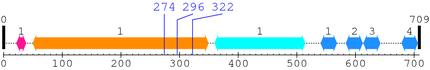
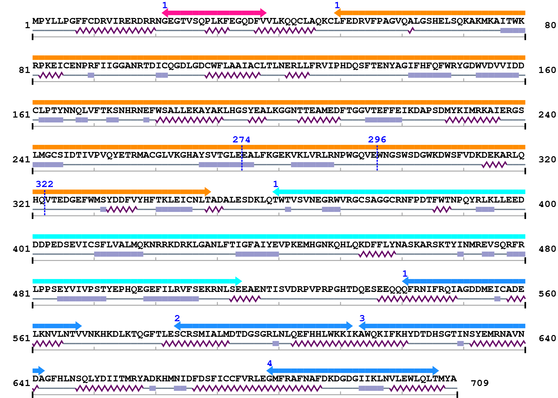
| # |
organism |
max score |
hits |
top seq |
| 1 |
Mus musculus |
85.10 |
3 |
crystallin, beta B2 |
| 2 |
Canis lupus familiaris |
85.10 |
1 |
crystallin, beta B2 |
| 3 |
Bos taurus |
84.00 |
2 |
crystallin, beta B2 |
| 4 |
Oryctolagus cuniculus |
84.00 |
1 |
crystallin, beta B2 |
| 5 |
N/A |
83.20 |
5 |
A |
| 6 |
Macaca mulatta |
82.00 |
2 |
PREDICTED: crystallin, beta B2 isoform 3 |
| 7 |
Homo sapiens |
80.90 |
3 |
crystallin, beta B2 |
| 8 |
Cavia porcellus |
80.90 |
1 |
betaB2-crystallin |
| 9 |
human, lens, Peptide, 204 aa |
79.00 |
1 |
A Chain A, Structure Of Native Human Beta B2 Cryst |
| 10 |
Rana catesbeiana |
58.20 |
1 |
CRBB2_RANCA Beta crystallin B2 (Beta-crystallin Bp |
| 11 |
Gallus gallus |
57.80 |
3 |
CRBB2_CHICK Beta crystallin B2 (Beta-crystallin Bp |
| 12 |
Xenopus laevis |
53.90 |
6 |
MGC84818 protein |
| 13 |
Monodelphis domestica |
48.50 |
2 |
PREDICTED: similar to betaB2-crystallin |
| 14 |
Danio rerio |
48.10 |
3 |
PREDICTED: beta B2-crystallin isoform 1 |
| 15 |
Pan troglodytes |
47.00 |
4 |
PREDICTED: crystallin, beta B3 isoform 3 |
| 16 |
Tetraodon nigroviridis |
45.40 |
2 |
unnamed protein product |
| 17 |
Canis familiaris |
38.50 |
1 |
PREDICTED: similar to crystallin, beta B1 |
| 18 |
Rattus norvegicus |
34.70 |
3 |
AF286652_1 betaB1-crystallin |
| 19 |
Xenopus tropicalis |
34.70 |
1 |
crystallin, beta B1 |
| 20 |
Sus scrofa |
34.70 |
1 |
beta crystallin subunit beta B1 |
| 21 |
Contains: Beta crystallin B1B |
34.70 |
1 |
CRBB1_RAT Beta crystallin B1 |
| 22 |
Dicentrarchus labrax |
33.50 |
1 |
crystallin beta B1 |
| 23 |
synthetic construct |
33.50 |
1 |
crystallin beta B1 |
| organism | matching |
|---|
| Mus musculus |
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
||||||||#||||||||||||||||||||||||||||||
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
|
| Canis lupus familiaris |
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
||||||||#||||||||||||||||||||||||||||||
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
|
| Bos taurus |
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
||||||||#||||||||||||||||||||||||||+|||
Sbjct 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELNGPC 38
|
| Oryctolagus cuniculus |
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
||||||||#||||||||||||||||||||||||||+|||
Sbjct 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELNGPC 38
|
| N/A |
Query 2 ASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
|||||||#||||||||||||||||||||||||||||||
Query 2 ASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
|
| Macaca mulatta |
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
||||||||#||||| ||||||||||||||||||||||||
Sbjct 1 MASDHQTQ#AGKPQSLNPKIIIFEQENFQGHSHELSGPC 38
|
| Homo sapiens |
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
||||||||#||||| ||||||||||||||||||||+|||
Sbjct 1 MASDHQTQ#AGKPQSLNPKIIIFEQENFQGHSHELNGPC 38
|
| Cavia porcellus |
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
||||||||#||| ||||||||||||||||||||||+|||
Sbjct 1 MASDHQTQ#AGKQQPLNPKIIIFEQENFQGHSHELNGPC 38
|
| human, lens, Peptide, 204 aa |
Query 2 ASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
|||||||#||||| ||||||||||||||||||||+|||
Sbjct 1 ASDHQTQ#AGKPQSLNPKIIIFEQENFQGHSHELNGPC 37
|
| Rana catesbeiana |
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
||||||+ #| | | + ||++|||||||| |||||||
Sbjct 1 MASDHQSP#ATKQQQPSSKIVLFEQENFQGRCHELSGPC 38
|
| Gallus gallus |
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
|||+|| #| | || +| | ||||||||| ||||| |
Sbjct 1 MASEHQMP#ASKQQPASPNIAIFEQENFQGRCHELSGAC 38
|
| Xenopus laevis |
Query 1 MASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
||||||| #+| | | |++|+||||||| ||||+| |
Sbjct 1 MASDHQT-#SGTKQQQNAKLVIYEQENFQGRSHELNGAC 37
|
| Monodelphis domestica |
Query 17 PKIIIFEQENFQGHSHELSGPC 38
|||+||||||||| ||||||||
Sbjct 396 PKIVIFEQENFQGRSHELSGPC 417
|
| Danio rerio |
Query 1 MASDHQTQ#AGKP-QPLNP--KIIIFEQENFQGHSHELSGPC 38
||+||| #| | ||+ |++|+||||||| |||+|||
Sbjct 1 MATDHQNP#ATKQKQPVASAFKLVIYEQENFQGRCHELTGPC 41
|
| Pan troglodytes |
Query 18 KIIIFEQENFQGHSHELSGPC 38
+||||||||||||||||+|||
Sbjct 205 RIIIFEQENFQGHSHELNGPC 225
|
| Tetraodon nigroviridis |
Query 1 MASDHQTQ#AGKPQPLNP---KIIIFEQENFQGHSHELSGPC 38
||+||| #| | | + |++|+|||||||| ||| |
Sbjct 25 MATDHQNP#ASKQQQPSASAFKLVIYEQENFQGHCQELSAAC 65
|
| Canis familiaris |
Query 7 TQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
|+#|| | + |+++|||||||| | || |
Sbjct 45 TK#AGDLPPGSYKLVVFEQENFQGRRVEFSGEC 76
|
| Rattus norvegicus |
Query 8 Q#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
+# |+ | + ++++|||||||| | || |
Sbjct 43 K#VGELPPGSYRLVVFEQENFQGRRVEFSGEC 73
|
| Xenopus tropicalis |
Query 7 TQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
|+# | | + ||++|||||||| || |
Sbjct 27 TR#TGDPMMGSFKIVLFEQENFQGRHVELLNEC 58
|
| Sus scrofa |
Query 10 GKPQPLNPKIIIFEQENFQGHSHELSGPC 38
| | + |+++|||||||| | || |
Sbjct 49 GDLPPGSYKLVVFEQENFQGRRVEFSGEC 77
|
| Contains: Beta crystallin B1B |
Query 8 Q#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
+# |+ | + ++++|||||||| | || |
Sbjct 48 K#VGELPPGSYRLVVFEQENFQGRRVEFSGEC 78
|
| Dicentrarchus labrax |
Query 2 ASDHQTQ#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
|| |+# |+| + +|++|+|||||| |+ |
Sbjct 25 ASSKATK#TGEPGMGSYRIMMFDQENFQGRMIEVQNEC 61
|
| synthetic construct |
Query 8 Q#AGKPQPLNPKIIIFEQENFQGHSHELSGPC 38
+#| + | | ++++|| ||||| | || |
Sbjct 50 K#AAELPPGNYRLVVFELENFQGRRAEFSGEC 80
|
 Ala
Ala

 Sequence conservation (by blast)
Sequence conservation (by blast)