SB0143 : [alpha]-actinin-1
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | alpha-actinin-1 isoform b [Homo sapiens]; alpha-actinin-1 isoform b; F-actin cross-linking protein; actinin 1 smooth muscle; non-muscle alpha-actinin-1; alpha-actinin cytoskeletal isoform; Alpha-actinin-1; Alpha-actinin cytoskeletal isoform; Non-muscle alpha-actinin-1 |
| Gene Names | ACTN1; actinin, alpha 1 |
| Gene Locus | 14q24; chromosome 14 |
| GO Function | Not available |
* Information From OMIM
Description: Alpha-actinin was initially isolated from rabbit skeletal muscle as a factor that induces the gelation of F-actin and promotes the superprecipitation of actomyosin. Subsequently, a number of different isoforms were isolated from both muscle and nonmuscle cells and from a wide variety of organisms. The native molecule is thought to be a homodimer of 97-kD subunits arranged in antiparallel fashion. In myofibrillar cells, alpha-actinin constitutes a major component of Z discs in striated muscle and of the functionally analogous dense bodies and dense plaques in smooth muscle. In nonmuscle cells, it is distributed along microfilament bundles and is thought to mediate their attachment to the membrane at adherens-type junctions (Youssoufian et al., 1990).
* Structure Information
1. Primary Information
Length: 892 aa
Average Mass: 103.056 kDa
Monoisotopic Mass: 102.993 kDa
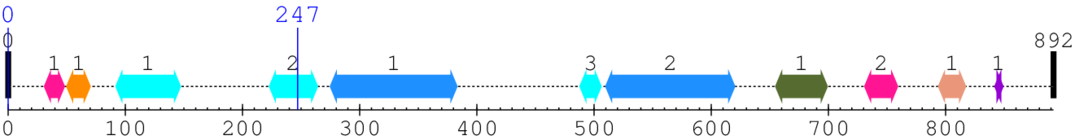
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Phage integrase SAM-like domain 1. | 31 | 48 | 77.0 | 0.0 |
| CAMSAP CH domain 1. | 50 | 70 | 11.0 | 0.0 |
| DAK2 domain 1. | 92 | 147 | 17.0 | 0.0 |
| DAK2 domain 2. | 223 | 264 | 85.0 | 0.1 |
| --- cleavage 247 (inside DAK2 domain 223..264) --- | ||||
| Spectrin repeat 1. | 275 | 383 | 2.0 | 0.4 |
| DAK2 domain 3. | 488 | 506 | 126.0 | 0.0 |
| Spectrin repeat 2. | 510 | 620 | 2.0 | 0.0 |
| Up-regulated During Septation 1. | 655 | 699 | 70.0 | 0.2 |
| Phage integrase SAM-like domain 2. | 731 | 759 | 45.0 | 0.1 |
| EF-hand domain pair 1. | 794 | 817 | 30.0 | 0.0 |
| EF hand 1. | 842 | 849 | 16.0 | 0.0 |
3. Sequence Information
Fasta Sequence: SB0143.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
2 [sites] cleaved by Calpain 1 and 2
Source Reference: [PubMed ID: 18258589] Sprague CR, Fraley TS, Jang HS, Lal S, Greenwood JA, Phosphoinositide binding to the substrate regulates susceptibility to proteolysis by calpain. J Biol Chem. 2008 Apr 4;283(14):9217-23. doi: 10.1074/jbc.M707436200. Epub 2008
Cleavage sites (±10aa)
[Site 1] MDHYDSQQTNDYMQPEEDWDRDLLLDPAWEKQQRKTFTAWCNSHLRKAGTQIENIEEDFRDGLKLMLLLEVISGERLAKPERGKMRVHKISNVNKALDFIASKGVKLVSIGAEEIVDGNVKMTLGMIWTIILRFAIQDISVEETSAKEGLLLWCQRKTAPYKNVNIQNFHISWKDGLGFCALIHRHRPELIDYGKLRKDDPLTNLNTAFDVAEKYLDIPKMLDAEDIVGTARPDEKAIMTYVSSFYHAFSGAQKAETAANRICKVLAVNQENEQLMEDYEKLASDLLEWIRRTIPWLENRVPENTMHAMQQKLEDFRDYRRLHKPPKVQEKCQLEINFNTLQTKLRLSNRPAFMPSEGRMVSDINNAWGCLEQVEKGYEEWLLNEIRRLERLDHLAEKFRQKASIHEAWTDGKEAMLRQKDYETATLSEIKALLKKHEAFESDLAAHQDRVEQIAAIAQELNELDYYDSPSVNARCQKICDQWDNLGALTQKRREALERTEKLLETIDQLYLEYAKRAAPFNNWMEGAMEDLQDTFIVHTIEEIQGLTTAHEQFKATLPDADKERLAILGIHNEVSKIVQTYHVNMAGTNPYTTITPQEINGKWDHVRQLVPRRDQALTEEHARQQHNERLRKQFGAQANVIGPWIQTKMEEIGRISIEMHGTLEDQLSHLRQYEKSIVNYKPKIDQLEGDHQLIQEALIFDNKHTNYTMEHIRVGWEQLLTTIARTINEVENQILTRDAKGISQEQMNEFRASFNHFDRDHSGTLGPEEFKACLISLGYDIGNDPQGEAEFARIMSIVDPNRLGVVTFQAFIDFMSRETADTDTADQVMASFKILAGDKNYITMDELRRELPPDQAEYCIARMAPYTGPDSVPGALDYMSFSTALYGESDL0-MDHYDSQQTN
Leu0  Met
Met
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Met1 | Asp2 | His3 | Tyr4 | Asp5 | Ser6 | Gln7 | Gln8 | Thr9 | Asn10 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
[Site 2] IMTYVSSFYH247-AFSGAQKAET
His247  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile238 | Met239 | Thr240 | Tyr241 | Val242 | Ser243 | Ser244 | Phe245 | Tyr246 | His247 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala248 | Phe249 | Ser250 | Gly251 | Ala252 | Gln253 | Lys254 | Ala255 | Glu256 | Thr257 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 24024966] Teumer A, Holtfreter B, Volker U, Petersmann A, Nauck M, Biffar R, Volzke H, Kroemer HK, Meisel P, Homuth G, Kocher T, Genome-wide association study of chronic periodontitis in a general German population. J Clin Periodontol. 2013 Nov;40(11):977-85. doi: 10.1111/jcpe.12154. Epub 2013