XSB1030 : PREDICTED: caspase 9 isoform 5 [Pan troglodytes]
[ CaMP Format ]
This entry is computationally expanded from SB0042
* Basic Information
| Organism | Pan troglodytes (chimpanzee) |
| Protein Names | caspase 9 isoform 5 |
| Gene Names | CASP9; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 12 mRNAs, 165 ESTs, 15 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 416 aa
Average Mass: 46.310 kDa
Monoisotopic Mass: 46.280 kDa
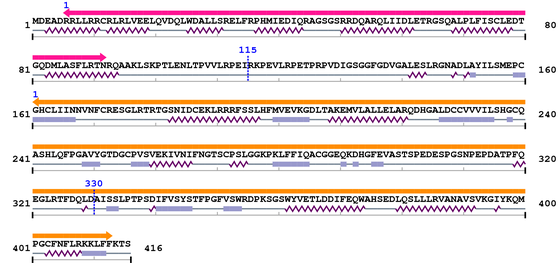
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| CARD 1. | 6 | 92 | 91.5 | 3e-24 |
| --- cleavage 115 --- | ||||
| Peptidase_C14 1. | 161 | 413 | 306.9 | 4.2e-89 |
| --- cleavage 330 (inside Peptidase_C14 161..413) --- | ||||
3. Sequence Information
Fasta Sequence: XSB1030.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
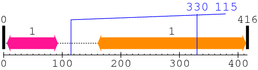
* Cleavage Information
2 [sites]
Cleavage sites (±10aa)
[Site 1] EGLRTFDQLD330-AISSLPTPSD
Asp330  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu321 | Gly322 | Leu323 | Arg324 | Thr325 | Phe326 | Asp327 | Gln328 | Leu329 | Asp330 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala331 | Ile332 | Ser333 | Ser334 | Leu335 | Pro336 | Thr337 | Pro338 | Ser339 | Asp340 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TSPEDESPGSNPEPDATPFQEGLRTFDQLDAISSLPTPSDIFVSYSTFPGFVSWRDPKSG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 129.00 | 29 | G |
| 2 | Homo sapiens | 129.00 | 13 | caspase 9, apoptosis-related cysteine protease |
| 3 | Pan troglodytes | 129.00 | 6 | PREDICTED: caspase 9 isoform 5 |
| 4 | synthetic construct | 129.00 | 6 | Homo sapiens caspase 9, apoptosis-related cysteine |
| 5 | Macaca mulatta | 117.00 | 6 | PREDICTED: similar to caspase 9 isoform alpha prep |
| 6 | Macaca fascicularis | 117.00 | 2 | caspase 9 |
| 7 | Felis catus | 107.00 | 3 | CASP9 |
| 8 | Canis familiaris | 105.00 | 3 | PREDICTED: caspase 9, apoptosis-related cysteine p |
| 9 | Canis lupus familiaris | 105.00 | 2 | caspase 9 |
| 10 | Monodelphis domestica | 97.40 | 3 | PREDICTED: similar to ICE-LAP6 |
| 11 | Rattus norvegicus | 96.30 | 5 | caspase 9 |
| 12 | Mus musculus | 91.30 | 12 | unnamed protein product |
| 13 | Gallus gallus | 73.90 | 2 | caspase 9 |
| 14 | Dicentrarchus labrax | 69.70 | 2 | caspase-9 |
| 15 | Xenopus laevis | 66.20 | 6 | caspase-9 |
| 16 | Danio rerio | 65.90 | 4 | Zgc:101776 protein |
| 17 | Tetraodon nigroviridis | 65.10 | 2 | unnamed protein product |
| 18 | Oncorhynchus mykiss | 60.80 | 1 | caspase-9 |
| 19 | Oryctolagus cuniculus | 52.00 | 3 | caspase 9 |
| 20 | Tribolium castaneum | 40.40 | 1 | PREDICTED: similar to Caspase precursor (drICE), p |
| 21 | Branchiostoma floridae | 40.00 | 1 | AF412335_1 amphiCASP-3/7 |
| 22 | Schistosoma japonicum | 39.30 | 4 | SJCHGC04214 protein |
| 23 | Anopheles gambiae str. PEST | 38.50 | 10 | ENSANGP00000008707 |
| 24 | Aedes aegypti | 38.10 | 2 | caspase-1 |
| 25 | Strongylocentrotus purpuratus | 37.70 | 4 | PREDICTED: similar to caspase-3, partial |
| 26 | Xenopus tropicalis | 37.70 | 2 | caspase 7, apoptosis-related cysteine protease |
| 27 | Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit | 36.60 | 3 | CASP3_CRILO Caspase-3 precursor (CASP-3) (Apopain) |
| 28 | Bos taurus | 35.80 | 4 | PREDICTED: similar to Mch3 isoform alpha |
| 29 | Geodia cydonium | 35.80 | 2 | caspase-3 |
| 30 | Equus caballus | 35.40 | 3 | caspase-3 |
| 31 | Gobius niger | 35.00 | 1 | caspase 3 |
| 32 | Sus scrofa | 34.70 | 1 | caspase 3 |
| 33 | Cricetulus griseus | 34.30 | 1 | caspase 3 |
| 34 | Contains: Caspase-7 subunit p20; Caspase-7 subunit p11 | 34.30 | 1 | CASP7_MESAU Caspase-7 precursor (CASP-7) (ICE-like |
| 35 | Salmo salar | 33.90 | 2 | caspase 7 |
| 36 | Ovis aries | 33.90 | 1 | caspase-3 |
| 37 | Drosophila melanogaster | 33.50 | 3 | drICE protein |
| 38 | Drosophila pseudoobscura | 33.50 | 2 | GA20588-PA |
| 39 | Apis mellifera | 33.50 | 1 | PREDICTED: similar to Caspase precursor (drICE) |
| 40 | Chrysiptera parasema | 33.10 | 1 | caspase 3 |
| 41 | Oryzias latipes | 33.10 | 1 | caspase 3A |
| 42 | Aspergillus clavatus NRRL 1 | 32.70 | 1 | pentatricopeptide repeat protein |
| 43 | Takifugu rubripes | 32.30 | 2 | caspase 3 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 |
| Homo sapiens | Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 |
| Pan troglodytes | Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 |
| synthetic construct | Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 |
| Macaca mulatta | Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 ||||||| ||||||||||||| ||||||||#|+||||||||||||||||||||| |||||| Sbjct 301 TSPEDESSGSNPEPDATPFQEDLRTFDQLD#AVSSLPTPSDIFVSYSTFPGFVSLRDPKSG 360 |
| Macaca fascicularis | Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 ||||||| ||||||||||||| ||||||||#|+||||||||||||||||||||| |||||| Sbjct 287 TSPEDESSGSNPEPDATPFQEDLRTFDQLD#AVSSLPTPSDIFVSYSTFPGFVSLRDPKSG 346 |
| Felis catus | Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 ||||| +|||+||||| |||| |||| |#|+|||||||||||||||||||||||| ||| Sbjct 254 TSPEDRTPGSDPEPDAVAFQEGPGTFDQPD#AVSSLPTPSDIFVSYSTFPGFVSWRDTKSG 313 |
| Canis familiaris | Query 302 SPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 |||| ||||+ |||| ||||| |||||#|+|||||||||||||||||||||||+ ||| Sbjct 301 SPEDRSPGSDSEPDAVPFQEGPGPFDQLD#AVSSLPTPSDIFVSYSTFPGFVSWRNTKSG 359 |
| Canis lupus familiaris | Query 302 SPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 |||| ||||+ |||| ||||| |||||#|+|||||||||||||||||||||||+ ||| Sbjct 332 SPEDRSPGSDSEPDAVPFQEGPGPFDQLD#AVSSLPTPSDIFVSYSTFPGFVSWRNTKSG 390 |
| Monodelphis domestica | Query 303 PEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 |||+||||+|| |||||| ||| |#|++|||||||| |||||||||||||||||| Sbjct 298 PEDKSPGSDPETDATPFQNN---FDQPD#AVASLPTPSDILVSYSTFPGFVSWRDPKSG 352 |
| Rattus norvegicus | Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 || +|++ |+ |||| |+||| || ||||#|+||||||||| |||||||||||||| ||| Sbjct 339 TSSQDKAFDSDSEPDAVPYQEGPRTLDQLD#AVSSLPTPSDILVSYSTFPGFVSWRDKKSG 398 |
| Mus musculus | Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 || + + |+ |||| |+||| | ||||#|+||||||||| |||||||||||||| ||| Sbjct 338 TSSQGRTLDSDSEPDAVPYQEGPRPLDQLD#AVSSLPTPSDILVSYSTFPGFVSWRDKKSG 397 |
| Gallus gallus | Query 302 SPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 ||+||+ + | || ||| |+ |#|++||||| || |||||||||||||| || Sbjct 289 SPQDETCRRSIESDAIPFQAPSGNEDEPD#AVASLPTPGDILVSYSTFPGFVSWRDKVSG 347 |
| Dicentrarchus labrax | Query 302 SPEDESP---GSNPEPDATPFQ---EGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWR 355 ||++ | |++ + || | + | | |+||#| +|||||||| ||||||||+|||| Sbjct 309 SPDEVEPSIGGADDQTDAIPTSSSSDSLSTSDELD#ARASLPTPSDILVSYSTFPGYVSWR 368 Query 356 DPKSG 360 | +|| Sbjct 369 DTQSG 373 |
| Xenopus laevis | Query 306 ESPGSNP-----EPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 |+| +| + |||| | |++|#|+|++|||||| ||||||||+||||| +| Sbjct 284 ETPPLSPTSTSLQSDATPVFSGEGDRDEVD#AVSNIPTPSDILVSYSTFPGYVSWRDKHTG 343 |
| Danio rerio | Query 302 SPEDESP---GSNPEPDATPFQEGLRTF----DQLD#AISSLPTPSDIFVSYSTFPGFVSW 354 ||+| | | + | || | + |+||#| +|||||||| ||||||||+||| Sbjct 310 SPDDVQPCIGGIDDEMDAIPMSSSSDSLSTASDELD#ARASLPTPSDILVSYSTFPGYVSW 369 Query 355 RDPKSG 360 || ++| Sbjct 370 RDTEAG 375 |
| Tetraodon nigroviridis | Query 302 SPEDESP---GSNPEPDATPFQ---EGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWR 355 ||++ | |++ + || | + | |+ |#| +|||| ||| ||||||||+|||| Sbjct 295 SPDEFEPPVVGADDQTDAIPVSSSSDSLSMSDEPD#ARASLPTSSDILVSYSTFPGYVSWR 354 Query 356 DPKSG 360 |||+| Sbjct 355 DPKAG 359 |
| Oncorhynchus mykiss | Query 302 SPEDESP---GSNPEPDATPFQEGLRTF----DQLD#AISSLPTPSDIFVSYSTFPGFVSW 354 +|+++ | |++ + || | + |+ |#| ++||||||| ||||||||+||| Sbjct 309 TPDEDRPCIGGTDDQTDAMPMSSSSDSLSTPSDEPD#ARATLPTPSDILVSYSTFPGYVSW 368 Query 355 RDPKSG 360 || ++| Sbjct 369 RDTQAG 374 |
| Oryctolagus cuniculus | Query 301 TSPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDI 341 || + + |+ |||| | ||| | +|||#|+||||||||| Sbjct 175 TSSQGRTLDSDSEPDAVPHQEGPRPLNQLD#AVSSLPTPSDI 215 |
| Tribolium castaneum | Query 330 D#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 |# ++| +|| | ||| || ||||||+| Sbjct 81 D#VSYTIPIMADILVMYSTVEGFYSWRDPKNG 111 |
| Branchiostoma floridae | Query 314 PDATPFQEGLRTFDQLD#A--ISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 ||| | + |+||#| ++|| +| ++||| ||+ |||+| | Sbjct 217 PDALPEVQ-----DELD#AGNKATLPAEADFLLAYSTVPGYYSWRNPGRG 260 |
| Schistosoma japonicum | Query 332 ISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 + || +|| ||||| ||+++||+ || Sbjct 183 VHKLPVEADILVSYSTVPGYIAWRNMTSG 211 |
| Anopheles gambiae str. PEST | Query 317 TPFQEGLR----TFDQLD#AISS-------LPTPSDIFVSYSTFPGFVSWRDPKSG 360 | | +|++ | +|#|+|| +|| +|+ | || | | |||+| | Sbjct 129 TKFDKGVKLTKIATDTVD#ALSSSSQRTCVIPTMADVLVMYSAFDGHYSWRNPTHG 183 |
| Aedes aegypti | Query 328 QLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 | # + |+| +|+ | |||+ |+ |||+|+ | Sbjct 184 QEQ#VLYSIPAMADLLVMYSTYDGYYSWRNPRQG 216 |
| Strongylocentrotus purpuratus | Query 333 SSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 | +|+ ||+ ++|+| |||||||+ + | Sbjct 347 SKVPSQSDMLLAYATVPGFVSWRNAERG 374 |
| Xenopus tropicalis | Query 313 EPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 | || | + | | +# +| +| +||| ||+ |||+| | Sbjct 207 ETDAGPSNDSLET--DAN#PRHKIPVEADFLYAYSTVPGYYSWRNPGRG 252 |
| Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit | Query 335 LPTPSDIFVSYSTFPGFVSWRDPKSG 360 +| +| +||| ||+ |||+|| | Sbjct 187 IPVEADFLYAYSTAPGYYSWRNPKDG 212 |
| Bos taurus | Query 335 LPTPSDIFVSYSTFPGFVSWRDPKSG 360 +| +| +||| ||+ |||+| || Sbjct 240 IPVEADFLFAYSTVPGYYSWRNPGSG 265 |
| Geodia cydonium | Query 334 SLPTPSDIFVSYSTFPGFVSWRDPKSG 360 +||| +| ++|+| ||+||||+ + | Sbjct 331 ALPTEADFVLAYATVPGYVSWRNSEYG 357 |
| Equus caballus | Query 317 TPFQEGLRTFDQLD---#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 | |+ | ++ #| +| +| +||| ||+ |||+ | | Sbjct 63 TELDSGIETDSGIEDDM#ACQKIPVEADFLYAYSTAPGYYSWRNSKDG 109 |
| Gobius niger | Query 317 TPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 | |+ | |# + +| +| +||| ||+ |||+ +| Sbjct 118 TDLDAGIETDSPDD#EGARIPVEADFLYAYSTAPGYYSWRNTMTG 161 |
| Sus scrofa | Query 331 AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 | +| +| +||| ||+ |||+ | | Sbjct 183 ACQKIPVEADFLYAYSTAPGYYSWRNSKDG 212 |
| Cricetulus griseus | Query 331 AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 | +| +| +||| ||+ |||+ | | Sbjct 183 ACHKIPVEADFLYAYSTAPGYYSWRNSKDG 212 |
| Contains: Caspase-7 subunit p20; Caspase-7 subunit p11 | Query 335 LPTPSDIFVSYSTFPGFVSWRDPKSG 360 +| +| +||| ||+ |||+| | Sbjct 213 IPVEADFLFAYSTVPGYYSWRNPGKG 238 |
| Salmo salar | Query 335 LPTPSDIFVSYSTFPGFVSWRDPKSG 360 +| +| +||| ||+ |||+| | Sbjct 155 IPVEADFLFAYSTVPGYYSWRNPGRG 180 |
| Ovis aries | Query 331 AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 | +| +| +||| ||+ |||+ | | Sbjct 152 ACQKIPVEADFLYAYSTAPGYFSWRNSKYG 181 |
| Drosophila melanogaster | Query 335 LPTPSDIFVSYSTFPGFVSWRDPKSG 360 +| +| ++||| ||| |||+ | Sbjct 239 IPVHADFLIAYSTVPGFYSWRNTTRG 264 |
| Drosophila pseudoobscura | Query 335 LPTPSDIFVSYSTFPGFVSWRDPKSG 360 +| +| ++||| ||| |||+ | Sbjct 236 IPVHADFLIAYSTVPGFYSWRNTTRG 261 |
| Apis mellifera | Query 320 QEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 +| | | #+ +|+ +| ++||| ||+ |||+ | Sbjct 186 KERTETDGQPA#STFRIPSHADFLIAYSTIPGYYSWRNTTRG 226 |
| Chrysiptera parasema | Query 317 TPFQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 | |+ | | +# + +| +| ++|| ||+ |||+ +| Sbjct 111 TDLDAGIET-DSGE#GTTKIPVEADFLYAFSTAPGYYSWRNTMTG 153 |
| Oryzias latipes | Query 319 FQEGLRTFDQLD#AISSLPTPSDIFVSYSTFPGFVSWRDPKSG 360 | ||+ + |# +| +| +||| |+ ||| +| Sbjct 181 FDEGIEHDAKDD#TSEKIPLEADFLFAYSTVSGYYSWRSTANG 222 |
| Aspergillus clavatus NRRL 1 | Query 302 SPEDESPGSNPEPDATPFQEGLRTFDQLD#AISSLPTPSDIFVSYS 346 +|+|+|| | ||||+ + | + # + |+| |+|| | | Sbjct 108 APQDQSPHSEYEPDASTPGSRRQAFGRFR#GVLSIPERSNIFGSGS 152 |
| Takifugu rubripes | Query 322 GLRTFDQLD#AISS-LPTPSDIFVSYSTFPGFVSWRDPKSG 360 |+ | |#+ ++ +| +| ++|| ||+ |||+ || Sbjct 177 GIETDSAAD#SSTTKIPVEADFLYAFSTAPGYYSWRNTTSG 216 |
[Site 2] LTPVVLRPEI115-RKPEVLRPET
Ile115  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu106 | Thr107 | Pro108 | Val109 | Val110 | Leu111 | Arg112 | Pro113 | Glu114 | Ile115 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg116 | Lys117 | Pro118 | Glu119 | Val120 | Leu121 | Arg122 | Pro123 | Glu124 | Thr125 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ASFLRTNRQAAKLSKPTLENLTPVVLRPEIRKPEVLRPETPRPVDIGSGGFGDVGALESL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 120.00 | 10 | caspase 9 isoform alpha preproprotein |
| 2 | Pan troglodytes | 120.00 | 4 | PREDICTED: caspase 9 isoform 5 |
| 3 | synthetic construct | 120.00 | 1 | Homo sapiens caspase 9, apoptosis-related cysteine |
| 4 | N/A | 119.00 | 1 | G |
| 5 | Macaca fascicularis | 113.00 | 2 | caspase 9 |
| 6 | Macaca mulatta | 113.00 | 1 | PREDICTED: similar to caspase 9 isoform alpha prep |
| 7 | Mus musculus | 59.30 | 8 | caspase 9 |
| 8 | Rattus norvegicus | 53.90 | 5 | caspase-9 CTD isoform |
| 9 | Canis familiaris | 48.90 | 2 | PREDICTED: caspase 9, apoptosis-related cysteine p |
| 10 | Canis lupus familiaris | 45.80 | 1 | caspase 9 |
| 11 | Monodelphis domestica | 40.40 | 1 | PREDICTED: similar to ICE-LAP6 |
| 12 | Chromobacterium violaceum ATCC 12472 | 38.90 | 1 | hypothetical protein CV2832 |
| 13 | Bacillus weihenstephanensis KBAB4 | 33.50 | 1 | Extensin-like protein:NEAr transporter |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 86 ASFLRTNRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDVGALESL 145 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 86 ASFLRTNRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDVGALESL 145 |
| Pan troglodytes | Query 86 ASFLRTNRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDVGALESL 145 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 86 ASFLRTNRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDVGALESL 145 |
| synthetic construct | Query 86 ASFLRTNRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDVGALESL 145 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 86 ASFLRTNRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDVGALESL 145 |
| N/A | Query 86 ASFLRTNRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDVGALESL 145 |||||||||| |||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 86 ASFLRTNRQAGKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDVGALESL 145 |
| Macaca fascicularis | Query 86 ASFLRTNRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDVGALESL 145 |||||||||| |||||||||||||||||||#|||||||||||||||||||| ||||| ||| Sbjct 72 ASFLRTNRQAVKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGCGDVGAPESL 131 |
| Macaca mulatta | Query 86 ASFLRTNRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDVGALESL 145 |||||||||| |||||||||||||||||||#|||||||||||||||||||| ||||| ||| Sbjct 86 ASFLRTNRQAVKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGCGDVGAPESL 145 |
| Mus musculus | Query 99 SKPTLENLTPVVLRPEI#R-----KPEVLRPETPRPVDIGSGGFGDV 139 |+| + ||||||| || # ||||||||||||||||||| || Sbjct 132 SQPAVGNLTPVVLGPEE#LWPARLKPEVLRPETPRPVDIGSGGAHDV 177 |
| Rattus norvegicus | Query 99 SKPTLENLTPVVLRPEI#R-----KPEVLRPETPRPVDIGSGGFGDV 139 |+| | ||||||| || # +|||| |||||||||||| || Sbjct 132 SQPALGNLTPVVLGPEE#LWPTRLRPEVLTPETPRPVDIGSGRAHDV 177 |
| Canis familiaris | Query 86 ASFLRTNRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDV 139 || | +||| || | | |||| | # +|+|+|| ||| | ||| | || Sbjct 86 ASCLTMSRQADP-SKLTPGKLAPVVLGPAE#LQPQVVRPSVPRPTDNGSGRFSDV 138 |
| Canis lupus familiaris | Query 99 SKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDV 139 || | | |||| | # +|+|+|| ||| | ||| | || Sbjct 129 SKLTPGKLAPVVLGPAE#LQPQVVRPSVPRPTDNGSGRFSDV 169 |
| Monodelphis domestica | Query 86 ASFLRTNRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRPVDIGSGGFGDVGALES 144 |+ | ||+| | +| | || | | || # +| +|+ | | | || +| | | Sbjct 84 AALLSENREAGKCLRPHL-NLGPPVSRPLC#DPEQVTKPQLPSPYHSGGGGCIFIGPLRS 141 |
| Chromobacterium violaceum ATCC 12472 | Query 96 AKLSKPTLENLTPVVLRPEI#RKPEVL-RPETPRPVDIGSGGFGDVG 140 | |||| |+|+ + ||| #+ |+| | |+| + | || +| Sbjct 111 AALSKPQLQNVFMAIERPET#KPGEILPGEEVPQPTPVNPGEFGSIG 156 |
| Bacillus weihenstephanensis KBAB4 | Query 92 NRQAAKLSKPTLENLTPVVLRPEI#RKPEVLRPETPRP 128 | + ++ || +| | | +||+# |||| +|| +| Sbjct 457 NEEKPEVEKPEVEK--PEVEKPEV#EKPEVEKPEVEKP 491 |
* References
None.