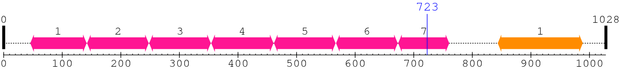
| # |
organism |
max score |
hits |
top seq |
| 1 |
Homo sapiens |
122.00 |
61 |
AF465439_1 beta I spectrin form betaI sigma3 |
| 2 |
N/A |
122.00 |
16 |
- |
| 3 |
Mus musculus |
114.00 |
47 |
mKIAA4219 protein |
| 4 |
Danio rerio |
79.30 |
9 |
spectrin, beta, erythrocytic |
| 5 |
Drosophila melanogaster |
76.30 |
8 |
beta Spectrin CG5870-PA |
| 6 |
Gallus gallus |
76.30 |
5 |
PREDICTED: similar to spectrin, beta, non-erythroc |
| 7 |
Aedes aegypti |
76.30 |
3 |
beta chain spectrin |
| 8 |
Drosophila pseudoobscura |
76.30 |
3 |
GA19192-PA |
| 9 |
Anopheles gambiae str. PEST |
75.50 |
3 |
ENSANGP00000012507 |
| 10 |
Canis familiaris |
75.10 |
18 |
PREDICTED: similar to spectrin, beta, non-erythroc |
| 11 |
Pan troglodytes |
75.10 |
14 |
PREDICTED: spectrin, beta, non-erythrocytic 1 isof |
| 12 |
Rattus norvegicus |
75.10 |
13 |
Spnb2 protein |
| 13 |
Macaca mulatta |
75.10 |
5 |
PREDICTED: spectrin, beta, non-erythrocytic 1 isof |
| 14 |
Bos taurus |
75.10 |
5 |
PREDICTED: similar to spectrin, beta, non-erythroc |
| 15 |
Macaca fascicularis |
75.10 |
2 |
unnamed protein product |
| 16 |
mice, brain, Peptide Partial, 332 aa |
75.10 |
1 |
beta-spectrin subunit |
| 17 |
Apis mellifera |
73.90 |
3 |
PREDICTED: similar to Spectrin beta chain |
| 18 |
Tribolium castaneum |
71.60 |
4 |
PREDICTED: similar to Spectrin beta chain isoform |
| 19 |
Tetraodon nigroviridis |
69.30 |
8 |
unnamed protein product |
| 20 |
Caenorhabditis briggsae |
64.70 |
3 |
Hypothetical protein CBG19038 |
| 21 |
Strongylocentrotus purpuratus |
64.30 |
5 |
PREDICTED: similar to spectrin beta 2 |
| 22 |
Caenorhabditis elegans |
63.20 |
5 |
UNCoordinated family member (unc-70) |
| 23 |
Brugia malayi |
63.20 |
1 |
beta-G spectrin |
| 24 |
Sus scrofa |
43.90 |
1 |
spectrin alpha non-erythrocytic 1 |
| 25 |
Monodelphis domestica |
36.60 |
4 |
PREDICTED: similar to spectrin, alpha, erythrocyti |
| 26 |
Xenopus tropicalis |
35.80 |
1 |
hypothetical protein LOC100036647 |
| 27 |
Mus sp. |
34.70 |
1 |
SPTB1_MOUSE Spectrin beta chain, erythrocyte (Beta |
| organism | matching |
|---|
| Homo sapiens |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
|
| N/A |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
|
| Mus musculus |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
|||||| |||||||||||||||||||||||#|||||||||||||||||||||| ||| ||
Sbjct 618 EPYLASRDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQTPERPTEE 677
|
| Danio rerio |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKE 743
|||+|| | | ||| ||||+||||||||||#|+| |||+|||+ ||||| |
Sbjct 2048 EPYVASKDVGQTVDEVEKLLKRHEAFEKST#ATWEERFSALERLTTLELLE 2097
|
| Drosophila melanogaster |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
|||| | + |||+| || |||+||||||| #|+ |||+|||+ || |||| + + |||
Sbjct 2038 EPYLLSSELGHTIDEVENLIKKHEAFEKSA#AAQEERFSALERLTTFELKEMKRRQELAEE 2097
|
| Gallus gallus |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKE 743
||||+| + | +|| |||||||||||||| #|+| |||||||+ ||||| |
Sbjct 2036 EPYLSSREIGQSVDEVEKLIKRHEAFEKSA#ATWDERFAALERLTTLELLE 2085
|
| Aedes aegypti |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
|||| | + |||+| || |||+||||||| #|+ |||+|||+ || |||| + + |||
Sbjct 2038 EPYLMSTELGHTIDEVENLIKKHEAFEKSA#AAQEERFSALERLTTFELKEMKRRQEAAEE 2097
|
| Drosophila pseudoobscura |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
|||| | + |||+| || |||+||||||| #|+ |||+|||+ || |||| + + |||
Sbjct 2038 EPYLLSSELGHTIDEVENLIKKHEAFEKSA#AAQEERFSALERLTTFELKEMKRRQELAEE 2097
|
| Anopheles gambiae str. PEST |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
|||| | + |||+| || |||+||||||| #|+ |||+|||+ || |||| + + |||
Sbjct 2038 EPYLMSTELGHTIDEVENLIKKHEAFEKSA#AAQEERFSALERLTTFELKEMKRRQDAAEE 2097
|
| Canis familiaris |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKE 743
||||+| + | +|| |||||||||||||| #|+| |||+|||+ ||||| |
Sbjct 2037 EPYLSSREIGQSVDEVEKLIKRHEAFEKSA#ATWDERFSALERLTTLELLE 2086
|
| Pan troglodytes |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKE 743
||||+| + | +|| |||||||||||||| #|+| |||+|||+ ||||| |
Sbjct 2037 EPYLSSREIGQSVDEVEKLIKRHEAFEKSA#ATWDERFSALERLTTLELLE 2086
|
| Rattus norvegicus |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKE 743
||||+| + | +|| |||||||||||||| #|+| |||+|||+ ||||| |
Sbjct 190 EPYLSSREIGQSVDEVEKLIKRHEAFEKSA#ATWDERFSALERLTTLELLE 239
|
| Macaca mulatta |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKE 743
||||+| + | +|| |||||||||||||| #|+| |||+|||+ ||||| |
Sbjct 2024 EPYLSSREIGQSVDEVEKLIKRHEAFEKSA#ATWDERFSALERLTTLELLE 2073
|
| Bos taurus |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKE 743
||||+| + | +|| |||||||||||||| #|+| |||+|||+ ||||| |
Sbjct 2036 EPYLSSREIGQSVDEVEKLIKRHEAFEKSA#ATWDERFSALERLTTLELLE 2085
|
| Macaca fascicularis |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKE 743
||||+| + | +|| |||||||||||||| #|+| |||+|||+ ||||| |
Sbjct 367 EPYLSSREIGQSVDEVEKLIKRHEAFEKSA#ATWDERFSALERLTTLELLE 416
|
| mice, brain, Peptide Partial, 332 aa |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKE 743
||||+| + | +|| |||||||||||||| #|+| |||+|||+ ||||| |
Sbjct 163 EPYLSSREIGQSVDEVEKLIKRHEAFEKSA#ATWDERFSALERLTTLELLE 212
|
| Apis mellifera |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
|||| | + |||+| || |||+||||||| #|+ |||+|| + || |||| + |+ ||
Sbjct 2036 EPYLMSQELGHTIDEVENLIKKHEAFEKSA#AAQEERFSALHRLTTFELKEMKRREQEREE 2095
|
| Tribolium castaneum |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQ 745
|||| | + |||+| || |||+||||||| #|+ |||+|||+ || ||+| |
Sbjct 2037 EPYLMSTELGHTIDDVENLIKKHEAFEKSA#AAQEERFSALERLTTFELREIQ 2088
|
| Tetraodon nigroviridis |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQ 745
||||+| + | +|| ||||+||||||||| #|+| |||+|||+ ||+ |+
Sbjct 1937 EPYLSSREVGRSVDQVEKLLKRHEAFEKSA#ATWEERFSALERLTTVSAGPRR 1988
|
| Caenorhabditis briggsae |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
|||| | ++| ++ ||||+||||||| # + ||| |||| || |||| | | ++
Sbjct 2024 EPYLISREYGRNLEETIKLIKKHEAFEKSA#LAQEERFLALEKLTTFELKENQHREEETQK 2083
|
| Strongylocentrotus purpuratus |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKE 743
| || | |+| ++| |||||||||||||+ # ||| |||| || ||+|
Sbjct 2026 EQYLRSDDYGDSLDEVEKLIKRHEAFEKAL#YCQEERFTALEKLTTFELRE 2075
|
| Caenorhabditis elegans |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAE 748
|||| | ++| ++ ||||+||||||| # + ||| |||| || |||| | |
Sbjct 2027 EPYLISKEYGRNLEETIKLIKKHEAFEKSA#FAQEERFLALEKLTTFELKETQHRE 2081
|
| Brugia malayi |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQ 745
|||| | ++| ++ ||||+||||||| # + ||| |||| || |||| |
Sbjct 2033 EPYLVSKEYGRNLEETIKLIKKHEAFEKSA#NAQEERFLALEKLTTFELKEMQ 2084
|
| Sus scrofa |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIA 747
| +| + | | ++|||| |+|+|| |||| #++ |+ ||++ | ++ |
Sbjct 24 EAFLLNEDLGDSLDSVEALLKKHEDFEKSL#SAQEEKITALDEFVTKLIQNNHYA 77
|
| Monodelphis domestica |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTT 738
| +| + | |+++ +|| |+++|| ||++ # + |+ |+| |
Sbjct 487 EAFLENEDLGNSLGTVEALLQKHEDFEEAF#GAQEEKITTLDKTAT 531
|
| Xenopus tropicalis |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAE 748
| +| + | | ++|||| |+|+|| |||| #++ |+ ||++ | ++ |+
Sbjct 487 EAFLLNEDLGDSLDSVEALLKKHEDFEKSL#SAQEEKITALDEFATKLIQNNHYAK 541
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALE 734
| +| | | | ++|||| |||+|| |+|+ # |+ |||+
Sbjct 1464 EAFLNSEDKGDSLDSVEALIKKHEDFDKAI#NVQEEKIAALQ 1504
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAAL 733
| +|| ||| + ||+ |+++|| |+ #|+ |+ ||
Sbjct 275 EQLMASDDFGRDLASVQALLRKHEGLERDL#AALEEKVKAL 314
|
| Mus sp. |
Query 694 EPYLASGDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQIAERPAEE 753
|||||| |||||||||||||||||||||||#|||||||||||||||||||||| ||| ||
Sbjct 2020 EPYLASRDFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALEKPTTLELKERQTPERPTEE 2079
Query 701 DFGHTVDSVEKLIKRHEAFEKST#ASWAERFAALE 734
+||+ + +|| |+||| | |#|++ || |||
Sbjct 441 NFGYDLAAVEAAKKKHEAIETDT#AAYEERVKALE 474
|
 Ala
Ala

 Sequence conservation (by blast)
Sequence conservation (by blast)