XSB0701 : PREDICTED: similar to Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119) isoform 1 [Canis familiaris]
[ CaMP Format ]
This entry is computationally expanded from SB0086
* Basic Information
| Organism | Canis lupus familiaris (dog) |
| Protein Names | similar to Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119) isoform 1 |
| Gene Names | LOC609429; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 156 ESTs, 67 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 319 aa
Average Mass: 34.126 kDa
Monoisotopic Mass: 34.104 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
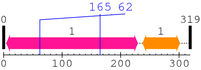
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Jun 1. | 5 | 229 | 436.7 | 3.6e-128 |
| --- cleavage 165 (inside Jun 5..229) --- | ||||
| --- cleavage 62 (inside Jun 5..229) --- | ||||
| bZIP_1 1. | 238 | 301 | 84.9 | 2.9e-22 |
3. Sequence Information
Fasta Sequence: XSB0701.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites]
Cleavage sites (±10aa)
[Site 1] GGGGFSAGLH165-SEPPVYANLS
His165  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly156 | Gly157 | Gly158 | Gly159 | Phe160 | Ser161 | Ala162 | Gly163 | Leu164 | His165 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser166 | Glu167 | Pro168 | Pro169 | Val170 | Tyr171 | Ala172 | Asn173 | Leu174 | Ser175 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PVSGAGMVAPAVASAAGGGGGGGGFSAGLHSEPPVYANLSNFNPGALSSGGGAPSYGAAA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 38.90 | 5 | PREDICTED: similar to Transcription factor AP-1 (A |
| 2 | N/A | 38.90 | 5 | J |
| 3 | Mus musculus | 38.90 | 4 | unnamed protein product |
| 4 | Homo sapiens | 38.90 | 4 | JUN |
| 5 | synthetic construct | 38.90 | 2 | v-jun sarcoma virus 17 oncogene-like |
| 6 | Bos taurus | 38.90 | 1 | PREDICTED: v-jun avian sarcoma virus 17 oncogene h |
| 7 | Pseudemys nelsoni | 38.90 | 1 | c-jun |
| 8 | Gallus gallus | 38.90 | 1 | jun oncogene |
| 9 | Crocodylus niloticus | 38.90 | 1 | c-jun |
| 10 | Monodelphis domestica | 38.90 | 1 | PREDICTED: similar to fc49b10 G-protein coupled re |
| 11 | Macaca mulatta | 38.90 | 1 | PREDICTED: similar to Transcription factor AP-1 (A |
| 12 | Serinus canaria | 38.90 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 13 | Sus scrofa | 38.90 | 1 | C-JUN protein |
| 14 | Rattus norvegicus | 38.90 | 1 | v-jun sarcoma virus 17 oncogene homolog |
| 15 | Oryctolagus cuniculus | 38.90 | 1 | c-jun transcription factor |
| 16 | Coturnix coturnix | 38.90 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 17 | Alligator mississippiensis | 38.90 | 1 | c-jun |
| 18 | Xenopus laevis | 37.70 | 3 | c-Jun protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| N/A | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Mus musculus | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Homo sapiens | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| synthetic construct | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Bos taurus | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Pseudemys nelsoni | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Gallus gallus | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Crocodylus niloticus | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Monodelphis domestica | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Macaca mulatta | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Serinus canaria | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Sus scrofa | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Rattus norvegicus | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Oryctolagus cuniculus | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Coturnix coturnix | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Alligator mississippiensis | Query 164 LH#SEPPVYANLSNFNP 179 ||#|||||||||||||| Query 164 LH#SEPPVYANLSNFNP 179 |
| Xenopus laevis | Query 164 LH#SEPPVYANLSNFNP 179 ||#+||||||||||||| Sbjct 157 LH#NEPPVYANLSNFNP 172 |
[Site 2] LRAKNSDLLT62-SPDVGLLKLA
Thr62  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu53 | Arg54 | Ala55 | Lys56 | Asn57 | Ser58 | Asp59 | Leu60 | Leu61 | Thr62 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser63 | Pro64 | Asp65 | Val66 | Gly67 | Leu68 | Leu69 | Lys70 | Leu71 | Ala72 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ILKQSMTLNLADPVGSLKPHLRAKNSDLLTSPDVGLLKLASPELERLIIQSSNGHITTTP |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 119.00 | 11 | unnamed protein product |
| 2 | Canis familiaris | 119.00 | 10 | PREDICTED: similar to Transcription factor AP-1 (A |
| 3 | Homo sapiens | 119.00 | 9 | jun oncogene |
| 4 | N/A | 119.00 | 7 | 1 |
| 5 | Bos taurus | 119.00 | 5 | jun oncogene |
| 6 | synthetic construct | 119.00 | 5 | v-jun sarcoma virus 17 oncogene-like |
| 7 | Macaca mulatta | 119.00 | 2 | PREDICTED: similar to Transcription factor AP-1 (A |
| 8 | Ovis aries | 119.00 | 2 | c-Jun protein |
| 9 | Macaca fuscata | 119.00 | 1 | transcription factor c-Jun |
| 10 | Rattus norvegicus | 117.00 | 5 | v-jun sarcoma virus 17 oncogene homolog |
| 11 | Sus scrofa | 114.00 | 1 | C-JUN protein |
| 12 | Monodelphis domestica | 106.00 | 2 | PREDICTED: similar to fc49b10 G-protein coupled re |
| 13 | Coturnix coturnix | 105.00 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 14 | Pseudemys nelsoni | 103.00 | 1 | c-jun |
| 15 | Tetraodon nigroviridis | 102.00 | 6 | unnamed protein product |
| 16 | Danio rerio | 102.00 | 5 | c-Jun |
| 17 | Ctenopharyngodon idella | 102.00 | 2 | c-Jun protein |
| 18 | Gallus gallus | 102.00 | 2 | jun oncogene |
| 19 | Serinus canaria | 102.00 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 20 | Alligator mississippiensis | 102.00 | 1 | c-jun |
| 21 | Carassius auratus | 102.00 | 1 | c-Jun protein |
| 22 | Crocodylus niloticus | 102.00 | 1 | c-jun |
| 23 | Takifugu rubripes | 101.00 | 6 | c-Jun protein |
| 24 | Xenopus laevis | 99.00 | 5 | c-Jun protein |
| 25 | Oncorhynchus mykiss | 96.30 | 2 | transcription factor AP-1 |
| 26 | Oryctolagus cuniculus | 78.60 | 1 | c-jun transcription factor |
| 27 | Xiphophorus maculatus | 71.20 | 2 | c-JUN |
| 28 | Pan troglodytes | 55.80 | 2 | PREDICTED: jun D proto-oncogene |
| 29 | Cyprinus carpio | 54.30 | 1 | JUNB_CYPCA Transcription factor jun-B gb |
| 30 | Strongylocentrotus purpuratus | 46.60 | 1 | PREDICTED: similar to transcription factor AP-1 |
| 31 | Apis mellifera | 37.40 | 1 | PREDICTED: similar to c-Jun protein |
| 32 | Aedes aegypti | 35.40 | 1 | jun |
| 33 | Taenia solium | 33.10 | 1 | c-jun |
| 34 | Taenia crassiceps | 33.10 | 1 | c-jun |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Canis familiaris | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Homo sapiens | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| N/A | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Bos taurus | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| synthetic construct | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Macaca mulatta | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Ovis aries | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Macaca fuscata | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Rattus norvegicus | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||+||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Sus scrofa | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||+|||||||++|||||#|||||||||||||||||||||||||||||| Sbjct 33 ILKQSMTLNLADPVGNLKPHLRAQDSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Monodelphis domestica | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +||||+|||||||| ||||||| || ||||#|||||||||||||||||||||||| ||||| Sbjct 33 VLKQSLTLNLADPVSSLKPHLRNKNPDLLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 92 |
| Coturnix coturnix | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||||||||+|| ||||||| ||+|+||#|||||||||||||||||||||||| ||||| Sbjct 32 VLKQSMTLNLSDPASSLKPHLRNKNADILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Pseudemys nelsoni | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||+|||||||| +|||||| ||+|+||#|||||||||||||||||||||||| ||||| Sbjct 33 VLKQNMTLNLADPSSNLKPHLRNKNADILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 92 |
| Tetraodon nigroviridis | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +||||| || |||||||||| |+|||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLCDPAGSLKPHLRAKASELLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Danio rerio | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +|||||+|| |+|||||||| ||+||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLSDPAGNLKPHLRAKASDILT#SPDVGLLKLASPELERLIIQSSNGMITTTP 91 |
| Ctenopharyngodon idella | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +||||| || |+|||||||| ||+||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLTDPAGNLKPHLRAKASDILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Gallus gallus | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||||||||+| ||||||| ||+|+||#|||||||||||||||||||||||| ||||| Sbjct 29 VLKQSMTLNLSDAASSLKPHLRNKNADILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 88 |
| Serinus canaria | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||+|||||+|| +|||||| ||+|+||#|||||||||||||||||||||||| ||||| Sbjct 33 VLKQNMTLNLSDPSSNLKPHLRNKNADILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 92 |
| Alligator mississippiensis | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||| +|||||| |++||||#|||||||||||||||||||||||| ||||| Sbjct 34 LKQSMTLNLADPASALKPHLRNKSADLLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 92 |
| Carassius auratus | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||+|||||| || |+|||||||| +|+||#|||||||||||||||||||||||| ||||| Sbjct 29 LKRSMTLNLTDPSGNLKPHLRAKANDILT#SPDVGLLKLASPELERLIIQSSNGMITTTP 87 |
| Crocodylus niloticus | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||| +|||||| |++||||#|||||||||||||||||||||||| ||||| Sbjct 34 LKQSMTLNLADPASALKPHLRNKSTDLLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 92 |
| Takifugu rubripes | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +|||||+|| |+|||||||| |+|||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLSDPTGTLKPHLRAKASELLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Xenopus laevis | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||||||||+|| ++||||| | ++|||#|||||||||||||||||||||||| ||||| Sbjct 31 VLKQSMTLNLSDPSSAIKPHLRNKAAELLT#SPDVGLLKLASPELERLIIQSSNGMITTTP 90 |
| Oncorhynchus mykiss | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || ||||||||| +|||||+|| ||+||#|||| ||||||||||||||||||| ||||| Sbjct 32 LKHSMTLNLADPTITLKPHLQAKASDILT#SPDVQLLKLASPELERLIIQSSNGLITTTP 90 |
| Oryctolagus cuniculus | Query 54 RAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||#|||||||||||||||||||||||||||||| Query 54 RAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Xiphophorus maculatus | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||+||||| || + || | + |+||#||||||||||||||||||||| +| | || Sbjct 37 LKQTMTLNLNDPK-NFKPQLGGR-LDILT#SPDVGLLKLASPELERLIIQSCSGLTTPTP 93 |
| Pan troglodytes | Query 35 KQSMTLNLADPVGS-LKPH-------LRAKNSD-------LLT#SPDVGLLKLASPELERL 79 | ++||+|++ | + ||| ||| + || #|||+||||||||||||| Sbjct 47 KDALTLSLSEQVAAALKPAAAPPPTPLRADGAPSAAPPDGLLA#SPDLGLLKLASPELERL 106 Query 80 IIQSSNGHITTTP 92 ||| ||| +|||| Sbjct 107 IIQ-SNGLVTTTP 118 |
| Cyprinus carpio | Query 35 KQSMTLNLADPVGSLKPHLRAKNSDLL--T#SPDVGLLKLASPELERLIIQSSNGHITT 90 |||| ||| +| +|| ||| +# | | ||||||||||||||+||| +|| Sbjct 33 KQSMNLNLTEPYRNLK-------SDLYHTS#GADSGSLKLASPELERLIIQTSNGVLTT 83 |
| Strongylocentrotus purpuratus | Query 35 KQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |+ |||+| +| | + ||+#+||| +|||||||||++|| | |+| ||| Sbjct 17 KKCMTLDLDVGMGK-----RKLTAALLS#TPDVQMLKLASPELEKMII-SQQGNICTTP 68 |
| Apis mellifera | Query 34 LKQSMTLNLAD-----------PVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQ 82 ||+++||+| + +| + | | + +|+#|||+ +|||+|||||+ || Sbjct 24 LKRNLTLDLNECQRQGPEAKKPRLGPIPPALN-NVTPILS#SPDLNMLKLSSPELEKFIIA 82 Query 83 SSNGHIT 89 + +| Sbjct 83 QQDSLVT 89 |
| Aedes aegypti | Query 60 LLT#SPDVGLLKLASPELERLIIQSS 84 ++|#|||+ +||| |||||++|| |+ Sbjct 64 VIT#SPDLQVLKLVSPELEKIIINSA 88 |
| Taenia solium | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASP 74 |||||||||||||+ | | || # || |+| Sbjct 73 ILKQSMTLNLADPL----PELSTAGEPLLL#GLVCGLQGTAAP 110 |
| Taenia crassiceps | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASP 74 |||||||||||||+ | | || # || |+| Sbjct 73 ILKQSMTLNLADPL----PELSTAGEPLLL#GLVCGLQGTAAP 110 |
* References
None.