SB0140 : TAR DNA-binding protein 43, TDP-43
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | TAR DNA-binding protein 43 [Homo sapiens]; TAR DNA-binding protein 43; TAR DNA-binding protein-43; TDP-43 |
| Gene Names | TARDBP; TDP43; TAR DNA binding protein |
| Gene Locus | 1p36.22; chromosome 1 |
| GO Function | Not available |
* Information From OMIM
Description: The TARDBP gene encodes the 43-kD TAR DNA-binding protein, which was originally identified as a transcriptional repressor that binds to TAR DNA of human immunodeficiency virus type 1. It is also involved in regulation of gene expression and splicing (summary by Benajiba et al., 2009).
Function: Functional analysis by Ou et al. (1995) indicated that TARDBP does not bind RNA. Gel retardation analysis followed by Western blot analysis (Shift-Western analysis) demonstrated that the RNP-binding motifs of TARDBP bind to the pyrimidine-rich motifs of TAR DNA. In an in vitro transcription analysis, increasing amounts of TARDBP, in the presence or absence of Tat, decreased the level of transcription from the HIV-1 LTR but not from the adenovirus major late promoter.
* Structure Information
1. Primary Information
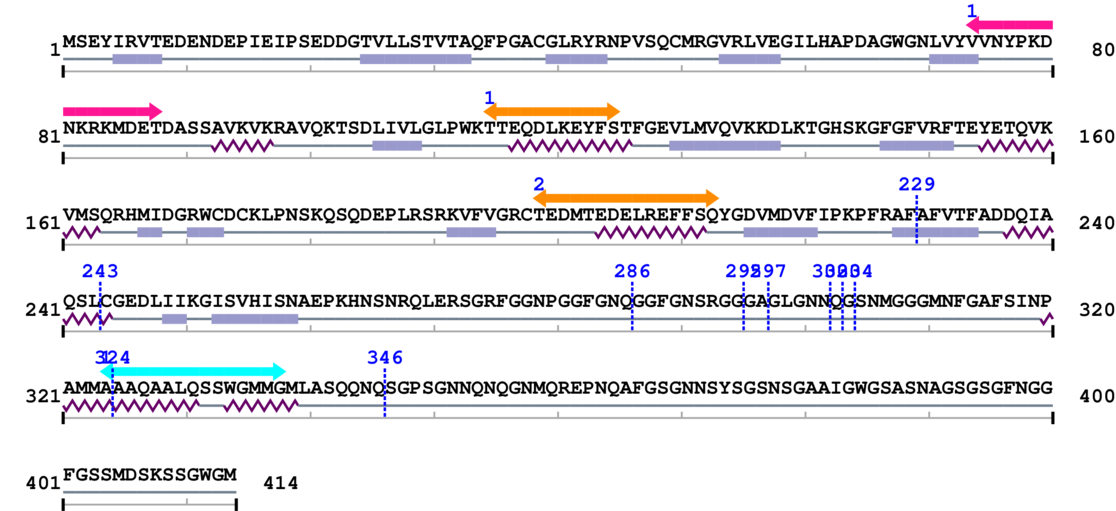
Length: 414 aa
Average Mass: 44.739 kDa
Monoisotopic Mass: 44.711 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| RNA recognition motif 1. | 74 | 88 | 29.0 | 0.0 |
| DNA ligase IV 1. | 115 | 125 | 20.0 | 0.2 |
| DNA ligase IV 2. | 199 | 213 | 17.0 | 0.1 |
| --- cleavage 229 --- | ||||
| --- cleavage 243 --- | ||||
| --- cleavage 286 --- | ||||
| --- cleavage 295 --- | ||||
| --- cleavage 297 --- | ||||
| --- cleavage 302 --- | ||||
| --- cleavage 303 --- | ||||
| --- cleavage 304 --- | ||||
| --- cleavage 324 --- | ||||
| RNA recognition motif (a.k.a. RRM, RBD, or RNP domain) 1. | 324 | 338 | 47.0 | 0.0 |
| --- cleavage 324 (inside RNA recognition motif (a.k.a. RRM, RBD, or RNP domain) 324..338) --- | ||||
3. Sequence Information
Fasta Sequence: SB0140.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
10 [sites] cleaved by Calpain 1
Source Reference: [PubMed ID: 23250437] Yamashita T, Hideyama T, Hachiga K, Teramoto S, Takano J, Iwata N, Saido TC, Kwak S, A role for calpain-dependent cleavage of TDP-43 in amyotrophic lateral sclerosis pathology. Nat Commun. 2012;3:1307. doi: 10.1038/ncomms2303.
Cleavage sites (±10aa)
[Site 1] VFIPKPFRAF229-AFVTFADDQI
Phe229  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Val220 | Phe221 | Ile222 | Pro223 | Lys224 | Pro225 | Phe226 | Arg227 | Ala228 | Phe229 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala230 | Phe231 | Val232 | Thr233 | Phe234 | Ala235 | Asp236 | Asp237 | Gln238 | Ile239 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EDMTEDELREFFSQYGDVMDVFIPKPFRAFAFVTFADDQIAQSLCGEDLIIKGISVHISN |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 123.00 | 4 | A Chain A, Solution Structure Of Rrm Domain In Ta |
| 2 | Bos taurus | 119.00 | 2 | TAR DNA binding protein |
| 3 | Tetraodon nigroviridis | 117.00 | 3 | unnamed protein product |
| 4 | Mus musculus | 116.00 | 15 | unnamed protein product |
| 5 | Xenopus laevis | 116.00 | 2 | mitotic phosphoprotein 39 |
| 6 | Danio rerio | 116.00 | 2 | TAR DNA binding protein |
| 7 | Danio rerio | 114.00 | 1 | TAR DNA binding protein, like |
| 8 | Rattus norvegicus | 111.00 | 1 | TAR DNA binding protein |
| 9 | Drosophila melanogaster | 102.00 | 3 | TAR-binding protein |
| 10 | Drosophila melanogaster | 102.00 | 2 | TBPH CG10327-PB, isoform B |
| 11 | Caenorhabditis briggsae AF16 | 48.10 | 2 | Hypothetical protein CBG03235 |
| 12 | Caenorhabditis elegans | 44.30 | 4 | Temporarily Assigned Gene name family member (tag |
| 13 | Arabidopsis thaliana | 42.70 | 7 | RNA-binding like protein |
| 14 | Schistosoma japonicum | 40.40 | 4 | RNA-binding protein |
| 15 | Arabidopsis thaliana | 39.70 | 14 | AC013483_15 putative RNA-binding protein |
| 16 | Physcomitrella patens | 39.30 | 1 | glycine-rich RNA-binding protein |
| 17 | Homo sapiens | 38.90 | 10 | similar to Splicing factor, arginine/serine-rich, |
| 18 | Aspergillus nidulans FGSC A4 | 38.90 | 1 | hypothetical protein AN6331.2 |
| 19 | Oryza sativa (japonica cultivar-group) | 38.50 | 3 | Os08g0492100 |
| 20 | Brassica oleracea | 38.50 | 1 | hypothetical protein 25.t00002 |
| 21 | Theileria parva strain Muguga | 38.10 | 2 | hypothetical protein TP03_0515 |
| 22 | Nicotiana tabacum | 38.10 | 2 | DNA binding protein ACBF |
| 23 | Yarrowia lipolytica | 38.10 | 1 | hypothetical protein |
| 24 | Ashbya gossypii ATCC 10895 | 38.10 | 1 | ADL160Wp |
| 25 | Solanum tuberosum | 38.10 | 1 | DNA binding protein ACBF-like |
| 26 | Gallus gallus | 38.10 | 1 | splicing factor, arginine/serine-rich 2 |
| 27 | Bombyx mori | 37.70 | 2 | Bmsqd-2 |
| 28 | Strongylocentrotus purpuratus | 37.70 | 1 | PREDICTED: similar to arginine/serine-rich splici |
| 29 | Solanum tuberosum | 37.70 | 1 | putative glycine-rich RNA binding protein-like |
| 30 | Macaca mulatta | 37.40 | 1 | PREDICTED: similar to FUS interacting protein (se |
| 31 | Medicago sativa | 36.60 | 1 | AF191305_1 glycine-rich RNA binding protein |
| 32 | Nitrococcus mobilis Nb-231 | 35.80 | 1 | RNA-binding protein |
| 33 | Coxiella burnetii RSA 493 | 35.80 | 1 | nucleic acid binding domain protein |
| 34 | Ciona intestinalis | 32.30 | 1 | Tardbp protein |
| 35 | Drosophila pseudoobscura | 32.30 | 1 | GA10247-PA |
| 36 | Xenopus tropicalis | 31.60 | 2 | TADBP_XENTR TAR DNA-binding protein 43 (TDP-43) g |
| 37 | Brassica oleracea | 31.60 | 1 | RNA recognition motif. (a.k.a. RRM, RBD, or RNP d |
| 38 | Oryza sativa Japonica Group | 31.20 | 1 | putative RNA-binding like protein |
| 39 | Canis familiaris | 31.20 | 1 | PREDICTED: similar to TAR DNA binding protein iso |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 201 DMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 201 DMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 |
| Bos taurus | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 |||| ||||+|| |||+|+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 200 EDMTADELRQFFCQYGEVVDVFIPKPFRAF#AFVTFADDQVAQSLCGEDLIIKGISVHISN 259 |
| Tetraodon nigroviridis | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 |||| |+||+|| |||+| |||||||||||#|||||||||+|||||||||||||+|||||| Sbjct 270 EDMTTDDLRQFFMQYGEVTDVFIPKPFRAF#AFVTFADDQVAQSLCGEDLIIKGVSVHISN 329 |
| Mus musculus | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 |||| +||++|| |||+|+|||||||||||#||||||||++|||||||||||||||||||| Sbjct 200 EDMTAEELQQFFCQYGEVVDVFIPKPFRAF#AFVTFADDKVAQSLCGEDLIIKGISVHISN 259 |
| Xenopus laevis | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHIS 258 |||+ +|||+||||||+|+|||||||||||#+||||||||+|||||||||||||+|||+| Sbjct 199 EDMSAEELRQFFSQYGEVVDVFIPKPFRAF#SFVTFADDQVAQSLCGEDLIIKGVSVHVS 257 |
| Danio rerio | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 |||| ||||+|| |||+| |||||||||||#|||||||||+| +||||||||||+|||||| Sbjct 206 EDMTADELRQFFMQYGEVTDVFIPKPFRAF#AFVTFADDQVAAALCGEDLIIKGVSVHISN 265 |
| Danio rerio | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 |||| ||||+|| |||+| |||||||||||#|||||||||+||||||||||||| |||||| Sbjct 206 EDMTADELRQFFMQYGEVTDVFIPKPFRAF#AFVTFADDQVAQSLCGEDLIIKGTSVHISN 265 |
| Rattus norvegicus | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 |||| +||++|| |||+|+|||||||||||#||||||||++|||||||||||||||||||| Sbjct 200 EDMTAEELQQFFCQYGEVVDVFIPKPFRAF#AFVTFADDKVAQSLCGEDLIIKGISVHISN 259 |
| Drosophila melanogaster | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 ||+ |+|||+||++|+| |||||+|||||#+|||| | +|||||||| ||||+|||+|| Sbjct 201 EDINSDDLREYFSKFGEVTDVFIPRPFRAF#SFVTFLDPDVAQSLCGEDHIIKGVSVHVSN 260 |
| Drosophila melanogaster | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 ||+ |+|||+||++|+| |||||+|||||#+|||| | +|||||||| ||||+|||+|| Sbjct 201 EDINSDDLREYFSKFGEVTDVFIPRPFRAF#SFVTFLDPDVAQSLCGEDHIIKGVSVHVSN 260 |
| Caenorhabditis briggsae AF16 | Query 204 EDELREFFS-------QYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGE-DLIIKGISV 255 |++||+ | + | ||||||||| |#|||| + + |+|+ + | + |+|| Sbjct 272 ENQLRKVFGDEAKVYVETAVVTDVFIPKPFRGF#AFVTLSSAEAAESIVSKGSLTVNGLSV 331 Query 256 HIS 258 +| Sbjct 332 GLS 334 |
| Caenorhabditis elegans | Query 200 EDMTEDELREFFS-------QYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGE-DLIIK 251 + + | +||+ | + | ||||||||| |#|||| + + |+ + + | + Sbjct 268 DKVDEHQLRKVFGDEAKSYIETAVVTDVFIPKPFRGF#AFVTLSSAEAAERIVSKGSLTVN 327 Query 252 GISVHIS 258 |+|| +| Sbjct 328 GLSVGLS 334 |
| Arabidopsis thaliana | Query 200 EDMTEDELREFFSQYGDVMDVFIPK-----PFRAF#AFVTFADDQIA-------QSLCGED 247 ++ + |+||++| ++| + | +||| | |# |||||++ +| +||++ Sbjct 193 QEASVDDLRDYFGRFGHIQDAYIPKDPKRSGHRGF#GFVTFAENGVADRVARRSHEICGQE 252 Query 248 LII 250 + | Sbjct 253 VAI 255 |
| Schistosoma japonicum | Query 203 TEDELREFFSQYGDVMDVFIPK-PF----RAF#AFVTFADDQIAQSLCGEDLIIKGISVH 256 | |+|| ||++|+| |++||+ |+ | |#||| + |+ | | |+|+ | Sbjct 27 TIDDLRRVFSRFGEVGDIYIPRDPYTFESRGF#AFVRYCTDREA------DCAIRGMDGH 79 |
| Arabidopsis thaliana | Query 201 DMTEDELREFFSQYGDVMDVFIPKP-----FRAF#AFVTFADDQIAQSLCGEDLIIKG 252 | |+ |+|+|| +|+|++ | | | |# || ||| +|+ + | | | Sbjct 16 DTNEERLKEYFSSFGEVIEAVILKDRTTGRARGF#GFVVFADPAVAEIVITEKHNIDG 72 |
| Physcomitrella patens | Query 203 TEDELREFFSQYGDVMDVFI-----PKPFRAF#AFVTFADDQIA----QSLCGEDLIIKGI 253 |+| ++| || +|+| +| | | |# ||||| || | |+| | || + | Sbjct 54 TDDNIKEAFSAFGEVTEVKIICDRDTGRSRGF#GFVTFATDQDAEAALQALDGRDLAGRTI 113 Query 254 SVHISN 259 |+ + Sbjct 114 RVNYAT 119 |
| Homo sapiens | Query 203 TEDELREFFSQYGDVMDVFIP-----KPFRAF#AFVTFADDQIAQ 241 + | || | +|| | ||+|| | | |#||| | | || Sbjct 26 SPDSLRRVFEKYGRVGDVYIPLEPHTKAPRGF#AFVRFHDRSDAQ 69 |
| Aspergillus nidulans FGSC A4 | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQS 242 + + | +||||||||++ |+ + +| | #| +|+|| |++ Sbjct 336 DKLDEFTVREFFSQYGEITDLQL-QPHRKL#ALITYADHATAKA 377 |
| Oryza sativa (japonica cultivar-group) | Query 200 EDMTEDELREFFSQYGDVMDVFI-----PKPFRAF#AFVTFADDQIAQSL 243 + +|||+ + || +|| |+| | | | |# |+ |+ ||+ | Sbjct 120 QALTEDDFKHFFQKYGPVVDHQIMRDHQTKRSRGF#GFIVFSSDQVVDDL 168 |
| Brassica oleracea | Query 201 DMTEDELREFFSQYGDVMDVFIPKP-----FRAF#AFVTFADDQIAQSL 243 + ||| ||++| +|+|++ | | | |# |+ ||| +|+ + Sbjct 16 ETTEDRLRDYFQSFGEVLEAVIMKDRSTGRARGF#GFLVFADPNVAERV 63 |
| Theileria parva strain Muguga | Query 207 LREFFSQYGDVMDVFIPK-----PFRAF#AFVTFA---------DDQIAQSLCGEDLIIKG 252 || +|||+|+|+||+||| + |# |+||| | + + | ++|+ Sbjct 354 LRTYFSQFGEVIDVYIPKDPHTQKGKGF#GFITFANKNSIHSALDPSLKHVVDGREIIVDY 413 Query 253 ISVH 256 || Sbjct 414 ADVH 417 |
| Nicotiana tabacum | Query 201 DMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQ 241 |+|++|||+ |+|+|+|+ | || + # || |+| || Sbjct 305 DVTDEELRQSFNQFGEVVSVKIPAG-KGC#GFVQFSDRSSAQ 344 |
| Yarrowia lipolytica | Query 201 DMTEDELREFFSQYGDVMDVFI------PKPFRAF#AFVTFADDQIAQSLCGEDLII 250 |+||++ ++|| ||| |+| + +| | |# |||| +| ++ + |+| Sbjct 93 DVTEEDYKQFFEQYGTVIDAQLMIDKDSGRP-RGF#GFVTFDSEQAMNTILAQPLLI 147 |
| Ashbya gossypii ATCC 10895 | Query 201 DMTEDELREFFSQYGDVMDVFIPKP-----FRAF#AFVTFAD 236 + ||| |||+||+||+| +| | + | |# |++||| Sbjct 173 ETTEDNLREYFSKYGNVTEVKIMRDGTTGRSRGF#GFLSFAD 213 |
| Solanum tuberosum | Query 201 DMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQ 241 |+|++|||+ |+|+|+|+ | || + # || |+| || Sbjct 254 DVTDEELRQSFTQFGEVVSVKIPAG-KGC#GFVQFSDRSSAQ 293 |
| Gallus gallus | Query 203 TEDELREFFSQYGDVMDVFIP-----KPFRAF#AFVTFADDQIAQ 241 + | || | +|| | ||+|| | | |#||| | | + |+ Sbjct 26 SPDTLRRVFEKYGRVGDVYIPRDRYTKESRGF#AFVRFHDKRDAE 69 |
| Bombyx mori | Query 201 DMTEDELREFFSQYGDVMDVFIP-----KPFRAF#AFVTFADDQIAQSL 243 ++++||+| |||++| +++| +| + |# |+|| +|+ | Sbjct 133 EISDDEIRNFFSEFGTILEVEMPFDKTKNQRKGF#CFITFESEQVVNDL 180 |
| Strongylocentrotus purpuratus | Query 203 TEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCG--EDLIIKGISVHI 257 + || | ||+|| | +|++ + |#||| | |++ | | +| + |+ | + Sbjct 28 SRTELEEAFSRYGRVKNVWVARNPPGF#AFVMFEDERDASDACKALDDRNVCGVRVRV 84 |
| Solanum tuberosum | Query 203 TEDELREFFSQYGDVMDVFI-----PKPFRAF#AFVTFADDQIA----QSLCGEDLIIKGI 253 |++ | | |||||+|++ | | |# |||| |+| + + |+|| + | Sbjct 18 TDNTLSEAFSQYGEVVESKIINDRETGRSRGF#GFVTFKDEQAMRDAIEGMNGQDLDGRNI 77 Query 254 SVH 256 +|+ Sbjct 78 TVN 80 |
| Macaca mulatta | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPF-----RAF#AFVTFADDQIAQS---------LCG 245 +| ++|| | +|| ++||++| | | |#|+| | | + |+ +|| Sbjct 19 DDTRSEDLRREFGRYGPIVDVYVPLDFYTRRPRGF#AYVQFEDVRDAEDALHNLDRKWICG 78 Query 246 EDLIIK 251 + |+ Sbjct 79 RQIEIQ 84 |
| Medicago sativa | Query 205 DELREFFSQYGDVMDVFI-----PKPFRAF#AFVTFADDQ----IAQSLCGEDLIIKGISV 255 | | + |||||+++| | | |# |||||+++ + +++ |+|| + |+| Sbjct 20 DALEKAFSQYGEIVDSKIINDRETGRSRGF#GFVTFANEKSMNDVIEAMNGQDLDGRNITV 79 Query 256 H 256 + Sbjct 80 N 80 |
| Nitrococcus mobilis Nb-231 | Query 203 TEDELREFFSQYGDVMDVFI------PKPFRAF#AFVTFADDQIA---QSLCGEDLIIKGI 253 +|||||| |+ ||+| | + +| | |# ||| +| | ++| |+| + + Sbjct 14 SEDELRELFAAYGNVDSVRLMTDRDTGRP-RGF#GFVTMSDTDAASAIEALDGKDFGGRNL 72 Query 254 SVH 256 |+ Sbjct 73 RVN 75 |
| Coxiella burnetii RSA 493 | Query 201 DMTEDELREFFSQYGD------VMDVFIPKPFRAF#AFVTFADDQIAQSLC----GEDLII 250 |+| |||+ || |||+ +|| + + |#||+|+ || | || Sbjct 14 DVTADELQSFFGQYGEIEEAKLIMDRETGRS-KGF#AFITYGTQDAAQEAVSKANGIDLQG 72 Query 251 KGISVHIS 258 + | |+|+ Sbjct 73 RKIRVNIA 80 |
| Ciona intestinalis | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHIS 258 | ||+||+|++| |+||| ||+|| |||||#||||| | +| +| ||| +| |+||+|+ Sbjct 232 EKMTKDEIRQYFEQFGDVDDVYIPIPFRAF#AFVTFRDSNVAANLIGEDQVINGVSVYIN 290 Query 202 MTEDELREFFSQYGDVMDVFIPK----PFRAF#AFVTFADDQIAQSLCGEDLII 250 + ||+|+|+|++||+++ | + + | +# |+ |+ ++ + + + || Sbjct 150 IAEDDLKEYFAKYGELVMVLVKRDENGKSRGY#GFIRFSSYEVQELVLSQRHII 202 |
| Drosophila pseudoobscura | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 ||+ |+|||+||++|+| |||||+|||||#+|||| | +|||||||| ||||+|||+|| Sbjct 201 EDINSDDLREYFSKFGEVTDVFIPRPFRAF#SFVTFFDPDVAQSLCGEDHIIKGVSVHVSN 260 Query 203 TEDELREFFSQYGDVMDVFIPKPFRA-----F#AFVTFA 235 ||| |||+| ||+|+ | | ++ |# || | Sbjct 119 TEDSLREYFESYGEVLMAQIKKDVKSGQSKGF#GFVRFG 156 |
| Xenopus tropicalis | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHIS 258 |||+ +|||+||||||+|+|||||||||||#|||||||||+|||||||||||||+|||+| Sbjct 200 EDMSAEELRQFFSQYGEVVDVFIPKPFRAF#AFVTFADDQVAQSLCGEDLIIKGVSVHVS 258 Query 203 TEDELREFFSQYGDVMDVFIPKPF-----RAF#AFVTFADDQIAQSLCGEDLIIKG 252 || +|+++|| +|+|+ | + | + |# || ||| + + + +| | Sbjct 117 TEQDLKDYFSTFGEVIMVQVKKDAKTGHSKGF#GFVRFADYETQVKVMSQRHMIDG 171 |
| Brassica oleracea | Query 201 DMTEDELREFFSQYGDVMDVFIPKP-----FRAF#AFVTFADDQIAQSL 243 + ||| |||+| +|+|++ | | | |# |+ ||| +|+ + Sbjct 15 ETTEDRLREYFQSFGEVLEAVIMKDRATGRARGF#GFLVFADPTVAERV 62 Query 202 MTEDELREFFSQYGDVMDVFI------PKPFRAF#AFVTFADDQ 238 +|| | +++|+|+| + || + +| | |# |++| ++ Sbjct 116 VTEAEFKKYFAQFGTITDVVVMYDHRTQRP-RGF#GFISFESEE 157 |
| Oryza sativa Japonica Group | Query 200 EDMTEDELREFFSQYGDVMDVFIPKP-----FRAF#AFVTFADDQIAQSLCGEDLIIKGIS 254 | ||| || +| ++| ++| +||| | |# ||||||+ +| + | | Sbjct 400 EANTED-LRHYFGKFGRIVDAYIPKDPKRSGHRGF#GFVTFADEGVADRVARRSHEILGHE 458 Query 255 VHI 257 | | Sbjct 459 VAI 461 Query 200 EDMTEDELREFFSQYGDVMDVFIPKP-----FRAF#AFVTFADDQIAQSLCGEDLIIKGIS 254 + + | | | ||++ |+++|| | # |+|| + |+ + + | + Sbjct 261 QSVDESMFRRHFEAYGEITDLYMPKEHGSKGHRGI#GFITFQSAESVDSIMQDSHELDGTT 320 Query 255 V 255 | Query 255 V 255 |
| Canis familiaris | Query 200 EDMTEDELREFFSQYGDVMDVFIPKPFRAF#AFVTFADDQIAQSLCGEDLIIKGISVHISN 259 |||| ||||+|| |||+|+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 323 EDMTADELRQFFCQYGEVVDVFIPKPFRAF#AFVTFADDQVAQSLCGEDLIIKGISVHISN 382 Query 203 TEDELREFFSQYGDVMDVFIPKPF-----RAF#AFVTFADDQIAQSLCGEDLIIKG 252 || +|+|+|| +|+|+ | + | + |# || | + + + + +| | Sbjct 239 TEQDLKEYFSTFGEVLMVQVKKDIKTGHSKGF#GFVRFTEYETQVKVMSQRHMIDG 293 |
[Site 2] FADDQIAQSL243-CGEDLIIKGI
Leu243  Cys
Cys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe234 | Ala235 | Asp236 | Asp237 | Gln238 | Ile239 | Ala240 | Gln241 | Ser242 | Leu243 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Cys244 | Gly245 | Glu246 | Asp247 | Leu248 | Ile249 | Ile250 | Lys251 | Gly252 | Ile253 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| YGDVMDVFIPKPFRAFAFVTFADDQIAQSLCGEDLIIKGISVHISNAEPKHNSNRQLERS |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 131.00 | 1 | TAR DNA binding protein |
| 2 | Canis familiaris | 127.00 | 1 | PREDICTED: similar to TAR DNA binding protein iso |
| 3 | Bos taurus | 125.00 | 1 | TAR DNA binding protein |
| 4 | Mus musculus | 124.00 | 8 | unnamed protein product |
| 5 | Rattus norvegicus | 123.00 | 1 | TAR DNA binding protein |
| 6 | Xenopus tropicalis | 119.00 | 2 | TADBP_XENTR TAR DNA-binding protein 43 (TDP-43) g |
| 7 | Xenopus laevis | 118.00 | 2 | mitotic phosphoprotein 39 |
| 8 | N/A | 115.00 | 2 | A Chain A, Solution Structure Of Rrm Domain In Ta |
| 9 | Tetraodon nigroviridis | 112.00 | 2 | unnamed protein product |
| 10 | Danio rerio | 112.00 | 1 | TAR DNA binding protein |
| 11 | Danio rerio | 106.00 | 1 | TAR DNA binding protein, like |
| 12 | Drosophila melanogaster | 90.50 | 3 | TAR-binding protein |
| 13 | Drosophila melanogaster | 90.50 | 2 | TBPH CG10327-PA, isoform A |
| 14 | Drosophila pseudoobscura | 88.20 | 1 | GA10247-PA |
| 15 | Ciona intestinalis | 71.60 | 1 | Tardbp protein |
| 16 | Schistosoma japonicum | 65.90 | 2 | SJCHGC08328 protein |
| 17 | Caenorhabditis briggsae AF16 | 51.20 | 1 | Hypothetical protein CBG03235 |
| 18 | Caenorhabditis elegans | 51.20 | 1 | Temporarily Assigned Gene name family member (tag |
| 19 | Oryza sativa (japonica cultivar-group) | 35.00 | 2 | Os08g0492100 |
| 20 | Arabidopsis thaliana | 33.10 | 4 | RNA-binding like protein |
| 21 | Arabidopsis thaliana | 32.30 | 13 | RNA recognition motif (RRM)-containing protein |
| 22 | Entamoeba histolytica HM-1:IMSS | 32.30 | 1 | RNA recognition motif domain containing protein |
| 23 | Oryza sativa Japonica Group | 31.60 | 1 | putative RNA-binding like protein |
| 24 | Brassica oleracea | 31.20 | 1 | hypothetical protein 25.t00002 |
| 25 | Aspergillus nidulans FGSC A4 | 30.40 | 1 | hypothetical protein AN6256.2 |
| 26 | Aspergillus oryzae RIB40 | 30.40 | 1 | hypothetical protein |
| 27 | Physcomitrella patens | 30.40 | 1 | glycine-rich RNA-binding protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLERS 273 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLERS 273 |
| Canis familiaris | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLERS 273 ||+|+||||||||||||||||||||+||||#|||||||||||||||||||||||||||||| Sbjct 337 YGEVVDVFIPKPFRAFAFVTFADDQVAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLERS 396 |
| Bos taurus | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLERS 273 ||+|+||||||||||||||||||||+||||#|||||||||||||||||||||||||||||| Sbjct 214 YGEVVDVFIPKPFRAFAFVTFADDQVAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLERS 273 |
| Mus musculus | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLERS 273 ||+|+|||||||||||||||||||++||||#|||||||||||||||||||||||||||||| Sbjct 214 YGEVVDVFIPKPFRAFAFVTFADDKVAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLERS 273 |
| Rattus norvegicus | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLERS 273 ||+|+|||||||||||||||||||++||||#|||||||||||||||||||||||||||||| Sbjct 214 YGEVVDVFIPKPFRAFAFVTFADDKVAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLERS 273 |
| Xenopus tropicalis | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLER 272 ||+|+||||||||||||||||||||+||||#|||||||||+|||+| ||||||+|||||| Sbjct 214 YGEVVDVFIPKPFRAFAFVTFADDQVAQSL#CGEDLIIKGVSVHVSTAEPKHNNNRQLER 272 |
| Xenopus laevis | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLER 272 ||+|+|||||||||||+||||||||+||||#|||||||||+|||+| ||||||+|||||| Sbjct 213 YGEVVDVFIPKPFRAFSFVTFADDQVAQSL#CGEDLIIKGVSVHVSTAEPKHNNNRQLER 271 |
| N/A | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSN 267 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||| Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSN 267 |
| Tetraodon nigroviridis | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNRQLERS 273 ||+| ||||||||||||||||||||+||||#|||||||||+||||||||||| ||| +|+ Sbjct 284 YGEVTDVFIPKPFRAFAFVTFADDQVAQSL#CGEDLIIKGVSVHISNAEPKH-GNRQYDRT 342 |
| Danio rerio | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNRQ-LER 272 ||+| ||||||||||||||||||||+| +|#|||||||||+||||||||||||+ || +|| Sbjct 220 YGEVTDVFIPKPFRAFAFVTFADDQVAAAL#CGEDLIIKGVSVHISNAEPKHNNTRQMMER 279 Query 273 S 273 + Sbjct 280 A 280 |
| Danio rerio | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNRQL 270 ||+| ||||||||||||||||||||+||||#||||||||| ||||||||||||+ | Sbjct 220 YGEVTDVFIPKPFRAFAFVTFADDQVAQSL#CGEDLIIKGTSVHISNAEPKHNNIHHL 276 |
| Drosophila melanogaster | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNR 268 +|+| |||||+|||||+|||| | +||||#|||| ||||+|||+||| || || Sbjct 215 FGEVTDVFIPRPFRAFSFVTFLDPDVAQSL#CGEDHIIKGVSVHVSNAAPKAEQNR 269 |
| Drosophila melanogaster | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNR 268 +|+| |||||+|||||+|||| | +||||#|||| ||||+|||+||| || || Sbjct 215 FGEVTDVFIPRPFRAFSFVTFLDPDVAQSL#CGEDHIIKGVSVHVSNAAPKAEQNR 269 |
| Drosophila pseudoobscura | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNR 268 +|+| |||||+|||||+|||| | +||||#|||| ||||+|||+||| || +| Sbjct 215 FGEVTDVFIPRPFRAFSFVTFFDPDVAQSL#CGEDHIIKGVSVHVSNAAPKAEQSR 269 |
| Ciona intestinalis | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPK 263 +||| ||+|| |||||||||| | +| +|# ||| +| |+||+|+ |+|| Sbjct 246 FGDVDDVYIPIPFRAFAFVTFRDSNVAANL#IGEDQVINGVSVYINTADPK 295 |
| Schistosoma japonicum | Query 214 YGDVMDVFIPKPFRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPK 263 +| |+|||||||||+||||+| | |+| |# |+| +|+|+++ | +| || Sbjct 112 FGRVIDVFIPKPFRSFAFVSFEDPQVATDL#LGKDQVIEGVTLSIGSAVPK 161 |
| Caenorhabditis briggsae AF16 | Query 217 VMDVFIPKPFRAFAFVTFADDQIAQSL#CGE-DLIIKGISVHISNAEPKHNS 266 | ||||||||| ||||| + + |+|+# + | + |+|| +| |+|+ + Sbjct 292 VTDVFIPKPFRGFAFVTLSSAEAAESI#VSKGSLTVNGLSVGLSIAQPREEN 342 |
| Caenorhabditis elegans | Query 217 VMDVFIPKPFRAFAFVTFADDQIAQSL#CGE-DLIIKGISVHISNAEPKHNSNRQL 270 | ||||||||| ||||| + + |+ +# + | + |+|| +| |+|+ +|+ + Sbjct 292 VTDVFIPKPFRGFAFVTLSSAEAAERI#VSKGSLTVNGLSVGLSIAQPREENNQSV 346 |
| Oryza sativa (japonica cultivar-group) | Query 214 YGDVMDVFI-----PKPFRAFAFVTFADDQIAQSL#C--GEDLIIKGISVHISNAEPKHNS 266 || |+| | | | | |+ |+ ||+ |# | + + | | | |||| +| Sbjct 134 YGPVVDHQIMRDHQTKRSRGFGFIVFSSDQVVDDL#LANGNMIDLAGAKVEIKKAEPKKSS 193 Query 267 N 267 | Query 267 N 267 |
| Arabidopsis thaliana | Query 214 YGDVMDVFIPKP-----FRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEP 262 +| + | +||| | | |||||++ +| +# | | | | +| | Sbjct 207 FGHIQDAYIPKDPKRSGHRGFGFVTFAENGVADRV#ARRSHEICGQEVAIDSATP 260 |
| Arabidopsis thaliana | Query 214 YGDVMDVFIPKP-----FRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEP 262 +| + | +||| | | |||||++ +| +# | | | | +| | Sbjct 263 FGHIQDAYIPKDPKRSGHRGFGFVTFAENGVADRV#ARRSHEICGQEVAIDSATP 316 Query 214 YGDVMDVFIPKPF-----RAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSN 267 ||++ |+++|| + | |+||+ + |# + + | +| + | || + + Sbjct 114 YGEITDLYMPKDYNSKQHRGIGFITFSSADSVEDL#MEDTHDLGGTTVAVDRATPKEDDH 172 |
| Entamoeba histolytica HM-1:IMSS | Query 214 YGDVMDVFIP-----KPFRAFAFVTFADDQIAQSL#CGE--DLIIKGISVHISNAEPKHNS 266 +|++ ++ +| | + | || | | | #| | + ||| ||++ |+ | Sbjct 210 FGNIAEIHLPIDKITKKSKGFGFVLFVVPQDAVKA#CNEMDNKFIKGRIVHVTYAKADPYS 269 Query 267 NRQLERS 273 |+|+ | Sbjct 270 NQQVGES 276 |
| Oryza sativa Japonica Group | Query 214 YGDVMDVFIPKP-----FRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEP 262 +| ++| +||| | | ||||||+ +| +# | | | | | | Sbjct 413 FGRIVDAYIPKDPKRSGHRGFGFVTFADEGVADRV#ARRSHEILGHEVAIDTAAP 466 Query 214 YGDVMDVFIPKPF-----RAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPKHNSNR 268 ||++ |+++|| | |+|| + |+# + + | +| + | || | Sbjct 275 YGEITDLYMPKEHGSKGHRGIGFITFQSAESVDSI#MQDSHELDGTTVVVDRATPKDEEVR 334 |
| Brassica oleracea | Query 214 YGDVMDVFIPKP-----FRAFAFVTFADDQIAQSL#CGEDLIIKGISVHISNAEPK--HNS 266 +|+|++ | | | | |+ ||| +|+ +# +| | | | |+ | | Sbjct 29 FGEVLEAVIMKDRSTGRARGFGFLVFADPNVAERV#VLLRHVIDGKLVEAKKAVPRDDHKS 88 Query 267 NRQLERS 273 | |+ | Sbjct 89 NSSLQGS 95 |
| Aspergillus nidulans FGSC A4 | Query 214 YGDVMDVFIPKP--------FRAFAFVTFADDQIAQ----SL#CGEDLIIKGISVHISNAE 261 +|+|+|+ +||| | | +| | + |+ ++# | +| | ++ ++ |+ Sbjct 32 FGEVVDITLPKPDLPNSNDNHRGFGYVEFDSPEDAKEAIDNM#DGSELY--GRTIKVAPAK 89 Query 262 PKHNSNRQL 270 |+ ++| | Sbjct 90 PQKDANEGL 98 |
| Aspergillus oryzae RIB40 | Query 214 YGDVMDVFIPKP--------FRAFAFVTFADDQIAQ----SL#CGEDLIIKGISVHISNAE 261 +|+|+|+ +||| | | +| | + |+ ++# | +| | ++ ++ |+ Sbjct 32 FGEVVDITLPKPDVPNSNELHRGFGYVEFEVPEDAKEAIDNM#DGSELY--GRTIKVAAAK 89 Query 262 PKHNSNRQL 270 |+ +|| | Sbjct 90 PQKDSNEGL 98 |
| Physcomitrella patens | Query 214 YGDVMDVFI-----PKPFRAFAFVTFADDQIA----QSL#CGEDLIIKGISVHISNAEPKH 264 +|+| +| | | | ||||| || | |+|# | || | ++ ++ | + Sbjct 65 FGEVTEVKIICDRDTGRSRGFGFVTFATDQDAEAALQAL#DGRDL--AGRTIRVNYATKQS 122 Query 265 NSNRQ 269 +|| Sbjct 123 PQDRQ 127 |
[Site 3] GGNPGGFGNQ286-GGFGNSRGGG
Gln286  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly277 | Gly278 | Asn279 | Pro280 | Gly281 | Gly282 | Phe283 | Gly284 | Asn285 | Gln286 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly287 | Gly288 | Phe289 | Gly290 | Asn291 | Ser292 | Arg293 | Gly294 | Gly295 | Gly296 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ISNAEPKHNSNRQLERSGRFGGNPGGFGNQGGFGNSRGGGAGLGNNQGSNMGGGMNFGAF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 116.00 | 1 | TAR DNA binding protein |
| 2 | Mus musculus | 113.00 | 8 | unnamed protein product |
| 3 | Canis familiaris | 55.50 | 1 | PREDICTED: similar to TAR DNA binding protein iso |
| 4 | Bos taurus | 54.30 | 1 | TAR DNA binding protein |
| 5 | Rattus norvegicus | 49.30 | 1 | TAR DNA binding protein |
| 6 | Xenopus tropicalis | 46.60 | 2 | TADBP_XENTR TAR DNA-binding protein 43 (TDP-43) g |
| 7 | Xenopus laevis | 45.10 | 1 | mitotic phosphoprotein 39 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 257 ISNAEPKHNSNRQLERSGRFGGNPGGFGNQ#GGFGNSRGGGAGLGNNQGSNMGGGMNFGAF 316 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 257 ISNAEPKHNSNRQLERSGRFGGNPGGFGNQ#GGFGNSRGGGAGLGNNQGSNMGGGMNFGAF 316 |
| Mus musculus | Query 257 ISNAEPKHNSNRQLERSGRFGGNPGGFGNQ#GGFGNSRGGGAGLGNNQGSNMGGGMNFGAF 316 ||||||||||||||||||||||||||||||#|||||||||||||||||| ||||||||||| Sbjct 257 ISNAEPKHNSNRQLERSGRFGGNPGGFGNQ#GGFGNSRGGGAGLGNNQGGNMGGGMNFGAF 316 |
| Canis familiaris | Query 257 ISNAEPKHNSNRQLERSGRFGGNPG 281 ||||||||||||||||||||||||| Query 257 ISNAEPKHNSNRQLERSGRFGGNPG 281 |
| Bos taurus | Query 257 ISNAEPKHNSNRQLERSGRFGGNP 280 |||||||||||||||||||||||| Query 257 ISNAEPKHNSNRQLERSGRFGGNP 280 |
| Rattus norvegicus | Query 257 ISNAEPKHNSNRQLERSGRFGGN 279 |||||||||||||||||||||| Sbjct 257 ISNAEPKHNSNRQLERSGRFGGK 279 |
| Xenopus tropicalis | Query 257 ISNAEPKHNSNRQLERSGRFGGNPGGFGNQ#GGFGNSRGGGAGL 299 +| ||||||+|||||| ||| | ||||# |+ ||| | Sbjct 257 VSTAEPKHNNNRQLERGGRFPG--PSFGNQ#-GYPNSRPSSGAL 296 |
| Xenopus laevis | Query 257 ISNAEPKHNSNRQLERSGRFGGNPGGFGNQ#GGFGNSRGGGA 297 +| ||||||+|||||| ||| | ||+|#| | || Sbjct 256 VSTAEPKHNNNRQLEREGRFPG--PSFGSQ#GYPSNRPSGGT 294 |
[Site 4] QGGFGNSRGG295-GAGLGNNQGS
Gly295  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gln286 | Gly287 | Gly288 | Phe289 | Gly290 | Asn291 | Ser292 | Arg293 | Gly294 | Gly295 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly296 | Ala297 | Gly298 | Leu299 | Gly300 | Asn301 | Asn302 | Gln303 | Gly304 | Ser305 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SNRQLERSGRFGGNPGGFGNQGGFGNSRGGGAGLGNNQGSNMGGGMNFGAFSINPAMMAA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 114.00 | 1 | TAR DNA binding protein |
| 2 | Mus musculus | 112.00 | 3 | TAR DNA binding protein isoform 1 |
| 3 | Xenopus tropicalis | 32.30 | 1 | TADBP_XENTR TAR DNA-binding protein 43 (TDP-43) g |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 266 SNRQLERSGRFGGNPGGFGNQGGFGNSRGG#GAGLGNNQGSNMGGGMNFGAFSINPAMMAA 325 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 266 SNRQLERSGRFGGNPGGFGNQGGFGNSRGG#GAGLGNNQGSNMGGGMNFGAFSINPAMMAA 325 |
| Mus musculus | Query 266 SNRQLERSGRFGGNPGGFGNQGGFGNSRGG#GAGLGNNQGSNMGGGMNFGAFSINPAMMAA 325 ||||||||||||||||||||||||||||||#||||||||| |||||||||||||||||||| Sbjct 266 SNRQLERSGRFGGNPGGFGNQGGFGNSRGG#GAGLGNNQGGNMGGGMNFGAFSINPAMMAA 325 |
| Xenopus tropicalis | Query 267 NRQLERSGRFGGNPGGFGNQGGFGNSRGG#GAGLGNNQGSNMGGGMNFG--AFSINP 320 |||||| ||| | ||||| + ||| # |||||| ||||| |||||| Sbjct 267 NRQLERGGRFPG--PSFGNQG-YPNSRPS#SGALGNNQGGNMGGGGGMNFGAFSINP 319 |
[Site 5] GFGNSRGGGA297-GLGNNQGSNM
Ala297  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly288 | Phe289 | Gly290 | Asn291 | Ser292 | Arg293 | Gly294 | Gly295 | Gly296 | Ala297 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly298 | Leu299 | Gly300 | Asn301 | Asn302 | Gln303 | Gly304 | Ser305 | Asn306 | Met307 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RQLERSGRFGGNPGGFGNQGGFGNSRGGGAGLGNNQGSNMGGGMNFGAFSINPAMMAAAQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 115.00 | 1 | TAR DNA binding protein |
| 2 | Mus musculus | 112.00 | 3 | TAR DNA binding protein isoform 1 |
| 3 | Xenopus tropicalis | 30.40 | 1 | hypothetical protein LOC394651 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 268 RQLERSGRFGGNPGGFGNQGGFGNSRGGGA#GLGNNQGSNMGGGMNFGAFSINPAMMAAAQ 327 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 268 RQLERSGRFGGNPGGFGNQGGFGNSRGGGA#GLGNNQGSNMGGGMNFGAFSINPAMMAAAQ 327 |
| Mus musculus | Query 268 RQLERSGRFGGNPGGFGNQGGFGNSRGGGA#GLGNNQGSNMGGGMNFGAFSINPAMMAAAQ 327 ||||||||||||||||||||||||||||||#||||||| |||||||||||||||||||||| Sbjct 268 RQLERSGRFGGNPGGFGNQGGFGNSRGGGA#GLGNNQGGNMGGGMNFGAFSINPAMMAAAQ 327 |
| Xenopus tropicalis | Query 268 RQLERSGRFGGNPGGFGNQGGFGNSRGGGA#GLGNNQGSNMGGGMNFG--AFSINP 320 ||||| ||| | ||||| + ||| # |||||| ||||| |||||| Sbjct 266 RQLERGGRFPG--PSFGNQG-YPNSRPSSG#ALGNNQGGNMGGGGGMNFGAFSINP 317 |
[Site 6] RGGGAGLGNN302-QGSNMGGGMN
Asn302  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg293 | Gly294 | Gly295 | Gly296 | Ala297 | Gly298 | Leu299 | Gly300 | Asn301 | Asn302 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln303 | Gly304 | Ser305 | Asn306 | Met307 | Gly308 | Gly309 | Gly310 | Met311 | Asn312 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SGRFGGNPGGFGNQGGFGNSRGGGAGLGNNQGSNMGGGMNFGAFSINPAMMAAAQAALQS |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 113.00 | 1 | TAR DNA binding protein |
| 2 | Mus musculus | 110.00 | 2 | unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 273 SGRFGGNPGGFGNQGGFGNSRGGGAGLGNN#QGSNMGGGMNFGAFSINPAMMAAAQAALQS 332 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 273 SGRFGGNPGGFGNQGGFGNSRGGGAGLGNN#QGSNMGGGMNFGAFSINPAMMAAAQAALQS 332 |
| Mus musculus | Query 273 SGRFGGNPGGFGNQGGFGNSRGGGAGLGNN#QGSNMGGGMNFGAFSINPAMMAAAQAALQS 332 ||||||||||||||||||||||||||||||#|| ||||||||||||||||||||||||||| Sbjct 273 SGRFGGNPGGFGNQGGFGNSRGGGAGLGNN#QGGNMGGGMNFGAFSINPAMMAAAQAALQS 332 |
[Site 7] GGGAGLGNNQ303-GSNMGGGMNF
Gln303  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly294 | Gly295 | Gly296 | Ala297 | Gly298 | Leu299 | Gly300 | Asn301 | Asn302 | Gln303 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly304 | Ser305 | Asn306 | Met307 | Gly308 | Gly309 | Gly310 | Met311 | Asn312 | Phe313 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GRFGGNPGGFGNQGGFGNSRGGGAGLGNNQGSNMGGGMNFGAFSINPAMMAAAQAALQSS |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 113.00 | 1 | TAR DNA binding protein |
| 2 | Mus musculus | 110.00 | 2 | unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 274 GRFGGNPGGFGNQGGFGNSRGGGAGLGNNQ#GSNMGGGMNFGAFSINPAMMAAAQAALQSS 333 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 274 GRFGGNPGGFGNQGGFGNSRGGGAGLGNNQ#GSNMGGGMNFGAFSINPAMMAAAQAALQSS 333 |
| Mus musculus | Query 274 GRFGGNPGGFGNQGGFGNSRGGGAGLGNNQ#GSNMGGGMNFGAFSINPAMMAAAQAALQSS 333 ||||||||||||||||||||||||||||||#| |||||||||||||||||||||||||||| Sbjct 274 GRFGGNPGGFGNQGGFGNSRGGGAGLGNNQ#GGNMGGGMNFGAFSINPAMMAAAQAALQSS 333 |
[Site 8] GGAGLGNNQG304-SNMGGGMNFG
Gly304  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly295 | Gly296 | Ala297 | Gly298 | Leu299 | Gly300 | Asn301 | Asn302 | Gln303 | Gly304 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser305 | Asn306 | Met307 | Gly308 | Gly309 | Gly310 | Met311 | Asn312 | Phe313 | Gly314 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RFGGNPGGFGNQGGFGNSRGGGAGLGNNQGSNMGGGMNFGAFSINPAMMAAAQAALQSSW |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 115.00 | 1 | TAR DNA binding protein |
| 2 | Mus musculus | 112.00 | 2 | unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 275 RFGGNPGGFGNQGGFGNSRGGGAGLGNNQG#SNMGGGMNFGAFSINPAMMAAAQAALQSSW 334 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 275 RFGGNPGGFGNQGGFGNSRGGGAGLGNNQG#SNMGGGMNFGAFSINPAMMAAAQAALQSSW 334 |
| Mus musculus | Query 275 RFGGNPGGFGNQGGFGNSRGGGAGLGNNQG#SNMGGGMNFGAFSINPAMMAAAQAALQSSW 334 ||||||||||||||||||||||||||||||# ||||||||||||||||||||||||||||| Sbjct 275 RFGGNPGGFGNQGGFGNSRGGGAGLGNNQG#GNMGGGMNFGAFSINPAMMAAAQAALQSSW 334 |
[Site 9] AFSINPAMMA324-AAQAALQSSW
Ala324  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala315 | Phe316 | Ser317 | Ile318 | Asn319 | Pro320 | Ala321 | Met322 | Met323 | Ala324 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala325 | Ala326 | Gln327 | Ala328 | Ala329 | Leu330 | Gln331 | Ser332 | Ser333 | Trp334 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GGAGLGNNQGSNMGGGMNFGAFSINPAMMAAAQAALQSSWGMMGMLASQQNQSGPSGNNQ |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 115.00 | 1 | TAR DNA binding protein |
| 2 | Mus musculus | 112.00 | 3 | TAR DNA binding protein isoform 1 |
| 3 | Xenopus tropicalis | 59.30 | 2 | TADBP_XENTR TAR DNA-binding protein 43 (TDP-43) g |
| 4 | Xenopus laevis | 57.40 | 1 | mitotic phosphoprotein 39 |
| 5 | Canis familiaris | 56.60 | 1 | PREDICTED: similar to glyceraldehyde-3-phosphate |
| 6 | Tetraodon nigroviridis | 48.10 | 1 | unnamed protein product |
| 7 | Danio rerio | 37.40 | 1 | TAR DNA binding protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 295 GGAGLGNNQGSNMGGGMNFGAFSINPAMMA#AAQAALQSSWGMMGMLASQQNQSGPSGNNQ 354 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 295 GGAGLGNNQGSNMGGGMNFGAFSINPAMMA#AAQAALQSSWGMMGMLASQQNQSGPSGNNQ 354 |
| Mus musculus | Query 295 GGAGLGNNQGSNMGGGMNFGAFSINPAMMA#AAQAALQSSWGMMGMLASQQNQSGPSGNNQ 354 |||||||||| |||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 295 GGAGLGNNQGGNMGGGMNFGAFSINPAMMA#AAQAALQSSWGMMGMLASQQNQSGPSGNNQ 354 |
| Xenopus tropicalis | Query 315 AFSINPAMMA#AAQAALQSSWGMMGMLASQQNQSGPSGNNQ 354 ||||||||||#||||||||||||||||||||||||| |+|| Sbjct 314 AFSINPAMMA#AAQAALQSSWGMMGMLASQQNQSGPQGSNQ 353 |
| Xenopus laevis | Query 315 AFSINPAMMA#AAQAALQSSWGMMGMLASQQNQSGPSGNN 353 ||||||||||#||||||||||||||||||||||||| |+| Sbjct 317 AFSINPAMMA#AAQAALQSSWGMMGMLASQQNQSGPQGSN 355 |
| Canis familiaris | Query 322 MMA#AAQAALQSSWGMMGMLASQQNQSGPS 350 | #|||||||||||||||||||||||||| Sbjct 1 MTV#AAQAALQSSWGMMGMLASQQNQSGPS 29 |
| Tetraodon nigroviridis | Query 312 NFGAFSINPAMMA#AAQAALQSSWGMMGMLASQQ 344 |||+||+||||||#|||||||||||||||||||| Sbjct 369 NFGSFSLNPAMMA#AAQAALQSSWGMMGMLASQQ 401 |
| Danio rerio | Query 316 FSINPAMMA#AAQAALQSSWGMMGMLASQQNQ 346 |++||||||#||||||||||||||||| |||| Sbjct 316 FNLNPAMMA#AAQAALQSSWGMMGMLA-QQNQ 345 |
[Site 10] MGMLASQQNQ346-SGPSGNNQNQ
Gln346  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Met337 | Gly338 | Met339 | Leu340 | Ala341 | Ser342 | Gln343 | Gln344 | Asn345 | Gln346 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser347 | Gly348 | Pro349 | Ser350 | Gly351 | Asn352 | Asn353 | Gln354 | Asn355 | Gln356 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SINPAMMAAAQAALQSSWGMMGMLASQQNQSGPSGNNQNQGNMQREPNQAFGSGNNSYSG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 118.00 | 1 | TAR DNA binding protein |
| 2 | Mus musculus | 114.00 | 3 | TAR DNA binding protein isoform 1 |
| 3 | Xenopus tropicalis | 73.60 | 2 | TADBP_XENTR TAR DNA-binding protein 43 (TDP-43) g |
| 4 | Xenopus laevis | 72.40 | 1 | mitotic phosphoprotein 39 |
| 5 | Canis familiaris | 57.00 | 1 | PREDICTED: similar to glyceraldehyde-3-phosphate |
| 6 | Tetraodon nigroviridis | 46.20 | 1 | unnamed protein product |
| 7 | Danio rerio | 40.40 | 1 | TAR DNA binding protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 317 SINPAMMAAAQAALQSSWGMMGMLASQQNQ#SGPSGNNQNQGNMQREPNQAFGSGNNSYSG 376 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 317 SINPAMMAAAQAALQSSWGMMGMLASQQNQ#SGPSGNNQNQGNMQREPNQAFGSGNNSYSG 376 |
| Mus musculus | Query 317 SINPAMMAAAQAALQSSWGMMGMLASQQNQ#SGPSGNNQNQGNMQREPNQAFGSGNNSYSG 376 ||||||||||||||||||||||||||||||#||||||||+||+|||||||||||||||||| Sbjct 317 SINPAMMAAAQAALQSSWGMMGMLASQQNQ#SGPSGNNQSQGSMQREPNQAFGSGNNSYSG 376 |
| Xenopus tropicalis | Query 317 SINPAMMAAAQAALQSSWGMMGMLASQQNQ#SGPSGNNQNQGNMQREPNQAFGSGNNSY 374 ||||||||||||||||||||||||||||||#||| |+|| ||| ||+ |+||| |||| Sbjct 316 SINPAMMAAAQAALQSSWGMMGMLASQQNQ#SGPQGSNQGQGNQQRDQPQSFGS-NNSY 372 |
| Xenopus laevis | Query 317 SINPAMMAAAQAALQSSWGMMGMLASQQNQ#SGPSGNNQNQGNMQREPNQAFGSGNNSY 374 ||||||||||||||||||||||||||||||#||| |+| ||| ||| |+||| |||| Sbjct 319 SINPAMMAAAQAALQSSWGMMGMLASQQNQ#SGPQGSNPGQGNQQREQPQSFGS-NNSY 375 |
| Canis familiaris | Query 322 MMAAAQAALQSSWGMMGMLASQQNQ#SGPS 350 | ||||||||||||||||||||||#|||| Sbjct 1 MTVAAQAALQSSWGMMGMLASQQNQ#SGPS 29 |
| Tetraodon nigroviridis | Query 317 SINPAMMAAAQAALQSSWGMMGMLASQQNQ#SGPSGNNQNQGNMQREPNQAFGSGNNSY 374 |+|||||||||||||||||||||||||| #+ ||+ + + |+ +|+||+||++| Sbjct 374 SLNPAMMAAAQAALQSSWGMMGMLASQQTS#N--SGSTSSGTSSSRDQSQSFGTGNSNY 429 |
| Danio rerio | Query 318 INPAMMAAAQAALQSSWGMMGMLASQQNQ#SGPSGNNQNQGNMQREPNQAFGSGNNSY 374 +||||||||||||||||||||||| ||||#|| || + + + |+ | + | |++| Sbjct 318 LNPAMMAAAQAALQSSWGMMGMLA-QQNQ#SGTSGTSTSGTSSSRDQAQTYSSANSNY 373 |
* References
[PubMed ID: 24440855] De Marco G, Lomartire A, Mandili G, Lupino E, Buccinna B, Ramondetti C, Moglia C, Novelli F, Piccinini M, Mostert M, Rinaudo MT, Chio A, Calvo A, Reduced cellular Ca(2+) availability enhances TDP-43 cleavage by apoptotic caspases. Biochim Biophys Acta. 2014 Apr;1843(4):725-34. doi: 10.1016/j.bbamcr.2014.01.010.
[PubMed ID: 24440310] ... Mompean M, Buratti E, Guarnaccia C, Brito RM, Chakrabartty A, Baralle FE, Laurents DV, "Structural characterization of the minimal segment of TDP-43 competent for aggregation". Arch Biochem Biophys. 2014 Mar 1;545:53-62. doi: 10.1016/j.abb.2014.01.007. Epub
[PubMed ID: 24507191] ... Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, Han SS, Kiskinis E, Winborn B, Freibaum BD, Kanagaraj A, Clare AJ, Badders NM, Bilican B, Chaum E, Chandran S, Shaw CE, Eggan KC, Maniatis T, Taylor JP, Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014 Feb 5;81(3):536-43. doi: 10.1016/j.neuron.2013.12.018.
[PubMed ID: 24507183] ... Jovicic A, Gitler AD, TDP-43 in ALS: stay on target...almost there. Neuron. 2014 Feb 5;81(3):463-5. doi: 10.1016/j.neuron.2014.01.034.
[PubMed ID: 17322306] ... Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y, Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics. 2007 Jun;6(6):1103-9. Epub 2007 Feb 23.
[PubMed ID: 17081983] ... Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M, Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006 Nov 3;127(3):635-48.
[PubMed ID: 11285240] ... Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE, Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001 Apr 2;20(7):1774-84.
[PubMed ID: 11256614] ... Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S, Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000 Sep;1(3):287-92.
[PubMed ID: 8599929] ... Tokai N, Fujimoto-Nishiyama A, Toyoshima Y, Yonemura S, Tsukita S, Inoue J, Yamamota T, Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO J. 1996 Feb 1;15(3):457-67.
[PubMed ID: 7745706] ... Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB, Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995 Jun;69(6):3584-96.
[PubMed ID: 20301761] ... Harms MM, Miller TM, Baloh RH, TARDBP-Related Amyotrophic Lateral Sclerosis
[PubMed ID: 20301623] ... Kinsley L, Siddique T, Amyotrophic Lateral Sclerosis Overview