XSB2269 : calpain 3, isoform CRA_a [Rattus norvegicus]
[ CaMP Format ]
This entry is computationally expanded from SB0104
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | calpain 3; isoform CRA_a |
| Gene Names | Capn3 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 516 aa
Average Mass: 58.604 kDa
Monoisotopic Mass: 58.567 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
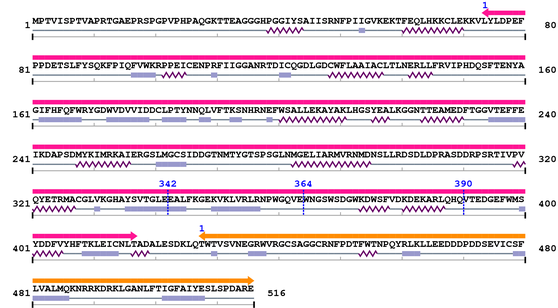
| Peptidase_C2 1. | 74 | 417 | 939.0 | 2.3e-279 |
| --- cleavage 342 (inside Peptidase_C2 74..417) --- | ||||
| --- cleavage 364 (inside Peptidase_C2 74..417) --- | ||||
| --- cleavage 390 (inside Peptidase_C2 74..417) --- | ||||
| Calpain_III 1. | 428 | 516 | 96.6 | 8.4e-26 |
3. Sequence Information
Fasta Sequence: XSB2269.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
3 [sites]
Cleavage sites (±10aa)
[Site 1] GHAYSVTGLE342-EALFKGEKVK
Glu342  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly333 | His334 | Ala335 | Tyr336 | Ser337 | Val338 | Thr339 | Gly340 | Leu341 | Glu342 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu343 | Ala344 | Leu345 | Phe346 | Lys347 | Gly348 | Glu349 | Lys350 | Val351 | Lys352 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PSRTIVPVQYETRMACGLVKGHAYSVTGLEEALFKGEKVKLVRLRNPWGQVEWNGSWSDG |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 130.00 | 39 | calpain 3 |
| 2 | Rattus norvegicus | 130.00 | 15 | calpain isoform Rt88' |
| 3 | N/A | 130.00 | 13 | 1 |
| 4 | Canis familiaris | 129.00 | 25 | PREDICTED: similar to Calpain-3 (Calpain L3) (Calp |
| 5 | Sus scrofa | 125.00 | 4 | calpain 3 |
| 6 | Macaca mulatta | 125.00 | 3 | PREDICTED: calpain 3 |
| 7 | Macaca fascicularis | 125.00 | 2 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) (C |
| 8 | Ovis aries | 125.00 | 2 | skeletal muscle-specific calpain p94 |
| 9 | Oryctolagus cuniculus | 122.00 | 1 | calpain 3, (p94) |
| 10 | Bos taurus | 121.00 | 12 | skeletal muscle-specific calpain p94 |
| 11 | Homo sapiens | 119.00 | 43 | calpain 3 isoform g |
| 12 | Gallus gallus | 107.00 | 9 | calpain 3, (p94) |
| 13 | Pan troglodytes | 98.20 | 13 | PREDICTED: similar to Calpain-3 (Calpain L3) (Calp |
| 14 | Oncorhynchus mykiss | 87.40 | 4 | gill-specific calpain |
| 15 | Tetraodon nigroviridis | 85.50 | 8 | unnamed protein product |
| 16 | Danio rerio | 83.60 | 23 | calpain 3, (p94) |
| 17 | Monodelphis domestica | 79.30 | 6 | PREDICTED: similar to digestive tract-specific cal |
| 18 | Xenopus laevis | 78.20 | 11 | MGC83034 protein |
| 19 | Xenopus tropicalis | 75.90 | 6 | calpain 5 |
| 20 | Bos grunniens | 72.40 | 1 | micromolar calcium-activated neutral protease 1 la |
| 21 | Caenorhabditis elegans | 71.60 | 1 | CaLPain family member (clp-6) |
| 22 | Coturnix coturnix | 70.90 | 1 | quail calpain |
| 23 | Pongo pygmaeus | 70.50 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 24 | synthetic construct | 70.10 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 25 | Strongylocentrotus purpuratus | 68.90 | 5 | PREDICTED: similar to calpain B isoform 2 |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 |
| Rattus norvegicus | Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 |
| N/A | Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 |
| Canis familiaris | Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 |+||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 293 PTRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 352 |
| Sus scrofa | Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 |+|||||||+||||||||||||||||||||#||||||||||||||||||||||||||||| Sbjct 313 PTRTIVPVQFETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 |
| Macaca mulatta | Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 |+|||||||||||||||||+|||||||||+#| |||||||||||||||||||||||||||| Sbjct 313 PTRTIVPVQYETRMACGLVRGHAYSVTGLD#EVLFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 |
| Macaca fascicularis | Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 |+|||||||||||||||||+|||||||||+#| |||||||||||||||||||||||||||| Sbjct 313 PTRTIVPVQYETRMACGLVRGHAYSVTGLD#EVLFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 |
| Ovis aries | Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 |+||||||||||||||||||||||||||||#|||+||||||||||||||||||||||||| Sbjct 27 PTRTIVPVQYETRMACGLVKGHAYSVTGLE#EALYKGEKVKLVRLRNPWGQVEWNGSWSD 85 |
| Oryctolagus cuniculus | Query 316 TIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 |||||||||||||||||||||||||||#| |||||||||||||||||||||||||||| Sbjct 248 TIVPVQYETRMACGLVKGHAYSVTGLE#ETLFKGEKVKLVRLRNPWGQVEWNGSWSDG 304 |
| Bos taurus | Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 |+| |||||+||||||||||||||||||||#|||+||||||||||||||||||||||||| Sbjct 27 PTRMIVPVQFETRMACGLVKGHAYSVTGLE#EALYKGEKVKLVRLRNPWGQVEWNGSWSD 85 |
| Homo sapiens | Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 |+|||+|||||||||||||+|||||||||+#| |||||||||||||||||||||||||| Sbjct 226 PTRTIIPVQYETRMACGLVRGHAYSVTGLD#EVPFKGEKVKLVRLRNPWGQVEWNGSWSD 284 |
| Gallus gallus | Query 313 PSRTIVPVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 |+ ||+|+||||||+|||||||||||| +|#| +||||++||||||||||||||| ||| Sbjct 306 PAWTIMPMQYETRMSCGLVKGHAYSVTAVE#ETTYKGEKMRLVRLRNPWGQVEWNGPWSD 364 |
| Pan troglodytes | Query 326 MACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 ||||||+|||||||||+#| ||||||||||||||||||||||||||| Sbjct 278 MACGLVRGHAYSVTGLD#EVLFKGEKVKLVRLRNPWGQVEWNGSWSD 323 |
| Oncorhynchus mykiss | Query 323 ETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 | + | |||||||||+||||#| ++|+||||+|+|||||||||||+||| Sbjct 229 EAQTASGLVKGHAYSITGLE#EVNYRGKKVKLIRIRNPWGQVEWNGAWSD 277 |
| Tetraodon nigroviridis | Query 316 TIVPVQYETRMACGLVKGHAYSVTGLE#E---ALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 ++|| ++||| | ||||||||||| +|#| + | ||+||||||||||||||| ||| Sbjct 249 SLVPARFETRTATGLVKGHAYSVTAVE#ECKPSQHKESKVRLVRLRNPWGQVEWNGPWSD 307 |
| Danio rerio | Query 316 TIVPVQYETRMACGLVKGHAYSVTGLE#E---ALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 ++|| ++||| | ||||||||||| +|#| + | +|+||||||||||||||| ||| Sbjct 249 SLVPARFETRTATGLVKGHAYSVTAVE#ECKQSQQKESRVRLVRLRNPWGQVEWNGPWSD 307 |
| Monodelphis domestica | Query 323 ETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 | + ||+|||||+|||+ #+ +||+||+|+|+||||||||||| ||| Sbjct 243 EAKTPFGLIKGHAYTVTGIN#QVNYKGQKVELIRVRNPWGQVEWNGPWSD 291 |
| Xenopus laevis | Query 323 ETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 | + |||||||||+| +# |+|+||||+|+|||||||||||+||| Sbjct 243 EAKTPQGLVKGHAYSITATD#VVNFQGQKVKLIRIRNPWGQVEWNGAWSD 291 |
| Xenopus tropicalis | Query 323 ETRMACGLVKGHAYSVTGLE#E--------ALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 | ||||||||||||+|| + #+ | || ||++++|+|||||+ |||| ||| Sbjct 241 EARMACGLVKGHAYAVTDVR#KVRLGHGLLAFFKSEKLEMIRMRNPWGEREWNGPWSD 297 |
| Bos grunniens | Query 330 LVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 ||||||||||| +#+ ++|+ | |+|+|||||+||| |+|||| Sbjct 232 LVKGHAYSVTGAK#QVNYQGQMVNLIRMRNPWGEVEWTGAWSDG 274 |
| Caenorhabditis elegans | Query 319 PVQYETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 | + | ++| |||+||||||||+ # ||| |+|+||||| |||| ||| Sbjct 354 PREKEAQLANGLVRGHAYSVTGVH#TVETDKEKVALLRIRNPWGDTEWNGDWSD 406 |
| Coturnix coturnix | Query 330 LVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSDG 372 |||||||||| +#+ ++|++ +|+|+||||||||| |+|||| Sbjct 261 LVKGHAYSVTAFK#DVNYRGQQEQLIRIRNPWGQVEWTGAWSDG 303 |
| Pongo pygmaeus | Query 330 LVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 ||||||||||| +#+ ++|+ | |+|+|||||+||| |+||| Sbjct 268 LVKGHAYSVTGAK#QVNYRGQMVSLIRMRNPWGEVEWTGAWSD 309 |
| synthetic construct | Query 330 LVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 ||||||||||| +#+ ++|+ | |+|+|||||+||| |+||| Sbjct 268 LVKGHAYSVTGAK#QVNYRGQVVSLIRMRNPWGEVEWTGAWSD 309 |
| Strongylocentrotus purpuratus | Query 323 ETRMACGLVKGHAYSVTGLE#EALFKGEKVKLVRLRNPWGQVEWNGSWSD 371 | ++ ||+ ||||++| ++#+ || |+|+|+||||| ||||| ||| Sbjct 224 EAKLPSGLIAGHAYTITDVK#KVNVKGRDVRLIRIRNPWGAVEWNGRWSD 272 |
[Site 2] RLRNPWGQVE364-WNGSWSDGWK
Glu364  Trp
Trp
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg355 | Leu356 | Arg357 | Asn358 | Pro359 | Trp360 | Gly361 | Gln362 | Val363 | Glu364 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Trp365 | Asn366 | Gly367 | Ser368 | Trp369 | Ser370 | Asp371 | Gly372 | Trp373 | Lys374 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| AYSVTGLEEALFKGEKVKLVRLRNPWGQVEWNGSWSDGWKDWSFVDKDEKARLQHQVTED |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 132.00 | 33 | mUp48 |
| 2 | N/A | 132.00 | 17 | 1 |
| 3 | Rattus norvegicus | 132.00 | 16 | calpain isoform Rt88' |
| 4 | Oryctolagus cuniculus | 131.00 | 1 | calpain 3, (p94) |
| 5 | Sus scrofa | 130.00 | 4 | lens-specific calpain Lp82 |
| 6 | Macaca mulatta | 130.00 | 4 | PREDICTED: calpain 3 |
| 7 | Macaca fascicularis | 130.00 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) (C |
| 8 | Canis familiaris | 129.00 | 25 | PREDICTED: similar to Calpain-3 (Calpain L3) (Calp |
| 9 | Bos taurus | 128.00 | 11 | lens-specific calpain Lp82 |
| 10 | Ovis aries | 128.00 | 2 | skeletal muscle-specific calpain p94 |
| 11 | Pan troglodytes | 127.00 | 12 | PREDICTED: similar to Calpain-3 (Calpain L3) (Calp |
| 12 | Homo sapiens | 124.00 | 47 | calpain 3 isoform g |
| 13 | Monodelphis domestica | 105.00 | 7 | PREDICTED: similar to ZP1-related protein |
| 14 | Gallus gallus | 98.60 | 7 | calpain 3, (p94) |
| 15 | Oncorhynchus mykiss | 86.30 | 4 | gill-specific calpain |
| 16 | Danio rerio | 83.60 | 24 | hypothetical protein LOC445107 |
| 17 | Tetraodon nigroviridis | 79.00 | 5 | unnamed protein product |
| 18 | Xenopus laevis | 76.60 | 12 | MGC83034 protein |
| 19 | Xenopus tropicalis | 76.30 | 6 | Unknown (protein for MGC:121640) |
| 20 | Coturnix coturnix | 71.60 | 1 | quail calpain |
| 21 | Pongo pygmaeus | 69.70 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 22 | Bos grunniens | 69.30 | 1 | micromolar calcium-activated neutral protease 1 la |
| 23 | synthetic construct | 69.30 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 24 | catalytic | 68.90 | 1 | Calpain 2, large |
| 25 | Caenorhabditis elegans | 68.20 | 3 | CaLPain family member (clp-6) |
| 26 | Stizostedion vitreum vitreum | 67.80 | 1 | putative calpain-like protein |
| 27 | Tribolium castaneum | 65.90 | 1 | PREDICTED: similar to calpain 8 |
| 28 | Strongylocentrotus purpuratus | 64.70 | 1 | PREDICTED: similar to calpain B isoform 2 |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |
| N/A | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |
| Rattus norvegicus | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |
| Oryctolagus cuniculus | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 ||||||||| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 267 AYSVTGLEETLFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 326 |
| Sus scrofa | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 ||||||||||||||||||||||||||||||#||||||| |||||||||||||||||||||| Sbjct 267 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDSWKDWSFVDKDEKARLQHQVTED 326 |
| Macaca mulatta | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |||||||+| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 335 AYSVTGLDEVLFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |
| Macaca fascicularis | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |||||||+| ||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 335 AYSVTGLDEVLFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |
| Canis familiaris | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 ||||||||||||||||||||||||||||||#||||||||||||||||| |||||||||||| Sbjct 292 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKAEKARLQHQVTED 351 |
| Bos taurus | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |||||||||||+||||||||||||||||||#||||||| |||||+|||||||||||||||| Sbjct 267 AYSVTGLEEALYKGEKVKLVRLRNPWGQVE#WNGSWSDSWKDWSYVDKDEKARLQHQVTED 326 |
| Ovis aries | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |||||||||||+||||||||||||||||||#||||||| |||||+|||||||||||||||| Sbjct 49 AYSVTGLEEALYKGEKVKLVRLRNPWGQVE#WNGSWSDSWKDWSYVDKDEKARLQHQVTED 108 |
| Pan troglodytes | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |||||||+| ||||||||||||||||||||#||||||| |||||||||||||||||||||| Sbjct 287 AYSVTGLDEVLFKGEKVKLVRLRNPWGQVE#WNGSWSDRWKDWSFVDKDEKARLQHQVTED 346 |
| Homo sapiens | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |||||||+| |||||||||||||||||||#||||||| |||||||||||||||||||||| Sbjct 248 AYSVTGLDEVPFKGEKVKLVRLRNPWGQVE#WNGSWSDRWKDWSFVDKDEKARLQHQVTED 307 |
| Monodelphis domestica | Query 346 FKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |||||||||||||||||||#|||||||||||| +||+||+||||||||| Sbjct 317 FKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWVLIDKEEKSRLQHQVTED 365 |
| Gallus gallus | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 ||||| +|| +||||++||||||||||||#||| ||| ++|+|+|++|| ||||++ || Sbjct 328 AYSVTAVEETTYKGEKMRLVRLRNPWGQVE#WNGPWSDKSEEWNFIDEEEKIRLQHKIAED 387 |
| Oncorhynchus mykiss | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |||+||||| ++|+||||+|+||||||||#|||+||| ++|+ +| || |+ +| Sbjct 241 AYSITGLEEVNYRGKKVKLIRIRNPWGQVE#WNGAWSDESREWNVIDSSEKKRILQNSMDD 300 |
| Danio rerio | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 ||||||+|| ++| ||+|+|+||||||||#||| ||| ++|+ +| || || +| Sbjct 253 AYSVTGVEEVNYRGAKVQLIRVRNPWGQVE#WNGPWSDNSREWTVIDSSEKKRLLQNSLDD 312 |
| Tetraodon nigroviridis | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQ 390 |||||| ++ ++|| |+|||+||||||||#|||+||| +| +| |++ || ++ Sbjct 269 AYSVTGADQVEYRGEAVQLVRIRNPWGQVE#WNGAWSDDSSEWRYVSSDDRERLTNR 324 |
| Xenopus laevis | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 |||+| + |+|+||||+|+||||||||#|||+||| +|+ + || || +| Sbjct 255 AYSITATDVVNFQGQKVKLIRIRNPWGQVE#WNGAWSDNSSEWNIIGAAEKNRLSQASLDD 314 |
| Xenopus tropicalis | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQ 390 |||||| || |++| + ||+|+|||||+||#| | ||| +|+++| || | | Sbjct 263 AYSVTGAEEVLYRGRQEKLIRVRNPWGEVE#WTGPWSDTAPEWNYIDPKVKAALDKQ 318 |
| Coturnix coturnix | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTE 393 ||||| ++ ++|++ +|+|+||||||||#| |+|||| +| +| ++ || ++ + Sbjct 266 AYSVTAFKDVNYRGQQEQLIRIRNPWGQVE#WTGAWSDGSPEWDNIDPSDREELQQKMED 324 |
| Pongo pygmaeus | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQ 388 |||||| ++ ++|+ | |+|+|||||+||#| |+||| +|+ || |+ +|+ Sbjct 273 AYSVTGAKQVNYRGQMVSLIRMRNPWGEVE#WTGAWSDSSSEWNNVDPYERDQLR 326 |
| Bos grunniens | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQ 388 |||||| ++ ++|+ | |+|+|||||+||#| |+|||| +|+ || + +|+ Sbjct 237 AYSVTGAKQVNYQGQMVNLIRMRNPWGEVE#WTGAWSDGSSEWNGVDPYMREQLR 290 |
| synthetic construct | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQ 388 |||||| ++ ++|+ | |+|+|||||+||#| |+||| +|+ || |+ +|+ Sbjct 273 AYSVTGAKQVNYRGQVVSLIRMRNPWGEVE#WTGAWSDSSSEWNNVDPYERDQLR 326 |
| catalytic | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARL 387 |||||| || | ||+|+|||||+||#| | |+| |+ +| +|+ || Sbjct 292 AYSVTGAEEVESNGSLQKLIRIRNPWGEVE#WTGRWNDNCPSWNTIDPEERERL 344 |
| Caenorhabditis elegans | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQVTED 394 ||||||+ ||| |+|+||||| |#||| ||| | ||++++ +++ ++ || Sbjct 370 AYSVTGVHTVETDKEKVALLRIRNPWGDTE#WNGDWSDKSSLWEQVDQEQREKMEFRIKED 429 |
| Stizostedion vitreum vitreum | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARLQHQ 390 |||||| + +| ||+|+||||||||#| |+||| | + +++ || |+ Sbjct 264 AYSVTGTAQVEHQGGTEKLIRIRNPWGQVE#WTGAWSDNSMQWRRISSEDRERLSHR 319 |
| Tribolium castaneum | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARL 387 |||+||| + | + |+|+||||| ||#||| ||| || | |+ | +| Sbjct 282 AYSITGLYTSQLKNVTIHLIRVRNPWGSVE#WNGPWSDNSVCWSLVPKETKQKL 334 |
| Strongylocentrotus purpuratus | Query 335 AYSVTGLEEALFKGEKVKLVRLRNPWGQVE#WNGSWSDGWKDWSFVDKDEKARL 387 ||++| +++ || |+|+|+||||| ||#||| ||| ++| +++ |+ | Sbjct 236 AYTITDVKKVNVKGRDVRLIRIRNPWGAVE#WNGRWSDNAREWDDINQRERKNL 288 |
[Site 3] KDEKARLQHQ390-VTEDGEFWMS
Gln390  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys381 | Asp382 | Glu383 | Lys384 | Ala385 | Arg386 | Leu387 | Gln388 | His389 | Gln390 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val391 | Thr392 | Glu393 | Asp394 | Gly395 | Glu396 | Phe397 | Trp398 | Met399 | Ser400 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GQVEWNGSWSDGWKDWSFVDKDEKARLQHQVTEDGEFWMSYDDFVYHFTKLEICNLTADA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 138.00 | 33 | mUp48 |
| 2 | N/A | 138.00 | 21 | 1 |
| 3 | Rattus norvegicus | 138.00 | 16 | calpain isoform Rt88' |
| 4 | Macaca mulatta | 136.00 | 4 | PREDICTED: calpain 3 |
| 5 | Macaca fascicularis | 136.00 | 1 | CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) (C |
| 6 | Oryctolagus cuniculus | 136.00 | 1 | calpain 3, (p94) |
| 7 | Sus scrofa | 135.00 | 4 | lens-specific calpain Lp82 |
| 8 | Canis familiaris | 134.00 | 22 | PREDICTED: similar to Calpain-3 (Calpain L3) (Calp |
| 9 | Bos taurus | 134.00 | 10 | lens-specific calpain Lp82 |
| 10 | Ovis aries | 134.00 | 2 | skeletal muscle-specific calpain p94 |
| 11 | Homo sapiens | 133.00 | 38 | calpain 3 isoform g |
| 12 | Pan troglodytes | 133.00 | 11 | PREDICTED: similar to Calpain-3 (Calpain L3) (Calp |
| 13 | Monodelphis domestica | 125.00 | 5 | PREDICTED: similar to ZP1-related protein |
| 14 | Gallus gallus | 100.00 | 7 | calpain 3, (p94) |
| 15 | Danio rerio | 89.70 | 19 | calpain 3, (p94) |
| 16 | Tetraodon nigroviridis | 84.70 | 6 | unnamed protein product |
| 17 | Oncorhynchus mykiss | 84.70 | 3 | calpain 1 catalytic subunit |
| 18 | Xenopus laevis | 84.00 | 9 | MGC83034 protein |
| 19 | Coturnix coturnix | 80.10 | 1 | quail calpain |
| 20 | Stizostedion vitreum vitreum | 79.70 | 1 | putative calpain-like protein |
| 21 | Caenorhabditis elegans | 79.30 | 4 | CaLPain family member (clp-7) |
| 22 | Caenorhabditis briggsae | 78.60 | 2 | Hypothetical protein CBG15134 |
| 23 | Xenopus tropicalis | 77.00 | 5 | CAPN2 protein |
| 24 | Pongo pygmaeus | 76.60 | 1 | CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 25 | catalytic | 76.60 | 1 | Calpain 2, large |
| 26 | synthetic construct | 76.60 | 1 | Homo sapiens calpain 1, (mu/I) large subunit |
| 27 | Bos taurus x Bos indicus | 75.10 | 1 | micromolar calcium activated neutral protease 1 |
| 28 | Bos grunniens | 74.70 | 2 | calpain 2 |
| 29 | Drosophila melanogaster | 73.60 | 7 | Calpain, without calmodulin-like domain |
| 30 | Schistosoma japonicum | 69.70 | 1 | SJCHGC01809 protein |
| 31 | Anopheles gambiae | 69.70 | 1 | calpain |
| 32 | Tribolium castaneum | 69.70 | 1 | PREDICTED: similar to CG8107-PA |
| 33 | Anopheles gambiae str. PEST | 69.70 | 1 | ENSANGP00000032073 |
| 34 | Aedes aegypti | 69.30 | 2 | calpain, putative |
| 35 | Homarus americanus | 69.30 | 1 | muscle-specific calpain |
| 36 | Gecarcinus lateralis | 67.80 | 2 | muscle-specific calpain |
| 37 | Drosophila pseudoobscura | 67.40 | 1 | GA20829-PA |
| 38 | Schistosoma haematobium | 67.00 | 1 | calpain |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 |
| N/A | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 |
| Rattus norvegicus | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 |
| Macaca mulatta | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||||||||||||||||||||||#|||||||||||+||+||||||||||||||| Sbjct 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYEDFIYHFTKLEICNLTADA 420 |
| Macaca fascicularis | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||||||||||||||||||||||#|||||||||||+||+||||||||||||||| Sbjct 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYEDFIYHFTKLEICNLTADA 420 |
| Oryctolagus cuniculus | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||||||||||||||||||||||#|||||||||||||||+|||||||||||||| Sbjct 293 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVFHFTKLEICNLTADA 352 |
| Sus scrofa | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||| ||||||||||||||||||#||||||||||||||+||||||||||||||| Sbjct 293 GQVEWNGSWSDSWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFIYHFTKLEICNLTADA 352 |
| Canis familiaris | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||||||||||||| ||||||||#||||||||||||||+||||||||||||||| Sbjct 318 GQVEWNGSWSDGWKDWSFVDKAEKARLQHQ#VTEDGEFWMSYDDFIYHFTKLEICNLTADA 377 |
| Bos taurus | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||| |||||+||||||||||||#||||||||||||||+||||||||||||||| Sbjct 293 GQVEWNGSWSDSWKDWSYVDKDEKARLQHQ#VTEDGEFWMSYDDFIYHFTKLEICNLTADA 352 |
| Ovis aries | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||| |||||+||||||||||||#||||||||||||||+||||||||||||||| Sbjct 75 GQVEWNGSWSDSWKDWSYVDKDEKARLQHQ#VTEDGEFWMSYDDFIYHFTKLEICNLTADA 134 |
| Homo sapiens | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||| ||||||||||||||||||#|||||||||||+||+||||||||||||||| Sbjct 274 GQVEWNGSWSDRWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYEDFIYHFTKLEICNLTADA 333 |
| Pan troglodytes | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||||||| ||||||||||||||||||#|||||||||||+||+||||||||||||||| Sbjct 313 GQVEWNGSWSDRWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYEDFIYHFTKLEICNLTADA 372 |
| Monodelphis domestica | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 |||||||||||||||| +||+||+|||||#|||||||||||+||+|+||||||||||||| Sbjct 332 GQVEWNGSWSDGWKDWVLIDKEEKSRLQHQ#VTEDGEFWMSYEDFMYNFTKLEICNLTADA 391 |
| Gallus gallus | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTAD 419 ||||||| ||| ++|+|+|++|| ||||+#+ ||||||+| +||+ ||||||||||| | Sbjct 354 GQVEWNGPWSDKSEEWNFIDEEEKIRLQHK#IAEDGEFWISLEDFMRHFTKLEICNLTPD 412 |
| Danio rerio | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||||| ||| |+| + | || +|| |# ||||||||++|| ++||+|||||| || Sbjct 297 GQVEWNGPWSDNSKEWESLSKAEKEKLQQQ#NAEDGEFWMSFEDFKKNYTKIEICNLTPDA 356 |
| Tetraodon nigroviridis | Query 361 GQVEWNGSWSD----------GWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTK 410 ||||||| ||| |+|| + | || +| ||# ||||||||++|| ++|| Sbjct 297 GQVEWNGPWSDKNRPFRLLRNSSKEWSTLSKAEKEKLHHQ#SAEDGEFWMSFEDFKKNYTK 356 Query 411 LEICNLTADA 420 +|||||| || Sbjct 357 IEICNLTPDA 366 |
| Oncorhynchus mykiss | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 |++|| |+||| ++| ||+ + |||++# +||||||||+ ||+ ||+|||||||||| Sbjct 294 GEIEWTGAWSDNSREWESVDRSVRGRLQNR#-SEDGEFWMSFSDFLREFTRLEICNLTADA 352 |
| Xenopus laevis | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 |||||||+||| +|+ + || || # +|||||| ++|| || |||||||| || Sbjct 281 GQVEWNGAWSDNSSEWNIIGAAEKNRLSQA#SLDDGEFWMDFEDFKRHFDKLEICNLTPDA 340 |
| Coturnix coturnix | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||| |+|||| +| +| ++ || +#+ ||||||||+ ||+ |++||||||| || Sbjct 292 GQVEWTGAWSDGSPEWDNIDPSDREELQQK#M-EDGEFWMSFRDFMREFSRLEICNLTPDA 350 |
| Stizostedion vitreum vitreum | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||| |+||| | + +++ || |+# +||||||||+ ||+ |+ +||||||| || Sbjct 290 GQVEWTGAWSDNSMQWRRISSEDRERLSHR#-SEDGEFWMSFADFLRHYDRLEICNLTPDA 348 |
| Caenorhabditis elegans | Query 364 EWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTAD 419 ||||+|||| +|| || ++ ++ |# +|||||||+|||+ +||++| |||+|| Sbjct 553 EWNGAWSDGSSEWSQVDPQQREQMGVQ#FAKDGEFWMSFDDFMTNFTQMECCNLSAD 608 |
| Caenorhabditis briggsae | Query 363 VEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTAD 419 +||||+|||| +|| +|+ | ++ |# ||||||++||| +||++|+|||||+ Sbjct 449 LEWNGAWSDGSMEWSKIDEATKKQIDVQ#FARDGEFWMNFDDFFANFTQMELCNLTAE 505 |
| Xenopus tropicalis | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||| |+||| +|+ || |+ || +#+ ||||||||+ +|+ |++||||||| || Sbjct 292 GQVEWTGAWSDNSSEWNEVDPSEQEDLQLK#M-EDGEFWMSFQEFLRQFSRLEICNLTPDA 350 |
| Pongo pygmaeus | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 |+||| |+||| +|+ || |+ +|+ +#+ ||||||||+ ||+ ||+||||||| || Sbjct 299 GEVEWTGAWSDSSSEWNNVDPYERDQLRVK#M-EDGEFWMSFRDFMREFTRLEICNLTPDA 357 |
| catalytic | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTAD 419 |+||| | |+| |+ +| +|+ || +# ||||||||+ ||+ |+++||||||| | Sbjct 318 GEVEWTGRWNDNCPSWNTIDPEERERLTRR#-HEDGEFWMSFSDFLRHYSRLEICNLTPD 375 |
| synthetic construct | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 |+||| |+||| +|+ || |+ +|+ +#+ ||||||||+ ||+ ||+||||||| || Sbjct 299 GEVEWTGAWSDSSSEWNNVDPYERDQLRVK#M-EDGEFWMSFRDFMREFTRLEICNLTPDA 357 |
| Bos taurus x Bos indicus | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTAD 419 |+||| |+|||| +|+ || + +|+ +#+ ||||||||+ ||+ ||+||||||| | Sbjct 15 GEVEWTGAWSDGSSEWNGVDPYMREQLRVK#M-EDGEFWMSFRDFMREFTRLEICNLTPD 72 |
| Bos grunniens | Query 361 GQVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTAD 419 |+||| | |+| +|+ || + + | |# ||||||||++||+ |+++||||||| | Sbjct 1 GEVEWTGQWNDNCPNWNTVDPEVRETLTRQ#-HEDGEFWMSFNDFLRHYSRLEICNLTPD 58 |
| Drosophila melanogaster | Query 362 QVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 + |||| ||| +| ++ +++|| + # |||||||+ ||+ || ++|||||+ |+ Sbjct 332 EAEWNGPWSDSSPEWRYIPEEQKAEIGLT#FDRDGEFWMSFQDFLNHFDRVEICNLSPDS 390 |
| Schistosoma japonicum | Query 364 EWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNL 416 || |+||| |+|| + +++ +| # +|||||||| ||| +| |||||+| Sbjct 368 EWKGAWSDKSKEWSLISPEQRQQLGLT#FDDDGEFWMSYQDFVSNFEKLEICHL 420 |
| Anopheles gambiae | Query 362 QVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 + |||| ||| +| ++ ++| | # |||||||| || +| ++|||||+ |+ Sbjct 15 EAEWNGPWSDKSPEWRYIPDEQKQELGLN#FDHDGEFWMSYRDFTRYFDRMEICNLSPDS 73 |
| Tribolium castaneum | Query 364 EWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||||+||| +| ++ +| | # |||||||+ || || ++||||| |+ Sbjct 701 EWNGAWSDHSPEWHYISDSDKEELGLN#FDADGEFWMSFKDFQQHFNRIEICNLNPDS 757 |
| Anopheles gambiae str. PEST | Query 362 QVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 + |||| ||| +| ++ ++| | # |||||||| || +| ++|||||+ |+ Sbjct 15 EAEWNGPWSDKSPEWRYIPDEQKQELGLN#FDHDGEFWMSYRDFTRYFDRMEICNLSPDS 73 |
| Aedes aegypti | Query 363 VEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTAD 419 +||||+||| ++| + | ++ +| #+ |||||| ++||+ +| ++|||||+ | Sbjct 307 IEWNGAWSDRSQEWQAIPKAQRKQLGLT#IDNDGEFWMDFEDFLRYFDRVEICNLSPD 363 |
| Homarus americanus | Query 362 QVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTAD 419 + || ||||| +|+ + +|| ||+ # +|||||||+ || +|| +|||++| + Sbjct 334 EAEWKGSWSDKSPEWNSITPEEKQRLKLN#FEDDGEFWMSFQDFASNFTTVEICDVTPE 391 |
| Gecarcinus lateralis | Query 362 QVEWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTAD 419 + || ||||| +|+ | |+ || # +|||||||| ||+ +|| +|||++ | Sbjct 334 ETEWKGSWSDKSPEWNAVPPGERQRLGLT#FNDDGEFWMSYQDFLKNFTTVEICDVNPD 391 |
| Drosophila pseudoobscura | Query 364 EWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNLTADA 420 ||+| ||| +| |+ + | + # |||||||+ ||+ || ++||||| |+ Sbjct 511 EWSGPWSDSSPEWRFIPEHVKEEIGLN#FDRDGEFWMSFQDFLNHFDRVEICNLNPDS 567 |
| Schistosoma haematobium | Query 364 EWNGSWSDGWKDWSFVDKDEKARLQHQ#VTEDGEFWMSYDDFVYHFTKLEICNL 416 || |+| || | + + || + # | ||||||||+||| ||++|+|+| Sbjct 344 EWKGAWCDGSPQWREISEQEKKNINLS#FTADGEFWMSYEDFVTCFTRVEVCHL 396 |
* References
[PubMed ID: 15632090] Florea L, Di Francesco V, Miller J, Turner R, Yao A, Harris M, Walenz B, Mobarry C, Merkulov GV, Charlab R, Dew I, Deng Z, Istrail S, Li P, Sutton G, Gene and alternative splicing annotation with AIR. Genome Res. 2005 Jan;15(1):54-66.