XSB0801 : unnamed protein product [Mus musculus]
[ CaMP Format ]
This entry is computationally expanded from SB0086
* Basic Information
| Organism | Mus musculus (house mouse) |
| Protein Names | Putative uncharacterized protein |
| Gene Names | Jun |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
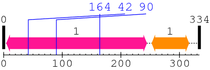
Length: 334 aa
Average Mass: 35.975 kDa
Monoisotopic Mass: 35.952 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Jun 1. | 5 | 244 | 444.2 | 2e-130 |
| --- cleavage 164 (inside Jun 5..244) --- | ||||
| --- cleavage 42 (inside Jun 5..244) --- | ||||
| --- cleavage 90 (inside Jun 5..244) --- | ||||
| bZIP_1 1. | 253 | 316 | 84.9 | 2.9e-22 |
3. Sequence Information
Fasta Sequence: XSB0801.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
3 [sites]
Cleavage sites (±10aa)
[Site 1] GGGGYSASLH164-SEPPVYANLS
His164  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly155 | Gly156 | Gly157 | Gly158 | Tyr159 | Ser160 | Ala161 | Ser162 | Leu163 | His164 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser165 | Glu166 | Pro167 | Pro168 | Val169 | Tyr170 | Ala171 | Asn172 | Leu173 | Ser174 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QPVSGAGMVAPAVASVAGAGGGGGYSASLHSEPPVYANLSNFNPGALSSGGGAPSYGAAG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 38.90 | 5 | PREDICTED: similar to Transcription factor AP-1 (A |
| 2 | N/A | 38.90 | 5 | 1 |
| 3 | Mus musculus | 38.90 | 4 | Jun oncogene |
| 4 | Homo sapiens | 38.90 | 4 | JUN |
| 5 | synthetic construct | 38.90 | 2 | v-jun sarcoma virus 17 oncogene-like |
| 6 | Bos taurus | 38.90 | 1 | PREDICTED: v-jun avian sarcoma virus 17 oncogene h |
| 7 | Pseudemys nelsoni | 38.90 | 1 | c-jun |
| 8 | Gallus gallus | 38.90 | 1 | jun oncogene |
| 9 | Crocodylus niloticus | 38.90 | 1 | c-jun |
| 10 | Monodelphis domestica | 38.90 | 1 | PREDICTED: similar to fc49b10 G-protein coupled re |
| 11 | Macaca mulatta | 38.90 | 1 | PREDICTED: similar to Transcription factor AP-1 (A |
| 12 | Serinus canaria | 38.90 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 13 | Sus scrofa | 38.90 | 1 | C-JUN protein |
| 14 | Rattus norvegicus | 38.90 | 1 | v-jun sarcoma virus 17 oncogene homolog |
| 15 | Oryctolagus cuniculus | 38.90 | 1 | c-jun transcription factor |
| 16 | Coturnix coturnix | 38.90 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 17 | Alligator mississippiensis | 38.90 | 1 | c-jun |
| 18 | Xenopus laevis | 37.70 | 3 | c-Jun protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| N/A | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Mus musculus | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Homo sapiens | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| synthetic construct | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Bos taurus | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Pseudemys nelsoni | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Gallus gallus | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Crocodylus niloticus | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Monodelphis domestica | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Macaca mulatta | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Serinus canaria | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Sus scrofa | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Rattus norvegicus | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Oryctolagus cuniculus | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Coturnix coturnix | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Alligator mississippiensis | Query 163 LH#SEPPVYANLSNFNP 178 ||#|||||||||||||| Query 163 LH#SEPPVYANLSNFNP 178 |
| Xenopus laevis | Query 163 LH#SEPPVYANLSNFNP 178 ||#+||||||||||||| Sbjct 157 LH#NEPPVYANLSNFNP 172 |
[Site 2] ILKQSMTLNL42-ADPVGSLKPH
Leu42  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile33 | Leu34 | Lys35 | Gln36 | Ser37 | Met38 | Thr39 | Leu40 | Asn41 | Leu42 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala43 | Asp44 | Pro45 | Val46 | Gly47 | Ser48 | Leu49 | Lys50 | Pro51 | His52 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ALNASFLQSESGAYGYSNPKILKQSMTLNLADPVGSLKPHLRAKNSDLLTSPDVGLLKLA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Canis familiaris | 119.00 | 6 | PREDICTED: similar to Transcription factor AP-1 (A |
| 2 | Mus musculus | 119.00 | 4 | unnamed protein product |
| 3 | Bos taurus | 119.00 | 3 | PREDICTED: v-jun avian sarcoma virus 17 oncogene h |
| 4 | Rattus norvegicus | 118.00 | 1 | v-jun sarcoma virus 17 oncogene homolog |
| 5 | Macaca fuscata | 117.00 | 1 | transcription factor c-Jun |
| 6 | Macaca mulatta | 117.00 | 1 | PREDICTED: similar to Transcription factor AP-1 (A |
| 7 | Ovis aries | 116.00 | 2 | c-Jun protein |
| 8 | N/A | 115.00 | 5 | 1 |
| 9 | Homo sapiens | 115.00 | 3 | JUN |
| 10 | synthetic construct | 115.00 | 2 | v-jun sarcoma virus 17 oncogene homolog (avian) |
| 11 | Sus scrofa | 114.00 | 1 | C-JUN protein |
| 12 | Monodelphis domestica | 100.00 | 1 | PREDICTED: similar to fc49b10 G-protein coupled re |
| 13 | Pseudemys nelsoni | 100.00 | 1 | c-jun |
| 14 | Crocodylus niloticus | 97.10 | 1 | c-jun |
| 15 | Alligator mississippiensis | 97.10 | 1 | c-jun |
| 16 | Gallus gallus | 94.70 | 1 | jun oncogene |
| 17 | Coturnix coturnix | 92.40 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 18 | Serinus canaria | 91.30 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 19 | Takifugu rubripes | 88.20 | 4 | c-Jun protein |
| 20 | Tetraodon nigroviridis | 84.00 | 2 | unnamed protein product |
| 21 | Carassius auratus | 83.60 | 1 | c-Jun protein |
| 22 | Danio rerio | 82.80 | 2 | c-Jun protein |
| 23 | Ctenopharyngodon idella | 80.90 | 2 | c-Jun protein |
| 24 | Xenopus laevis | 78.20 | 5 | c-Jun protein |
| 25 | Oncorhynchus mykiss | 75.90 | 1 | transcription factor AP-1 |
| 26 | Xiphophorus maculatus | 57.40 | 2 | c-JUN |
| 27 | Taenia solium | 40.00 | 1 | c-jun |
| 28 | Taenia crassiceps | 40.00 | 1 | c-jun |
| 29 | Oryctolagus cuniculus | 39.70 | 1 | c-jun transcription factor |
Top-ranked sequences
| organism | matching |
|---|---|
| Canis familiaris | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Mus musculus | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Bos taurus | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Rattus norvegicus | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Macaca fuscata | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| ||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 13 ALNASFLPSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Macaca mulatta | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| ||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 13 ALNASFLPSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Ovis aries | Query 15 NASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 15 NASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| N/A | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| |||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Homo sapiens | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| |||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| synthetic construct | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| |||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Sus scrofa | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||+|||||||++||||||||||||||| Sbjct 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAQDSDLLTSPDVGLLKLA 72 |
| Monodelphis domestica | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||+|+| ||| ||||+ |+||||+||||#|||| ||||||| || |||||||||||||| Sbjct 13 ALNAAFVQPESGGYGYSHAKVLKQSLTLNL#ADPVSSLKPHLRNKNPDLLTSPDVGLLKLA 72 |
| Pseudemys nelsoni | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||+|+| ||| |||+|||+|||+|||||#||| +|||||| ||+|+|||||||||||| Sbjct 13 ALNATFVQPESGGYGYNNPKVLKQNMTLNL#ADPSSNLKPHLRNKNADILTSPDVGLLKLA 72 |
| Crocodylus niloticus | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||+| ||| |||+||| |||||||||#||| +|||||| |++|||||||||||||| Sbjct 13 ALNAAFAPPESGGYGYNNPKALKQSMTLNL#ADPASALKPHLRNKSTDLLTSPDVGLLKLA 72 |
| Alligator mississippiensis | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||+| ||| |||+||| |||||||||#||| +|||||| |++|||||||||||||| Sbjct 13 ALNAAFAPPESGGYGYNNPKALKQSMTLNL#ADPASALKPHLRNKSADLLTSPDVGLLKLA 72 |
| Gallus gallus | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |||||| ||| |||+| |+|||||||||#+| ||||||| ||+|+|||||||||||| Sbjct 9 ALNASFAPPESGGYGYNNAKVLKQSMTLNL#SDAASSLKPHLRNKNADILTSPDVGLLKLA 68 |
| Coturnix coturnix | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |||||| || ||||+| |+|||||||||#+|| ||||||| ||+|+|||||||||||| Sbjct 13 ALNASFAPPES-AYGYNNAKVLKQSMTLNL#SDPASSLKPHLRNKNADILTSPDVGLLKLA 71 |
| Serinus canaria | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||+| | ||| |||+| |+|||+|||||#+|| +|||||| ||+|+|||||||||||| Sbjct 13 ALSAGFAPPESGGYGYNNAKVLKQNMTLNL#SDPSSNLKPHLRNKNADILTSPDVGLLKLA 72 |
| Takifugu rubripes | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 +||| | | ++ ||||||| || +|||||#+|| |+|||||||| |+||||||||||||| Sbjct 13 SLNA-FSQHDTAGYGYSNPKALKHNMTLNL#SDPTGTLKPHLRAKASELLTSPDVGLLKLA 71 |
| Tetraodon nigroviridis | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 +||| | | ++ +|| +|| || +|||||# || |||||||||| |+||||||||||||| Sbjct 13 SLNA-FSQHDTAGFGYGHPKALKHNMTLNL#CDPAGSLKPHLRAKASELLTSPDVGLLKLA 71 |
| Carassius auratus | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 +||++| | +| |+|| | | ||+||||||# || |+|||||||| +|+|||||||||||| Sbjct 9 SLNSAFSQHDSAAFGY-NHKALKRSMTLNL#TDPSGNLKPHLRAKANDILTSPDVGLLKLA 67 |
| Danio rerio | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 +||++| | +| +|| | | || +|||||#+|| |+|||||||| ||+|||||||||||| Sbjct 13 SLNSAFSQHDSATFGY-NHKALKHNMTLNL#SDPAGNLKPHLRAKASDILTSPDVGLLKLA 71 |
| Ctenopharyngodon idella | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ++|++| | +| +|| | | || +|||||# || |+|||||||| ||+|||||||||||| Sbjct 13 SVNSAFSQHDSAPFGY-NHKALKHNMTLNL#TDPAGNLKPHLRAKASDILTSPDVGLLKLA 71 |
| Xenopus laevis | Query 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||+|+| | ++ +|||+ |+|||||||||#+|| ++||||| | ++||||||||||||| Sbjct 13 ALSAAFAQHDTVSYGYN--KVLKQSMTLNL#SDPSSAIKPHLRNKAAELLTSPDVGLLKLA 70 |
| Oncorhynchus mykiss | Query 17 SFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 +| | ++ ||| ||| || ||||||#||| +|||||+|| ||+|||||| ||||| Sbjct 16 AFSQHQNAGYGY-NPKALKHSMTLNL#ADPTITLKPHLQAKASDILTSPDVQLLKLA 70 |
| Xiphophorus maculatus | Query 13 ALNASFLQSESGA-YGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKL 71 |+||| | + | ||+ ||| |||+|||||# || + || | + |+||||||||||| Sbjct 16 AVNASNSQHDGAAVYGF-NPKTLKQTMTLNL#NDP-KNFKPQLGGR-LDILTSPDVGLLKL 72 Query 72 A 72 | Query 72 A 72 |
| Taenia solium | Query 19 LQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGL 68 | +| | | |||||||||||||#|||+ | | || || Sbjct 61 LTAEEGTVG--NPKILKQSMTLNL#ADPL----PELSTAGEPLLLGLVCGL 104 |
| Taenia crassiceps | Query 19 LQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGL 68 | +| | | |||||||||||||#|||+ | | || || Sbjct 61 LTAEEGTVG--NPKILKQSMTLNL#ADPL----PELSTAGEPLLLGLVCGL 104 |
| Oryctolagus cuniculus | Query 54 RAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||| Query 54 RAKNSDLLTSPDVGLLKLA 72 |
[Site 3] IQSSNGHITT90-TPTPTQFLCP
Thr90  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile81 | Gln82 | Ser83 | Ser84 | Asn85 | Gly86 | His87 | Ile88 | Thr89 | Thr90 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr91 | Pro92 | Thr93 | Pro94 | Thr95 | Gln96 | Phe97 | Leu98 | Cys99 | Pro100 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LTSPDVGLLKLASPELERLIIQSSNGHITTTPTPTQFLCPKNVTDEQEGFAEGFVRALAE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 122.00 | 11 | Jun oncogene |
| 2 | Homo sapiens | 122.00 | 10 | jun oncogene |
| 3 | Canis familiaris | 122.00 | 10 | PREDICTED: similar to Transcription factor AP-1 (A |
| 4 | N/A | 122.00 | 7 | 1 |
| 5 | synthetic construct | 122.00 | 5 | v-jun sarcoma virus 17 oncogene-like |
| 6 | Rattus norvegicus | 122.00 | 5 | v-jun sarcoma virus 17 oncogene homolog |
| 7 | Bos taurus | 122.00 | 5 | PREDICTED: v-jun avian sarcoma virus 17 oncogene h |
| 8 | Macaca mulatta | 122.00 | 2 | PREDICTED: similar to Transcription factor AP-1 (A |
| 9 | Sus scrofa | 122.00 | 1 | C-JUN protein |
| 10 | Oryctolagus cuniculus | 122.00 | 1 | c-jun transcription factor |
| 11 | Macaca fuscata | 122.00 | 1 | transcription factor c-Jun |
| 12 | Danio rerio | 118.00 | 5 | c-Jun protein |
| 13 | Ctenopharyngodon idella | 118.00 | 2 | c-Jun protein |
| 14 | Monodelphis domestica | 118.00 | 2 | PREDICTED: similar to fc49b10 G-protein coupled re |
| 15 | Gallus gallus | 118.00 | 2 | jun oncogene |
| 16 | Pseudemys nelsoni | 118.00 | 1 | c-jun |
| 17 | Alligator mississippiensis | 118.00 | 1 | c-jun |
| 18 | Coturnix coturnix | 118.00 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 19 | Crocodylus niloticus | 118.00 | 1 | c-jun |
| 20 | Serinus canaria | 118.00 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 21 | Xenopus laevis | 117.00 | 7 | c-Jun protein |
| 22 | Carassius auratus | 117.00 | 1 | c-Jun protein |
| 23 | Tetraodon nigroviridis | 115.00 | 6 | unnamed protein product |
| 24 | Oncorhynchus mykiss | 114.00 | 2 | transcription factor AP-1 |
| 25 | Takifugu rubripes | 112.00 | 6 | c-Jun protein |
| 26 | Xiphophorus maculatus | 104.00 | 2 | c-JUN |
| 27 | Ovis aries | 91.70 | 2 | c-Jun protein |
| 28 | Pan troglodytes | 80.90 | 2 | PREDICTED: jun D proto-oncogene |
| 29 | Cyprinus carpio | 79.00 | 1 | JUNB_CYPCA Transcription factor jun-B gb |
| 30 | Strongylocentrotus purpuratus | 78.60 | 1 | PREDICTED: similar to transcription factor AP-1 |
| 31 | Apis mellifera | 65.50 | 1 | PREDICTED: similar to c-Jun protein |
| 32 | Aedes aegypti | 58.50 | 1 | jun |
| 33 | Anopheles gambiae str. PEST | 52.80 | 1 | ENSANGP00000020405 |
| 34 | Xenopus tropicalis | 45.40 | 1 | jun B proto-oncogene |
| 35 | Ciona intestinalis | 44.30 | 1 | transcription factor protein |
| 36 | Tribolium castaneum | 42.00 | 1 | PREDICTED: similar to Transcription factor AP-1 (J |
| 37 | Poecilia reticulata | 37.40 | 1 | transcription factor JunB |
| 38 | Drosophila pseudoobscura | 37.40 | 1 | GA15338-PA |
| 39 | Schistosoma japonicum | 35.00 | 1 | SJCHGC03684 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Homo sapiens | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Canis familiaris | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| N/A | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| synthetic construct | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Rattus norvegicus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Bos taurus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Macaca mulatta | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Sus scrofa | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Oryctolagus cuniculus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Macaca fuscata | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Danio rerio | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGMITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 119 |
| Ctenopharyngodon idella | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 119 |
| Monodelphis domestica | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 61 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Gallus gallus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 57 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 116 |
| Pseudemys nelsoni | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 61 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Alligator mississippiensis | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 61 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Coturnix coturnix | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 119 |
| Crocodylus niloticus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 61 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Serinus canaria | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 61 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Xenopus laevis | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||+|||||||| Sbjct 59 LTSPDVGLLKLASPELERLIIQSSNGMITT#TPTPTQFLCPKNVTDEQEGFADGFVRALAE 118 |
| Carassius auratus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||| ||||| Sbjct 56 LTSPDVGLLKLASPELERLIIQSSNGMITT#TPTPTQFLCPKNVTDEQEGFAEGFARALAE 115 |
| Tetraodon nigroviridis | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||+||||||+||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFICPKNVTEEQEGFAEGFVRALAE 119 |
| Oncorhynchus mykiss | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||| ||||||||||||||||||| |||#||||||||||+||||||||||||||||||| Sbjct 59 LTSPDVQLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPRNVTDEQEGFAEGFVRALAE 118 |
| Takifugu rubripes | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||+ |||||+||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFISPKNVTEEQEGFAEGFVRALAE 119 |
| Xiphophorus maculatus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||| +| |#|||||||+|||||+|||||||||||||||| Sbjct 62 LTSPDVGLLKLASPELERLIIQSCSG--LT#TPTPTQFICPKNVSDEQEGFAEGFVRALAE 119 |
| Ovis aries | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVT 104 ||||||||||||||||||||||||||||||#|||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVT 104 |
| Pan troglodytes | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 | |||+||||||||||||||||| || +||#||| +||| || |++ ||||||+|| + Sbjct 88 LASPDLGLLKLASPELERLIIQS-NGLVTT#TPTSSQFLYPKVAASEEQEFAEGFVKALED 146 |
| Cyprinus carpio | Query 65 DVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 | | ||||||||||||||+||| + |#|||| |+| + +||||||||||||+|| Sbjct 58 DSGSLKLASPELERLIIQTSNG-VLT#TPTPGQYLYSRGITDEQEGFAEGFVKAL 110 |
| Strongylocentrotus purpuratus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 |++||| +|||||||||++|| | |+| |#|||| ||+ |||||+|| ||+||| || Sbjct 38 LSTPDVQMLKLASPELEKMII-SQQGNICT#TPTPGQFISPKNVTEEQAAFAQGFVEAL 94 |
| Apis mellifera | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |+|||+ +|||+|||||+ || + +| # |||| | || ||+ || +| ||| || | Sbjct 61 LSSPDLNMLKLSSPELEKFIIAQQDSLVTN#LVTPTQILFPKAVTEAQELYARGFVDALNE 120 |
| Aedes aegypti | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 +||||+ +||| |||||++|| |+ #||||+ | | | ||| ||+|| || Sbjct 65 ITSPDLQVLKLVSPELEKIIINSA---ALP#TPTPSSILYPTKATTEQEQFAKGFDEAL 119 |
| Anopheles gambiae str. PEST | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 | |||+ +||| |||||++| | #||||+ + | + | ||+ ||+|| || Sbjct 14 LPSPDLQMLKLVSPELEKII---STNATLP#TPTPSAIIFPPSATSEQQQFAKGFEDAL 68 |
| Xenopus tropicalis | Query 64 PDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKN--VTDEQEGFAEGFVRAL 118 | ||+|+|||||+|| | | |+ # + + +|+||||| +|||+|| Sbjct 61 PAPSSLKMAAPELERMIIHSGPGVISG#PSGGQLYYSSRGNGITEEQEGFVDGFVKAL 117 |
| Ciona intestinalis | Query 61 LTSPD-VGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 |++|| | +||+ ||+ +|+++ || + #| +|| +| | ||| +|+|||| | Sbjct 134 LSTPDLVNMLKVGSPDFDRMLVHSSQAN---#TQNNSQFF-QRNATQEQEIYAQGFVREL 188 |
| Tribolium castaneum | Query 63 SPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 +|| |||+ +| +| +|+ ++ | #||+|+ + |++|| ||| || ||| || Sbjct 32 TPD--LLKVDTPTIENIILANNISH---#TPSPS-LIFPRDVTVEQEKFAGGFVEAL 81 |
| Poecilia reticulata | Query 96 QFLCPKNVTDEQEGFAEGFVRALAE 120 |+ + +||||||||||||+|| | Sbjct 1 QYFYNRGITDEQEGFAEGFVKALDE 25 |
| Drosophila pseudoobscura | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPK--NVTDEQEGFAEGFVRAL 118 + |||+ + +|+||++++ + |+ #|| | + | || ||| | +|| || Sbjct 82 INSPDLQAKTVNTPDLEKILLSN---HMMQ#TPQPGKVFPTKVGPVTSEQEAFGKGFEEAL 138 |
| Schistosoma japonicum | Query 87 HITT#TP-TPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |+ #+| || || ||+||+||| || | + | | Sbjct 91 HLKA#SPQTPGSFLNPKHVTEEQERFANVFTQKLNE 125 |
* References
[PubMed ID: 10349636] Carninci P, Hayashizaki Y, High-efficiency full-length cDNA cloning. Methods Enzymol. 1999;303:19-44.
[PubMed ID: 11042159] ... Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y, Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 2000 Oct;10(10):1617-30.
[PubMed ID: 11076861] ... Shibata K, Itoh M, Aizawa K, Nagaoka S, Sasaki N, Carninci P, Konno H, Akiyama J, Nishi K, Kitsunai T, Tashiro H, Itoh M, Sumi N, Ishii Y, Nakamura S, Hazama M, Nishine T, Harada A, Yamamoto R, Matsumoto H, Sakaguchi S, Ikegami T, Kashiwagi K, Fujiwake S, Inoue K, Togawa Y, RIKEN integrated sequence analysis (RISA) system--384-format sequencing pipeline with 384 multicapillary sequencer. Genome Res. 2000 Nov;10(11):1757-71.
[PubMed ID: 11217851] ... Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, Adachi J, Fukuda S, Aizawa K, Izawa M, Nishi K, Kiyosawa H, Kondo S, Yamanaka I, Saito T, Okazaki Y, Gojobori T, Bono H, Kasukawa T, Saito R, Kadota K, Matsuda H, Ashburner M, Batalov S, Casavant T, Fleischmann W, Gaasterland T, Gissi C, King B, Kochiwa H, Kuehl P, Lewis S, Matsuo Y, Nikaido I, Pesole G, Quackenbush J, Schriml LM, Staubli F, Suzuki R, Tomita M, Wagner L, Washio T, Sakai K, Okido T, Furuno M, Aono H, Baldarelli R, Barsh G, Blake J, Boffelli D, Bojunga N, Carninci P, de Bonaldo MF, Brownstein MJ, Bult C, Fletcher C, Fujita M, Gariboldi M, Gustincich S, Hill D, Hofmann M, Hume DA, Kamiya M, Lee NH, Lyons P, Marchionni L, Mashima J, Mazzarelli J, Mombaerts P, Nordone P, Ring B, Ringwald M, Rodriguez I, Sakamoto N, Sasaki H, Sato K, Schonbach C, Seya T, Shibata Y, Storch KF, Suzuki H, Toyo-oka K, Wang KH, Weitz C, Whittaker C, Wilming L, Wynshaw-Boris A, Yoshida K, Hasegawa Y, Kawaji H, Kohtsuki S, Hayashizaki Y, Functional annotation of a full-length mouse cDNA collection. Nature. 2001 Feb 8;409(6821):685-90.
[PubMed ID: 12466851] ... Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y, Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002 Dec 5;420(6915):563-73.
[PubMed ID: 16141072] ... Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y, The transcriptional landscape of the mammalian genome. Science. 2005 Sep 2;309(5740):1559-63.
[PubMed ID: 16141073] ... Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C, Antisense transcription in the mammalian transcriptome. Science. 2005 Sep 2;309(5740):1564-6.