XSB2348 : jun oncogene [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0086
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Transcription factor AP-1; Activator protein 1; AP1; Proto-oncogene c-jun; V-jun avian sarcoma virus 17 oncogene homolog; p39; jun oncogene; Jun activation domain binding protein; v-jun sarcoma virus 17 oncogene homolog; enhancer-binding protein AP1 |
| Gene Names | JUN; jun oncogene |
| Gene Locus | 1p32-p31; chromosome 1 |
| GO Function | Not available |
* Information From OMIM
Description: The oncogene JUN is the putative transforming gene of avian sarcoma virus 17; it appears to be derived from a gene of the chicken genome and has homologs in several other vertebrate species. (The name JUN comes from the Japanese 'ju-nana,' meaning the number 17.) JUN was originally thought to be identical to the transcription factor AP1. However, it is now known that AP1 is not a single protein, but constitutes a group of related dimeric basic region-leucine zipper proteins that belong to the JUN, FOS (OMIM:164810), MAF (OMIM:177075), and ATF (see OMIM:603148) subfamilies. The various dimers recognize either 12-O-tetradecanoylphorbol-13-acetate (TPA) response elements or cAMP response elements. JUN is the most potent transcriptional activator in its group, and its transcriptional activity is attenuated and sometimes antagonized by JUNB (OMIM:165161). For a review of the structure and function of the AP1 transcription complexes, see Shaulian and Karin (2002).
Function: Lamph et al. (1988) investigated regulation of murine c-jun gene transcription and found that both serum and phorbol-ester TPA induced c-jun gene expression.
Function: Marx (1988) reviewed information indicating that the protein encoded by the JUN gene acts directly to activate gene transcription in response to cell stimulation; that the product of the FOS oncogene cooperates with the JUN product in fostering gene transcription; that there is a structural and functional similarity between JUN and GCN4, which induces the activity of a large set of genes needed for amino acid synthesis in yeast; that specifically both have a DNA-binding domain on the carboxyl end essential for the activation of genes; that the JUN and FOS proteins are held together in a complex by a leucine zipper; and that there are other JUN genes in addition to the original one.
Function: Shaulian et al. (2000) found that in mouse fibroblasts, Jun was necessary for cell cycle reentry of ultraviolet (UV)-irradiated cells but did not participate in the response to ionizing radiation. Cells lacking Jun underwent prolonged cell cycle arrest but resisted apoptosis, whereas cells that expressed Jun constitutively did not arrest and undergo apoptosis. This function of Jun was exerted through negative regulation of p53 (OMIM:191170) association with the p21 (OMIM:116899) promoter. Cells lacking Jun exhibited prolonged p21 induction, whereas constitutive Jun inhibited UV-mediated p21 induction.
Function: Whitfield et al. (2001) noted that apoptosis induced in rat sympathetic neurons by nerve growth factor (NGF; see OMIM:162030) withdrawal can be blocked by inhibitors of RNA and protein synthesis. They presented experimental evidence that activation of the JNK (see OMIM:601158)/JUN pathway and increased expression of BIM (OMIM:603827) are key events required for cytochrome c release and apoptosis following NGF withdrawal.
Function: Sphingosylphosphocholine (SPC) is a deacylated derivative of sphingomyelin known to accumulate in Niemann-Pick disease type A (OMIM:257200). SPC is a potent mitogen that increases intracellular free Ca(2+) and free arachidonate through pathways that are only partly protein kinase C-dependent. Berger et al. (1995) showed that SPC increases specific DNA-binding activity of transcription activator AP1 in electrophoretic mobility-shift assays.
Function: Using a Drosophila model synapse, Sanyal et al. (2002) analyzed cellular functions and regulation of the immediate-early transcription factor AP1, a heterodimer of the basic leucine zipper proteins FOS and JUN. They observed that AP1 positively regulates synaptic strength and synapse number, thus showing a greater range of influence than CREB (OMIM:123810). Observations from genetic epistasis and RNA quantification experiments indicate that AP1 acts upstream of CREB, regulates levels of CREB mRNA, and functions at the top of the hierarchy of transcription factors known to regulate long-term plasticity. A JUN-kinase signaling module provided a CREB-independent route for neuronal AP1 activation; thus, CREB regulation of AP1 expression may, in some neurons, constitute a positive feedback loop rather than the primary step in AP1 activation.
Function: Mathas et al. (2002) found AP1 constitutively activated, with robust JUN and JUNB overexpression, in all cell lines derived from patients with classical Hodgkin lymphoma (OMIM:236000) and anaplastic large cell lymphoma (ALCL), but not in other lymphoma types. AP1 supported proliferation of Hodgkin cells, but suppressed apoptosis of ALCL cells. Mathas et al. (2002) noted that, whereas JUN is upregulated by an autoregulatory process, JUNB is under the control of nuclear factor kappa-B (NFKB; OMIM:164011). They found that AP1 and NFKB cooperate and stimulate expression of the cell cycle regulator cyclin D2 (OMIM:123833), the protooncogene MET (OMIM:164860), and the lymphocyte homing receptor CCR7 (OMIM:600242), which are all strongly expressed in primary Hodgkin/Reed-Sternberg (HRS) cells.
Function: Wertz et al. (2004) reported that human DET1 (OMIM:608727) promotes ubiquitination and degradation of the protooncogenic transcription factor c-Jun by assembling a multisubunit ubiquitin ligase containing DNA damage-binding protein-1 (DDB1; OMIM:600045), cullin 4A (CUL4A; OMIM:603137), regulator of cullins-1 (ROC1; OMIM:603814), and constitutively photomorphogenic-1 (COP1; OMIM:608067). Ablation of any subunit by RNA interference stabilized c-Jun and increased c-Jun-activated transcription. Wertz et al. (2004) concluded that their findings characterized a c-Jun ubiquitin ligase and define a specific function for DET1 in mammalian cells.
Function: JUN and N-terminal kinases (JNK) are essential for neuronal microtubule assembly and apoptosis. Phosphorylation of the activating protein 1 (AP1) transcription factor c-Jun, at multiple sites within its transactivation domain, is required for JNK-induced neurotoxicity. Nateri et al. (2004) reported that in neurons the stability of c-Jun is regulated by the E3 ligase SCF(Fbw7) (FBXW7; OMIM:606278), which ubiquitinates phosphorylated c-Jun and facilitates c-Jun degradation. Fbxw7 depletion resulted in accumulation of phosphorylated c-Jun, stimulation of AP1 activity, and neuronal apoptosis. SCF-7 therefore antagonizes the apoptotic c-Jun-dependent effector arm of JNK signaling, allowing neurons to tolerate potentially neurotoxic JNK activity.
Function: Fang and Kerppola (2004) found evidence that JUN proteins ubiquitinated by ITCH (OMIM:606409) are targeted to lysosomes for degradation. Mutation of the ITCH recognition motif in the N terminus of JUN eliminated its ubiquitination and increased its stability.
Function: Ikeda et al. (2004) generated transgenic mice expressing dominant-negative c-Jun specifically in the osteoclast lineage and found that they developed severe osteopetrosis due to impaired osteoclastogenesis. Blockade of c-Jun signaling also markedly inhibited soluble RANKL (OMIM:602642)-induced osteoclast differentiation in vitro. Overexpression of nuclear factor of activated T cells 1 (NFATC2; OMIM:600490) or NFATC1 (OMIM:600489) promoted differentiation of osteoclast precursor cells into tartrate-resistant acid phosphatase-positive (TRAP-positive) multinucleated osteoclast-like cells even in the absence of RANKL. These osteoclastogenic activities of NFAT were abrogated by overexpression of dominant-negative c-Jun. Ikeda et al. (2004) concluded that c-Jun signaling in cooperation with NFAT is crucial for RANKL-regulated osteoclast differentiation.
Function: Gao et al. (2004) found in the case of c-JUN and JUNB that extracellular stimuli modulate protein turnover by regulating the activity of an E3 ligase by means of its phosphorylation. Activation of the Jun amino-terminal kinase (JNK; see OMIM:601158) mitogen-activated protein kinase (MAPK) cascade after T cell stimulation accelerated degradation of c-JUN and JUNB through phosphorylation-dependent activation of the E3 ligase ITCH. Gao et al. (2004) found that this pathway modulates cytokine production by effector T cells.
Function: Nateri et al. (2005) showed that phosphorylated c-JUN interacts with the HMG-box transcription factor TCF4 (TCF7L2; OMIM:602228) to form a ternary complex containing c-JUN, TCF4, and beta-catenin (see OMIM:116806). Chromatin immunoprecipitation assays revealed JNK-dependent c-JUN-TCF4 interaction on the c-JUN promoter, and c-JUN and TCF4 cooperatively activated the c-JUN promoter in reporter assays in a beta-catenin-dependent manner. In the Apc(Min) mouse model of intestinal cancer (see OMIM:611731), genetic abrogation of c-JUN N-terminal phosphorylation or gut-specific conditional c-JUN inactivation reduced tumor number and size and prolonged life span. Therefore, Nateri et al. (2005) concluded that the phosphorylation-dependent interaction between c-JUN and TCF4 regulates intestinal tumorigenesis by integrating JNK and APC/beta-catenin, 2 distinct pathways activating WNT signaling.
Function: Koyama-Nasu et al. (2007) showed that FBL10 (FBXL10; OMIM:609078) interacted with JUN and repressed JUN-mediated transcription in human cell lines. Chromatin immunoprecipitation assays demonstrated that FBL10 was present at the JUN promoter and that JUN was required for recruitment of FBL10. FBL10 bound unmethylated CpG sequences in the JUN promoter through its CxxC zinc finger and tethered transcriptional repressor complexes. Suppression of FBL10 expression by RNA interference induced transcription of JUN and JUN target genes and caused aberrant cell cycle progression and increased UV-induced cell death. Furthermore, FBL10 protein and mRNA were downregulated in response to UV in an inverse correlation with JUN. Koyama-Nasu et al. (2007) concluded that FBL10 is a key regulator of JUN function.
* Structure Information
1. Primary Information
Length: 331 aa
Average Mass: 35.676 kDa
Monoisotopic Mass: 35.653 kDa
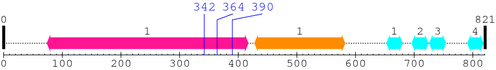
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Jun 1. | 5 | 241 | 447.5 | 2e-131 |
| --- cleavage 90 (inside Jun 5..241) --- | ||||
| --- cleavage 62 (inside Jun 5..241) --- | ||||
| --- cleavage 42 (inside Jun 5..241) --- | ||||
| bZIP_1 1. | 250 | 313 | 84.9 | 2.9e-22 |
3. Sequence Information
Fasta Sequence: XSB2348.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
3 [sites]
Cleavage sites (±10aa)
[Site 1] IQSSNGHITT90-TPTPTQFLCP
Thr90  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile81 | Gln82 | Ser83 | Ser84 | Asn85 | Gly86 | His87 | Ile88 | Thr89 | Thr90 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr91 | Pro92 | Thr93 | Pro94 | Thr95 | Gln96 | Phe97 | Leu98 | Cys99 | Pro100 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LTSPDVGLLKLASPELERLIIQSSNGHITTTPTPTQFLCPKNVTDEQEGFAEGFVRALAE |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 122.00 | 11 | Jun oncogene |
| 2 | Homo sapiens | 122.00 | 10 | jun oncogene |
| 3 | Canis familiaris | 122.00 | 10 | PREDICTED: similar to Transcription factor AP-1 (A |
| 4 | N/A | 122.00 | 7 | 1 |
| 5 | synthetic construct | 122.00 | 5 | v-jun sarcoma virus 17 oncogene-like |
| 6 | Rattus norvegicus | 122.00 | 5 | v-jun sarcoma virus 17 oncogene homolog |
| 7 | Bos taurus | 122.00 | 5 | PREDICTED: v-jun avian sarcoma virus 17 oncogene h |
| 8 | Macaca mulatta | 122.00 | 2 | PREDICTED: similar to Transcription factor AP-1 (A |
| 9 | Sus scrofa | 122.00 | 1 | C-JUN protein |
| 10 | Oryctolagus cuniculus | 122.00 | 1 | c-jun transcription factor |
| 11 | Macaca fuscata | 122.00 | 1 | transcription factor c-Jun |
| 12 | Danio rerio | 118.00 | 5 | c-Jun protein |
| 13 | Ctenopharyngodon idella | 118.00 | 2 | c-Jun protein |
| 14 | Monodelphis domestica | 118.00 | 2 | PREDICTED: similar to fc49b10 G-protein coupled re |
| 15 | Gallus gallus | 118.00 | 2 | jun oncogene |
| 16 | Pseudemys nelsoni | 118.00 | 1 | c-jun |
| 17 | Alligator mississippiensis | 118.00 | 1 | c-jun |
| 18 | Coturnix coturnix | 118.00 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 19 | Crocodylus niloticus | 118.00 | 1 | c-jun |
| 20 | Serinus canaria | 118.00 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 21 | Xenopus laevis | 117.00 | 7 | c-Jun protein |
| 22 | Carassius auratus | 117.00 | 1 | c-Jun protein |
| 23 | Tetraodon nigroviridis | 115.00 | 6 | unnamed protein product |
| 24 | Oncorhynchus mykiss | 114.00 | 2 | transcription factor AP-1 |
| 25 | Takifugu rubripes | 112.00 | 6 | c-Jun protein |
| 26 | Xiphophorus maculatus | 104.00 | 2 | c-JUN |
| 27 | Ovis aries | 91.70 | 2 | c-Jun protein |
| 28 | Pan troglodytes | 80.90 | 2 | PREDICTED: jun D proto-oncogene |
| 29 | Cyprinus carpio | 79.00 | 1 | JUNB_CYPCA Transcription factor jun-B gb |
| 30 | Strongylocentrotus purpuratus | 78.60 | 1 | PREDICTED: similar to transcription factor AP-1 |
| 31 | Apis mellifera | 65.50 | 1 | PREDICTED: similar to c-Jun protein |
| 32 | Aedes aegypti | 58.50 | 1 | jun |
| 33 | Anopheles gambiae str. PEST | 52.80 | 1 | ENSANGP00000020405 |
| 34 | Xenopus tropicalis | 45.40 | 1 | jun B proto-oncogene |
| 35 | Ciona intestinalis | 44.30 | 1 | transcription factor protein |
| 36 | Tribolium castaneum | 42.00 | 1 | PREDICTED: similar to Transcription factor AP-1 (J |
| 37 | Poecilia reticulata | 37.40 | 1 | transcription factor JunB |
| 38 | Drosophila pseudoobscura | 37.40 | 1 | GA15338-PA |
| 39 | Schistosoma japonicum | 35.00 | 1 | SJCHGC03684 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Homo sapiens | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Canis familiaris | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| N/A | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| synthetic construct | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Rattus norvegicus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Bos taurus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Macaca mulatta | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Sus scrofa | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Oryctolagus cuniculus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Macaca fuscata | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Danio rerio | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGMITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 119 |
| Ctenopharyngodon idella | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 119 |
| Monodelphis domestica | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 61 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Gallus gallus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 57 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 116 |
| Pseudemys nelsoni | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 61 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Alligator mississippiensis | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 61 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Coturnix coturnix | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 119 |
| Crocodylus niloticus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 61 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Serinus canaria | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||||||||| Sbjct 61 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |
| Xenopus laevis | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||+|||||||| Sbjct 59 LTSPDVGLLKLASPELERLIIQSSNGMITT#TPTPTQFLCPKNVTDEQEGFADGFVRALAE 118 |
| Carassius auratus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||||||||||||||||||| ||||| Sbjct 56 LTSPDVGLLKLASPELERLIIQSSNGMITT#TPTPTQFLCPKNVTDEQEGFAEGFARALAE 115 |
| Tetraodon nigroviridis | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||+||||||+||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFICPKNVTEEQEGFAEGFVRALAE 119 |
| Oncorhynchus mykiss | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||| ||||||||||||||||||| |||#||||||||||+||||||||||||||||||| Sbjct 59 LTSPDVQLLKLASPELERLIIQSSNGLITT#TPTPTQFLCPRNVTDEQEGFAEGFVRALAE 118 |
| Takifugu rubripes | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |||||||||||||||||||||||||| |||#|||||||+ |||||+||||||||||||||| Sbjct 60 LTSPDVGLLKLASPELERLIIQSSNGLITT#TPTPTQFISPKNVTEEQEGFAEGFVRALAE 119 |
| Xiphophorus maculatus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 ||||||||||||||||||||||| +| |#|||||||+|||||+|||||||||||||||| Sbjct 62 LTSPDVGLLKLASPELERLIIQSCSG--LT#TPTPTQFICPKNVSDEQEGFAEGFVRALAE 119 |
| Ovis aries | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVT 104 ||||||||||||||||||||||||||||||#|||||||||||||| Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVT 104 |
| Pan troglodytes | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 | |||+||||||||||||||||| || +||#||| +||| || |++ ||||||+|| + Sbjct 88 LASPDLGLLKLASPELERLIIQS-NGLVTT#TPTSSQFLYPKVAASEEQEFAEGFVKALED 146 |
| Cyprinus carpio | Query 65 DVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 | | ||||||||||||||+||| + |#|||| |+| + +||||||||||||+|| Sbjct 58 DSGSLKLASPELERLIIQTSNG-VLT#TPTPGQYLYSRGITDEQEGFAEGFVKAL 110 |
| Strongylocentrotus purpuratus | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 |++||| +|||||||||++|| | |+| |#|||| ||+ |||||+|| ||+||| || Sbjct 38 LSTPDVQMLKLASPELEKMII-SQQGNICT#TPTPGQFISPKNVTEEQAAFAQGFVEAL 94 |
| Apis mellifera | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |+|||+ +|||+|||||+ || + +| # |||| | || ||+ || +| ||| || | Sbjct 61 LSSPDLNMLKLSSPELEKFIIAQQDSLVTN#LVTPTQILFPKAVTEAQELYARGFVDALNE 120 |
| Aedes aegypti | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 +||||+ +||| |||||++|| |+ #||||+ | | | ||| ||+|| || Sbjct 65 ITSPDLQVLKLVSPELEKIIINSA---ALP#TPTPSSILYPTKATTEQEQFAKGFDEAL 119 |
| Anopheles gambiae str. PEST | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 | |||+ +||| |||||++| | #||||+ + | + | ||+ ||+|| || Sbjct 14 LPSPDLQMLKLVSPELEKII---STNATLP#TPTPSAIIFPPSATSEQQQFAKGFEDAL 68 |
| Xenopus tropicalis | Query 64 PDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKN--VTDEQEGFAEGFVRAL 118 | ||+|+|||||+|| | | |+ # + + +|+||||| +|||+|| Sbjct 61 PAPSSLKMAAPELERMIIHSGPGVISG#PSGGQLYYSSRGNGITEEQEGFVDGFVKAL 117 |
| Ciona intestinalis | Query 61 LTSPD-VGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 |++|| | +||+ ||+ +|+++ || + #| +|| +| | ||| +|+|||| | Sbjct 134 LSTPDLVNMLKVGSPDFDRMLVHSSQAN---#TQNNSQFF-QRNATQEQEIYAQGFVREL 188 |
| Tribolium castaneum | Query 63 SPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPKNVTDEQEGFAEGFVRAL 118 +|| |||+ +| +| +|+ ++ | #||+|+ + |++|| ||| || ||| || Sbjct 32 TPD--LLKVDTPTIENIILANNISH---#TPSPS-LIFPRDVTVEQEKFAGGFVEAL 81 |
| Poecilia reticulata | Query 96 QFLCPKNVTDEQEGFAEGFVRALAE 120 |+ + +||||||||||||+|| | Sbjct 1 QYFYNRGITDEQEGFAEGFVKALDE 25 |
| Drosophila pseudoobscura | Query 61 LTSPDVGLLKLASPELERLIIQSSNGHITT#TPTPTQFLCPK--NVTDEQEGFAEGFVRAL 118 + |||+ + +|+||++++ + |+ #|| | + | || ||| | +|| || Sbjct 82 INSPDLQAKTVNTPDLEKILLSN---HMMQ#TPQPGKVFPTKVGPVTSEQEAFGKGFEEAL 138 |
| Schistosoma japonicum | Query 87 HITT#TP-TPTQFLCPKNVTDEQEGFAEGFVRALAE 120 |+ #+| || || ||+||+||| || | + | | Sbjct 91 HLKA#SPQTPGSFLNPKHVTEEQERFANVFTQKLNE 125 |
[Site 2] LRAKNSDLLT62-SPDVGLLKLA
Thr62  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu53 | Arg54 | Ala55 | Lys56 | Asn57 | Ser58 | Asp59 | Leu60 | Leu61 | Thr62 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser63 | Pro64 | Asp65 | Val66 | Gly67 | Leu68 | Leu69 | Lys70 | Leu71 | Ala72 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ILKQSMTLNLADPVGSLKPHLRAKNSDLLTSPDVGLLKLASPELERLIIQSSNGHITTTP |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 119.00 | 11 | unnamed protein product |
| 2 | Canis familiaris | 119.00 | 10 | PREDICTED: similar to Transcription factor AP-1 (A |
| 3 | Homo sapiens | 119.00 | 9 | jun oncogene |
| 4 | N/A | 119.00 | 7 | 1 |
| 5 | Bos taurus | 119.00 | 5 | PREDICTED: v-jun avian sarcoma virus 17 oncogene h |
| 6 | synthetic construct | 119.00 | 5 | v-jun sarcoma virus 17 oncogene-like |
| 7 | Macaca mulatta | 119.00 | 2 | PREDICTED: similar to Transcription factor AP-1 (A |
| 8 | Ovis aries | 119.00 | 2 | c-Jun protein |
| 9 | Macaca fuscata | 119.00 | 1 | transcription factor c-Jun |
| 10 | Rattus norvegicus | 117.00 | 5 | v-jun sarcoma virus 17 oncogene homolog |
| 11 | Sus scrofa | 114.00 | 1 | C-JUN protein |
| 12 | Monodelphis domestica | 106.00 | 2 | PREDICTED: similar to fc49b10 G-protein coupled re |
| 13 | Coturnix coturnix | 105.00 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 14 | Pseudemys nelsoni | 103.00 | 1 | c-jun |
| 15 | Tetraodon nigroviridis | 102.00 | 6 | unnamed protein product |
| 16 | Danio rerio | 102.00 | 5 | c-Jun |
| 17 | Ctenopharyngodon idella | 102.00 | 2 | c-Jun protein |
| 18 | Gallus gallus | 102.00 | 2 | jun oncogene |
| 19 | Serinus canaria | 102.00 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 20 | Alligator mississippiensis | 102.00 | 1 | c-jun |
| 21 | Carassius auratus | 102.00 | 1 | c-Jun protein |
| 22 | Crocodylus niloticus | 102.00 | 1 | c-jun |
| 23 | Takifugu rubripes | 101.00 | 6 | c-Jun protein |
| 24 | Xenopus laevis | 99.00 | 5 | c-Jun protein |
| 25 | Oncorhynchus mykiss | 96.30 | 2 | transcription factor AP-1 |
| 26 | Oryctolagus cuniculus | 78.60 | 1 | c-jun transcription factor |
| 27 | Xiphophorus maculatus | 71.20 | 2 | c-JUN |
| 28 | Pan troglodytes | 55.80 | 2 | PREDICTED: jun D proto-oncogene |
| 29 | Cyprinus carpio | 54.30 | 1 | JUNB_CYPCA Transcription factor jun-B gb |
| 30 | Strongylocentrotus purpuratus | 46.60 | 1 | PREDICTED: similar to transcription factor AP-1 |
| 31 | Apis mellifera | 37.40 | 1 | PREDICTED: similar to c-Jun protein |
| 32 | Aedes aegypti | 35.40 | 1 | jun |
| 33 | Taenia solium | 33.10 | 1 | c-jun |
| 34 | Taenia crassiceps | 33.10 | 1 | c-jun |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Canis familiaris | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Homo sapiens | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| N/A | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Bos taurus | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| synthetic construct | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Macaca mulatta | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Ovis aries | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Macaca fuscata | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Rattus norvegicus | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||+||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 33 ILKQSMTLNLADPVGNLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Sus scrofa | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||||||+|||||||++|||||#|||||||||||||||||||||||||||||| Sbjct 33 ILKQSMTLNLADPVGNLKPHLRAQDSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Monodelphis domestica | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +||||+|||||||| ||||||| || ||||#|||||||||||||||||||||||| ||||| Sbjct 33 VLKQSLTLNLADPVSSLKPHLRNKNPDLLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 92 |
| Coturnix coturnix | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||||||||+|| ||||||| ||+|+||#|||||||||||||||||||||||| ||||| Sbjct 32 VLKQSMTLNLSDPASSLKPHLRNKNADILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Pseudemys nelsoni | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||+|||||||| +|||||| ||+|+||#|||||||||||||||||||||||| ||||| Sbjct 33 VLKQNMTLNLADPSSNLKPHLRNKNADILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 92 |
| Tetraodon nigroviridis | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +||||| || |||||||||| |+|||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLCDPAGSLKPHLRAKASELLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Danio rerio | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +|||||+|| |+|||||||| ||+||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLSDPAGNLKPHLRAKASDILT#SPDVGLLKLASPELERLIIQSSNGMITTTP 91 |
| Ctenopharyngodon idella | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +||||| || |+|||||||| ||+||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLTDPAGNLKPHLRAKASDILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Gallus gallus | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||||||||+| ||||||| ||+|+||#|||||||||||||||||||||||| ||||| Sbjct 29 VLKQSMTLNLSDAASSLKPHLRNKNADILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 88 |
| Serinus canaria | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||+|||||+|| +|||||| ||+|+||#|||||||||||||||||||||||| ||||| Sbjct 33 VLKQNMTLNLSDPSSNLKPHLRNKNADILT#SPDVGLLKLASPELERLIIQSSNGLITTTP 92 |
| Alligator mississippiensis | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||| +|||||| |++||||#|||||||||||||||||||||||| ||||| Sbjct 34 LKQSMTLNLADPASALKPHLRNKSADLLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 92 |
| Carassius auratus | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 ||+|||||| || |+|||||||| +|+||#|||||||||||||||||||||||| ||||| Sbjct 29 LKRSMTLNLTDPSGNLKPHLRAKANDILT#SPDVGLLKLASPELERLIIQSSNGMITTTP 87 |
| Crocodylus niloticus | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||||| +|||||| |++||||#|||||||||||||||||||||||| ||||| Sbjct 34 LKQSMTLNLADPASALKPHLRNKSTDLLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 92 |
| Takifugu rubripes | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || +|||||+|| |+|||||||| |+|||#|||||||||||||||||||||||| ||||| Sbjct 33 LKHNMTLNLSDPTGTLKPHLRAKASELLT#SPDVGLLKLASPELERLIIQSSNGLITTTP 91 |
| Xenopus laevis | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 +|||||||||+|| ++||||| | ++|||#|||||||||||||||||||||||| ||||| Sbjct 31 VLKQSMTLNLSDPSSAIKPHLRNKAAELLT#SPDVGLLKLASPELERLIIQSSNGMITTTP 90 |
| Oncorhynchus mykiss | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 || ||||||||| +|||||+|| ||+||#|||| ||||||||||||||||||| ||||| Sbjct 32 LKHSMTLNLADPTITLKPHLQAKASDILT#SPDVQLLKLASPELERLIIQSSNGLITTTP 90 |
| Oryctolagus cuniculus | Query 54 RAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||||||||#|||||||||||||||||||||||||||||| Query 54 RAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |
| Xiphophorus maculatus | Query 34 LKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |||+||||| || + || | + |+||#||||||||||||||||||||| +| | || Sbjct 37 LKQTMTLNLNDPK-NFKPQLGGR-LDILT#SPDVGLLKLASPELERLIIQSCSGLTTPTP 93 |
| Pan troglodytes | Query 35 KQSMTLNLADPVGS-LKPH-------LRAKNSD-------LLT#SPDVGLLKLASPELERL 79 | ++||+|++ | + ||| ||| + || #|||+||||||||||||| Sbjct 47 KDALTLSLSEQVAAALKPAAAPPPTPLRADGAPSAAPPDGLLA#SPDLGLLKLASPELERL 106 Query 80 IIQSSNGHITTTP 92 ||| ||| +|||| Sbjct 107 IIQ-SNGLVTTTP 118 |
| Cyprinus carpio | Query 35 KQSMTLNLADPVGSLKPHLRAKNSDLL--T#SPDVGLLKLASPELERLIIQSSNGHITT 90 |||| ||| +| +|| ||| +# | | ||||||||||||||+||| +|| Sbjct 33 KQSMNLNLTEPYRNLK-------SDLYHTS#GADSGSLKLASPELERLIIQTSNGVLTT 83 |
| Strongylocentrotus purpuratus | Query 35 KQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQSSNGHITTTP 92 |+ |||+| +| | + ||+#+||| +|||||||||++|| | |+| ||| Sbjct 17 KKCMTLDLDVGMGK-----RKLTAALLS#TPDVQMLKLASPELEKMII-SQQGNICTTP 68 |
| Apis mellifera | Query 34 LKQSMTLNLAD-----------PVGSLKPHLRAKNSDLLT#SPDVGLLKLASPELERLIIQ 82 ||+++||+| + +| + | | + +|+#|||+ +|||+|||||+ || Sbjct 24 LKRNLTLDLNECQRQGPEAKKPRLGPIPPALN-NVTPILS#SPDLNMLKLSSPELEKFIIA 82 Query 83 SSNGHIT 89 + +| Sbjct 83 QQDSLVT 89 |
| Aedes aegypti | Query 60 LLT#SPDVGLLKLASPELERLIIQSS 84 ++|#|||+ +||| |||||++|| |+ Sbjct 64 VIT#SPDLQVLKLVSPELEKIIINSA 88 |
| Taenia solium | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASP 74 |||||||||||||+ | | || # || |+| Sbjct 73 ILKQSMTLNLADPL----PELSTAGEPLLL#GLVCGLQGTAAP 110 |
| Taenia crassiceps | Query 33 ILKQSMTLNLADPVGSLKPHLRAKNSDLLT#SPDVGLLKLASP 74 |||||||||||||+ | | || # || |+| Sbjct 73 ILKQSMTLNLADPL----PELSTAGEPLLL#GLVCGLQGTAAP 110 |
[Site 3] ILKQSMTLNL42-ADPVGSLKPH
Leu42  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile33 | Leu34 | Lys35 | Gln36 | Ser37 | Met38 | Thr39 | Leu40 | Asn41 | Leu42 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala43 | Asp44 | Pro45 | Val46 | Gly47 | Ser48 | Leu49 | Lys50 | Pro51 | His52 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ALNASFLPSESGPYGYSNPKILKQSMTLNLADPVGSLKPHLRAKNSDLLTSPDVGLLKLA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 121.00 | 5 | 1 |
| 2 | Homo sapiens | 121.00 | 3 | JUN |
| 3 | synthetic construct | 121.00 | 2 | v-jun sarcoma virus 17 oncogene-like |
| 4 | Macaca fuscata | 118.00 | 1 | transcription factor c-Jun |
| 5 | Macaca mulatta | 118.00 | 1 | PREDICTED: similar to Transcription factor AP-1 (A |
| 6 | Canis familiaris | 115.00 | 6 | PREDICTED: similar to Transcription factor AP-1 (A |
| 7 | Mus musculus | 115.00 | 4 | unnamed protein product |
| 8 | Bos taurus | 115.00 | 3 | PREDICTED: v-jun avian sarcoma virus 17 oncogene h |
| 9 | Rattus norvegicus | 114.00 | 1 | v-jun sarcoma virus 17 oncogene homolog |
| 10 | Ovis aries | 112.00 | 2 | c-Jun protein |
| 11 | Sus scrofa | 110.00 | 1 | C-JUN protein |
| 12 | Alligator mississippiensis | 99.40 | 1 | c-jun |
| 13 | Crocodylus niloticus | 99.40 | 1 | c-jun |
| 14 | Pseudemys nelsoni | 97.80 | 1 | c-jun |
| 15 | Monodelphis domestica | 97.40 | 1 | PREDICTED: similar to fc49b10 G-protein coupled re |
| 16 | Gallus gallus | 97.10 | 1 | jun oncogene |
| 17 | Coturnix coturnix | 94.00 | 1 | JUN_COTJA Transcription factor AP-1 (Proto-oncogen |
| 18 | Serinus canaria | 93.60 | 1 | JUN_SERCA Transcription factor AP-1 (Proto-oncogen |
| 19 | Takifugu rubripes | 85.10 | 5 | c-Jun protein |
| 20 | Ctenopharyngodon idella | 81.60 | 2 | c-Jun protein |
| 21 | Tetraodon nigroviridis | 80.90 | 2 | unnamed protein product |
| 22 | Danio rerio | 80.10 | 2 | c-Jun |
| 23 | Carassius auratus | 79.30 | 1 | c-Jun protein |
| 24 | Xenopus laevis | 78.60 | 5 | c-Jun protein |
| 25 | Oncorhynchus mykiss | 72.80 | 1 | transcription factor AP-1 |
| 26 | Xiphophorus maculatus | 53.10 | 2 | c-JUN |
| 27 | Oryctolagus cuniculus | 39.70 | 1 | c-jun transcription factor |
| 28 | Taenia solium | 39.70 | 1 | c-jun |
| 29 | Taenia crassiceps | 39.70 | 1 | c-jun |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Homo sapiens | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| synthetic construct | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Macaca fuscata | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |||||||||||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 13 ALNASFLPSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Macaca mulatta | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |||||||||||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 13 ALNASFLPSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Canis familiaris | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| |||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Mus musculus | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| |||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Bos taurus | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| |||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Rattus norvegicus | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| |||| |||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAKNSDLLTSPDVGLLKLA 72 |
| Ovis aries | Query 15 NASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||| |||| |||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 1 NASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 58 |
| Sus scrofa | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||||| |||| |||||||||||||||||#|||||+|||||||++||||||||||||||| Sbjct 13 ALNASFLQSESGAYGYSNPKILKQSMTLNL#ADPVGNLKPHLRAQDSDLLTSPDVGLLKLA 72 |
| Alligator mississippiensis | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||+| | ||| |||+||| |||||||||#||| +|||||| |++|||||||||||||| Sbjct 13 ALNAAFAPPESGGYGYNNPKALKQSMTLNL#ADPASALKPHLRNKSADLLTSPDVGLLKLA 72 |
| Crocodylus niloticus | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||+| | ||| |||+||| |||||||||#||| +|||||| |++|||||||||||||| Sbjct 13 ALNAAFAPPESGGYGYNNPKALKQSMTLNL#ADPASALKPHLRNKSTDLLTSPDVGLLKLA 72 |
| Pseudemys nelsoni | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||+|+ ||| |||+|||+|||+|||||#||| +|||||| ||+|+|||||||||||| Sbjct 13 ALNATFVQPESGGYGYNNPKVLKQNMTLNL#ADPSSNLKPHLRNKNADILTSPDVGLLKLA 72 |
| Monodelphis domestica | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||||+|+ ||| ||||+ |+||||+||||#|||| ||||||| || |||||||||||||| Sbjct 13 ALNAAFVQPESGGYGYSHAKVLKQSLTLNL#ADPVSSLKPHLRNKNPDLLTSPDVGLLKLA 72 |
| Gallus gallus | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |||||| | ||| |||+| |+|||||||||#+| ||||||| ||+|+|||||||||||| Sbjct 9 ALNASFAPPESGGYGYNNAKVLKQSMTLNL#SDAASSLKPHLRNKNADILTSPDVGLLKLA 68 |
| Coturnix coturnix | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 |||||| | || |||+| |+|||||||||#+|| ||||||| ||+|+|||||||||||| Sbjct 13 ALNASFAPPESA-YGYNNAKVLKQSMTLNL#SDPASSLKPHLRNKNADILTSPDVGLLKLA 71 |
| Serinus canaria | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||+| | | ||| |||+| |+|||+|||||#+|| +|||||| ||+|+|||||||||||| Sbjct 13 ALSAGFAPPESGGYGYNNAKVLKQNMTLNL#SDPSSNLKPHLRNKNADILTSPDVGLLKLA 72 |
| Takifugu rubripes | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 +||| | ++ ||||||| || +|||||#+|| |+|||||||| |+||||||||||||| Sbjct 13 SLNA-FSQHDTAGYGYSNPKALKHNMTLNL#SDPTGTLKPHLRAKASELLTSPDVGLLKLA 71 |
| Ctenopharyngodon idella | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ++|++| +| |+|| | | || +|||||# || |+|||||||| ||+|||||||||||| Sbjct 13 SVNSAFSQHDSAPFGY-NHKALKHNMTLNL#TDPAGNLKPHLRAKASDILTSPDVGLLKLA 71 |
| Tetraodon nigroviridis | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 +||| | ++ +|| +|| || +|||||# || |||||||||| |+||||||||||||| Sbjct 13 SLNA-FSQHDTAGFGYGHPKALKHNMTLNL#CDPAGSLKPHLRAKASELLTSPDVGLLKLA 71 |
| Danio rerio | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 +||++| +| +|| | | || +|||||#+|| |+|||||||| ||+|||||||||||| Sbjct 13 SLNSAFSQHDSATFGY-NHKALKHNMTLNL#SDPAGNLKPHLRAKASDILTSPDVGLLKLA 71 |
| Carassius auratus | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 +||++| +| +|| | | ||+||||||# || |+|||||||| +|+|||||||||||| Sbjct 9 SLNSAFSQHDSAAFGY-NHKALKRSMTLNL#TDPSGNLKPHLRAKANDILTSPDVGLLKLA 67 |
| Xenopus laevis | Query 13 ALNASFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 ||+|+| ++ ||||+ |+|||||||||#+|| ++||||| | ++||||||||||||| Sbjct 13 ALSAAFAQHDATPYGYN--KVLKQSMTLNL#SDPSSAIKPHLRNKAAELLTSPDVGLLKLA 70 |
| Oncorhynchus mykiss | Query 17 SFLPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKLA 72 +| ++ ||| ||| || ||||||#||| +|||||+|| ||+|||||| ||||| Sbjct 16 AFSQHQNAGYGY-NPKALKHSMTLNL#ADPTITLKPHLQAKASDILTSPDVQLLKLA 70 |
| Xiphophorus maculatus | Query 13 ALNASFLPSESGP-YGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGLLKL 71 |+||| + ||+ ||| |||+|||||# || + || | + |+||||||||||| Sbjct 16 AVNASNSQHDGAAVYGF-NPKTLKQTMTLNL#NDP-KNFKPQLGGR-LDILTSPDVGLLKL 72 Query 72 A 72 | Query 72 A 72 |
| Oryctolagus cuniculus | Query 54 RAKNSDLLTSPDVGLLKLA 72 ||||||||||||||||||| Query 54 RAKNSDLLTSPDVGLLKLA 72 |
| Taenia solium | Query 19 LPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGL 68 | +| | | |||||||||||||#|||+ | | || || Sbjct 61 LTAEEGTVG--NPKILKQSMTLNL#ADPL----PELSTAGEPLLLGLVCGL 104 |
| Taenia crassiceps | Query 19 LPSESGPYGYSNPKILKQSMTLNL#ADPVGSLKPHLRAKNSDLLTSPDVGL 68 | +| | | |||||||||||||#|||+ | | || || Sbjct 61 LTAEEGTVG--NPKILKQSMTLNL#ADPL----PELSTAGEPLLLGLVCGL 104 |
* References
[PubMed ID: 19219074] Laner-Plamberger S, Kaser A, Paulischta M, Hauser-Kronberger C, Eichberger T, Frischauf AM, Cooperation between GLI and JUN enhances transcription of JUN and selected GLI target genes. Oncogene. 2009 Apr 2;28(13):1639-51. Epub 2009 Feb 16.
[PubMed ID: 19258923] ... Siezen CL, Bont L, Hodemaekers HM, Ermers MJ, Doornbos G, Van't Slot R, Wijmenga C, Houwelingen HC, Kimpen JL, Kimman TG, Hoebee B, Janssen R, Genetic susceptibility to respiratory syncytial virus bronchiolitis in preterm children is associated with airway remodeling genes and innate immune genes. Pediatr Infect Dis J. 2009 Apr;28(4):333-5.
[PubMed ID: 19281909] ... Gutzmer R, Mommert S, Gschwandtner M, Zwingmann K, Stark H, Werfel T, The histamine H4 receptor is functionally expressed on T(H)2 cells. J Allergy Clin Immunol. 2009 Mar;123(3):619-25.
[PubMed ID: 19249008] ... Wang K, Zhang H, Kugathasan S, Annese V, Bradfield JP, Russell RK, Sleiman PM, Imielinski M, Glessner J, Hou C, Wilson DC, Walters T, Kim C, Frackelton EC, Lionetti P, Barabino A, Van Limbergen J, Guthery S, Denson L, Piccoli D, Li M, Dubinsky M, Silverberg M, Griffiths A, Grant SF, Satsangi J, Baldassano R, Hakonarson H, Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009 Mar;84(3):399-405. Epub 2009 Feb 26.
[PubMed ID: 19334540] ... Piruzian ES, Ishkin AA, Nikol'skaia TA, Abdeev RM, Bruskin SA, [The comparative analysis of psoriasis and Crohn disease molecular-genetical processes under pathological conditions] Mol Biol (Mosk). 2009 Jan-Feb;43(1):175-9.
[PubMed ID: 15302935] ... Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP, Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004 Aug 17;101(33):12130-5. Epub 2004 Aug 9.
[PubMed ID: 11689449] ... Vries RG, Prudenziati M, Zwartjes C, Verlaan M, Kalkhoven E, Zantema A, A specific lysine in c-Jun is required for transcriptional repression by E1A and is acetylated by p300. EMBO J. 2001 Nov 1;20(21):6095-103.
[PubMed ID: 10637231] ... Barila D, Mangano R, Gonfloni S, Kretzschmar J, Moro M, Bohmann D, Superti-Furga G, A nuclear tyrosine phosphorylation circuit: c-Jun as an activator and substrate of c-Abl and JNK. EMBO J. 2000 Jan 17;19(2):273-81.
[PubMed ID: 8654373] ... Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Derijard B, Davis RJ, Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996 Jun 3;15(11):2760-70.
[PubMed ID: 8137421] ... Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ, JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994 Mar 25;76(6):1025-37.
[PubMed ID: 8464713] ... Bannister AJ, Gottlieb TM, Kouzarides T, Jackson SP, c-Jun is phosphorylated by the DNA-dependent protein kinase in vitro; definition of the minimal kinase recognition motif. Nucleic Acids Res. 1993 Mar 11;21(5):1289-95.
[PubMed ID: 1448082] ... Oehler T, Angel P, A common intermediary factor (p52/54) recognizing "acidic blob"-type domains is required for transcriptional activation by the Jun proteins. Mol Cell Biol. 1992 Dec;12(12):5508-15.
[PubMed ID: 1516134] ... Lin A, Frost J, Deng T, Smeal T, al-Alawi N, Kikkawa U, Hunter T, Brenner D, Karin M, Casein kinase II is a negative regulator of c-Jun DNA binding and AP-1 activity. Cell. 1992 Sep 4;70(5):777-89.
[PubMed ID: 1516134] ... Lin A, Frost J, Deng T, Smeal T, al-Alawi N, Kikkawa U, Hunter T, Brenner D, Karin M, Casein kinase II is a negative regulator of c-Jun DNA binding and AP-1 activity. Cell. 1992 Sep 4;70(5):777-89.
[PubMed ID: 1631061] ... Chevray PM, Nathans D, Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5789-93.
[PubMed ID: 1310896] ... Bengal E, Ransone L, Scharfmann R, Dwarki VJ, Tapscott SJ, Weintraub H, Verma IM, Functional antagonism between c-Jun and MyoD proteins: a direct physical association. Cell. 1992 Feb 7;68(3):507-19.
[PubMed ID: 1651323] ... Alvarez E, Northwood IC, Gonzalez FA, Latour DA, Seth A, Abate C, Curran T, Davis RJ, Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate protein phosphorylation. Characterization of the phosphorylation of c-myc and c-jun proteins by an epidermal growth factor receptor threonine 669 protein kinase. J Biol Chem. 1991 Aug 15;266(23):15277-85.