X-ray; 211 A; A/B/C/D=223-462), 4N8R (X-ray; 203 A; A/B/C/D=223-462), 4NQA (X-ray; 310 A; A/H=98-462), 4OC7 (X-ray; 250 A; A=223-462), 4POH (X-ray; 230 A; A=228-458), 4POJ (X-ray; 200 A; A=228-458), 4PP3 (X-ray; 200 A; A=228-458), 4PP5 (X-ray; 200 A; A=228-458)
* Cleavage Information
2 [sites] cleaved by Calpain 2
Source Reference: [PubMed ID: 23389291] Gao W, Liu J, Hu M, Huang M, Cai S, Zeng Z, Lin B, Cao X, Chen J, Zeng JZ, Zhou H, Zhang XK, Regulation of proteolytic cleavage of retinoid X receptor-alpha by GSK-3beta. Carcinogenesis. 2013 Jun;34(6):1208-15. doi: 10.1093/carcin/bgt043. Epub 2013 Feb
Cleavage sites (±10aa)
[Site 1] TTPTLGFSTG90-SPQLSSPMNP
Gly90  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Thr81 | Thr82 | Pro83 | Thr84 | Leu85 | Gly86 | Phe87 | Ser88 | Thr89 | Gly90 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser91 | Pro92 | Gln93 | Leu94 | Ser95 | Ser96 | Pro97 | Met98 | Asn99 | Pro100 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
[Site 2] PPLGLNGVLK118-VPAHPSGNMA
Lys118  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro109 | Pro110 | Leu111 | Gly112 | Leu113 | Asn114 | Gly115 | Val116 | Leu117 | Lys118 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val119 | Pro120 | Ala121 | His122 | Pro123 | Ser124 | Gly125 | Asn126 | Met127 | Ala128 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24513686] Zhang J, Ma X, Wang H, Ma D, Huang G, Elevated methylation of the RXRA promoter region may be responsible for its downregulated expression in the myocardium of patients with TOF. Pediatr Res. 2014 May;75(5):588-94. doi: 10.1038/pr.2014.17. Epub 2014 Feb 10.