XSB0479 : p53 transformation suppressor [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0053
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Cellular tumor antigen p53; Tumor suppressor p53; Phosphoprotein p53; Antigen NY-CO-13; p53 transformation suppressor |
| Gene Names | p53; P53 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 393 aa
Average Mass: 43.714 kDa
Monoisotopic Mass: 43.686 kDa
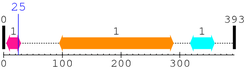
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| P53_TAD 1. | 5 | 29 | 46.4 | 1.1e-10 |
| --- cleavage 25 (inside P53_TAD 5..29) --- | ||||
| P53 1. | 95 | 289 | 504.2 | 1.7e-148 |
| P53_tetramer 1. | 318 | 359 | 85.5 | 1.9e-22 |
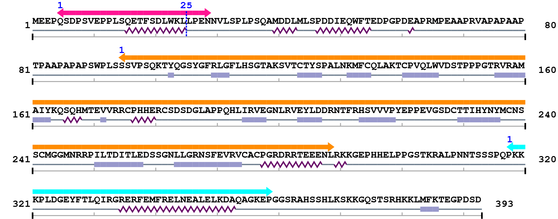
3. Sequence Information
Fasta Sequence: XSB0479.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 1A1U (NMR; -; A/C=324-358), 1AIE (X-ray; 150 A; A=326-356), 1C26 (X-ray; 170 A; A=325-356), 1DT7 (NMR; -; X/Y=367-388), 1GZH (X-ray; 260 A; A=95-292, C=95-292), 1H26 (X-ray; 224 A; E=376-386), 1HS5 (NMR; -; A/B=324-357), 1JSP (NMR; -; A=367-386), 1KZY (X-ray; 250 A; A/B=95-289), 1MA3 (X-ray; 200 A; B=372-389), 1OLG (NMR; -; A/B/C/D=319-360), 1OLH (NMR; -; A/B/C/D=319-360), 1PES (NMR; -; A/B/C/D=325-355), 1PET (NMR; -; A/B/C/D=325-355), 1SAE (NMR; -; A/B/C/D=319-360), 1SAF (NMR; -; A/B/C/D=319-360), 1SAH (NMR; -; A/B/C/D=319-360), 1SAJ (NMR; -; A/B/C/D=319-360), 1SAK (NMR; -; A/B/C/D=319-360), 1SAL (NMR; -; A/B/C/D=319-360), 1TSR (X-ray; 220 A; A/B/C=94-312), 1TUP (X-ray; 220 A; A/B/C=94-312), 1UOL (X-ray; 190 A; A/B=94-312), 1XQH (X-ray; 175 A; B/F=369-377), 1YC5 (X-ray; 140 A; B=372-389), 1YCQ (X-ray; 230 A; B=13-29), 1YCR (X-ray; 260 A; B=15-29), 1YCS (X-ray; 220 A; A=94-292), 2AC0 (X-ray; 180 A; A/B/C/D=94-293), 2ADY (X-ray; 250 A; A/B=94-293), 2AHI (X-ray; 185 A; A/B/C/D=94-293), 2ATA (X-ray; 220 A; A/B/C/D=94-293), 2B3G (X-ray; 160 A; B=33-60), 2BIM (X-ray; 198 A; A/B=94-312), 2BIN (X-ray; 190 A; A=94-312), 2BIO (X-ray; 190 A; A=94-312), 2BIP (X-ray; 180 A; A=94-312), 2BIQ (X-ray; 180 A; A=94-312), 2F1X (X-ray; 230 A; A/B=360-368), 2FEJ (NMR; -; A=94-297), 2FOJ (X-ray; 160 A; B=363-367), 2FOO (X-ray; 220 A; B=358-363), 2GS0 (NMR; -; B=20-71), 2H1L (X-ray; 316 A; M/N/O/P/Q/R/S/T/U/V/W/X=92-292), 2H2D (X-ray; 170 A; B=372-389), 2H2F (X-ray; 220 A; B=372-389), 2H4F (X-ray; 200 A; D=372-389), 2H4H (X-ray; 199 A; B=372-389), 2H4J (X-ray; 210 A; D=372-389), 2H59 (X-ray; 190 A; D/E=372-389), 2J0Z (NMR; -; A/B/C/D=326-356), 2J10 (NMR; -; A/B/C/D=326-356), 2J11 (NMR; -; A/B/C/D=332-356), 2J1W (X-ray; 180 A; A/B=94-312), 2J1X (X-ray; 165 A; A/B=94-312), 2J1Y (X-ray; 169 A; A/B/C/D=94-293), 2J1Z (X-ray; 180 A; A/B=94-312), 2J20 (X-ray; 180 A; A/B=94-312), 2J21 (X-ray; 160 A; A/B=94-312), 2K8F (NMR; -; B=1-39), 2OCJ (X-ray; 205 A; A/B/C/D=94-312), 2PCX (X-ray; 154 A; A=94-292), 2QVQ (X-ray; 200 A; A=94-289), 2QXA (X-ray; 150 A; A/B/C/D=95-289), 2QXB (X-ray; 250 A; A/B/C/D=95-289), 2QXC (X-ray; 185 A; A/B/C/D=95-289), 2VUK (X-ray; 150 A; A/B=94-312), 2Z5S (X-ray; 230 A; P/Q/R=15-29), 2Z5T (X-ray; 230 A; P/Q/R=15-29), 3D05 (X-ray; 170 A; A=94-293), 3D06 (X-ray; 120 A; A=94-293), 3D07 (X-ray; 220 A; A/B=94-293), 3D08 (X-ray; 140 A; A=94-293), 3D09 (X-ray; 190 A; A=94-293), 3D0A (X-ray; 180 A; A/B/C/D=94-293), 3DAB (X-ray; 190 A; B/D/F/H=15-29), 3DAC (X-ray; 180 A; B/P=17-37), 3SAK (NMR; -; A/B/C/D=319-360)
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] QETFSDLWKL25-LPENNVLSPL
Leu25  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gln16 | Glu17 | Thr18 | Phe19 | Ser20 | Asp21 | Leu22 | Trp23 | Lys24 | Leu25 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu26 | Pro27 | Glu28 | Asn29 | Asn30 | Val31 | Leu32 | Ser33 | Pro34 | Leu35 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 1915267] Farrell PJ, Allan GJ, Shanahan F, Vousden KH, Crook T, p53 is frequently mutated in Burkitt's lymphoma cell lines. EMBO J. 1991 Oct;10(10):2879-87.