XSB2374 : PREDICTED: similar to integrin beta-1 subunit isoform 2 [Ornithorhynchus anatinus]
[ CaMP Format ]
This entry is computationally expanded from SB0079
* Basic Information
| Organism | Ornithorhynchus anatinus (platypus) |
| Protein Names | similar to integrin beta-1 subunit isoform 2 |
| Gene Names | LOC100073959; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 2 ESTs, 27 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001508202 | XM_001508152 | 100073959 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 802 aa
Average Mass: 88.443 kDa
Monoisotopic Mass: 88.385 kDa
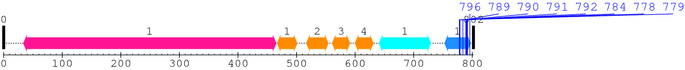
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Integrin_beta 1. | 34 | 465 | 1123.6 | 0 |
| EGF_2 1. | 467 | 501 | 12.0 | 0.13 |
| EGF_2 2. | 517 | 554 | 21.5 | 0.0036 |
| EGF_2 3. | 561 | 591 | 21.4 | 0.0038 |
| EGF_2 4. | 600 | 631 | 36.0 | 1.5e-07 |
| Integrin_B_tail 1. | 641 | 729 | 156.8 | 6.3e-44 |
| Integrin_b_cyt 1. | 753 | 797 | 96.5 | 9.4e-26 |
| --- cleavage 796 (inside Integrin_b_cyt 753..797) --- | ||||
| --- cleavage 789 (inside Integrin_b_cyt 753..797) --- | ||||
| --- cleavage 790 (inside Integrin_b_cyt 753..797) --- | ||||
| --- cleavage 791 (inside Integrin_b_cyt 753..797) --- | ||||
| --- cleavage 792 (inside Integrin_b_cyt 753..797) --- | ||||
| --- cleavage 784 (inside Integrin_b_cyt 753..797) --- | ||||
| --- cleavage 778 (inside Integrin_b_cyt 753..797) --- | ||||
| --- cleavage 779 (inside Integrin_b_cyt 753..797) --- | ||||
3. Sequence Information
Fasta Sequence: XSB2374.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
8 [sites]
Cleavage sites (±10aa)
[Site 1] PINKFKNPNY796-GRKAGL
Tyr796  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro787 | Ile788 | Asn789 | Lys790 | Phe791 | Lys792 | Asn793 | Pro794 | Asn795 | Tyr796 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly797 | Arg798 | Lys799 | Ala800 | Gly801 | Leu802 | - | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| FEKEKMNAKWDTQENPIYKSPINKFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 79.70 | 17 | S |
| 2 | Homo sapiens | 79.70 | 10 | integrin beta 1 isoform 1D precursor |
| 3 | Mus musculus | 79.70 | 10 | beta 1 integrin |
| 4 | Canis familiaris | 79.70 | 8 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 5 | Bos taurus | 79.70 | 7 | ITGB1 protein |
| 6 | Gallus gallus | 79.70 | 6 | integrin beta1D |
| 7 | Monodelphis domestica | 79.70 | 4 | PREDICTED: similar to integrin beta-1 subunit isof |
| 8 | Drosophila melanogaster | 53.50 | 1 | myospheroid CG1560-PA |
| 9 | Biomphalaria glabrata | 53.50 | 1 | beta integrin subunit |
| 10 | Pan troglodytes | 52.00 | 10 | PREDICTED: hypothetical protein |
| 11 | Rattus norvegicus | 52.00 | 7 | integrin beta 1 |
| 12 | Tetraodon nigroviridis | 52.00 | 5 | unnamed protein product |
| 13 | Sus scrofa | 52.00 | 5 | integrin beta-1 subunit |
| 14 | Xenopus laevis | 52.00 | 5 | integrin beta 1 |
| 15 | synthetic construct | 52.00 | 3 | hypothetical protein |
| 16 | Oryctolagus cuniculus | 52.00 | 2 | integrin beta 1 |
| 17 | Pongo pygmaeus | 52.00 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 18 | Felis catus | 52.00 | 1 | integrin beta 1 |
| 19 | Xenopus tropicalis | 52.00 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 20 | Danio rerio | 51.60 | 8 | integrin beta1 subunit-like protein 1 |
| 21 | Ostrinia furnacalis | 51.20 | 1 | integrin beta 1 precursor |
| 22 | Pseudoplusia includens | 51.20 | 1 | integrin beta 1 |
| 23 | Anopheles gambiae str. PEST | 50.80 | 1 | ENSANGP00000021373 |
| 24 | Anopheles gambiae | 50.80 | 1 | integrin beta subunit |
| 25 | Aedes aegypti | 50.80 | 1 | integrin beta subunit |
| 26 | Ictalurus punctatus | 50.10 | 1 | beta-1 integrin |
| 27 | Tribolium castaneum | 49.30 | 2 | PREDICTED: similar to CG1560-PA |
| 28 | Podocoryne carnea | 48.50 | 1 | AF308652_1 integrin beta chain |
| 29 | Crassostrea gigas | 48.10 | 1 | integrin beta cgh |
| 30 | Caenorhabditis briggsae | 47.00 | 1 | Hypothetical protein CBG03601 |
| 31 | Caenorhabditis elegans | 47.00 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Strongylocentrotus purpuratus | 44.30 | 5 | integrin beta-C subunit |
| 33 | Lytechinus variegatus | 44.30 | 1 | beta-C integrin subunit |
| 34 | Acropora millepora | 39.30 | 1 | integrin subunit betaCn1 |
| 35 | Macaca mulatta | 38.50 | 8 | PREDICTED: similar to integrin beta chain, beta 3 |
| 36 | Equus caballus | 38.50 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 37 | rats, Peptide Partial, 723 aa | 38.50 | 1 | beta 3 integrin, GPIIIA |
| 38 | Canis lupus familiaris | 38.50 | 1 | integrin beta chain, beta 3 |
| 39 | Ophlitaspongia tenuis | 38.50 | 1 | integrin subunit betaPo1 |
| 40 | Halocynthia roretzi | 37.40 | 2 | integrin beta Hr1 precursor |
| 41 | Ovis aries | 37.40 | 1 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 42 | mice, Peptide Partial, 680 aa | 37.00 | 1 | beta 3 integrin, GPIIIA |
| 43 | Apis mellifera | 35.80 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 44 | Suberites domuncula | 34.70 | 1 | integrin beta subunit |
| 45 | Mus sp. | 33.50 | 1 | beta 7 integrin |
| 46 | Geodia cydonium | 33.50 | 1 | integrin beta subunit |
| 47 | Schistosoma japonicum | 32.70 | 1 | SJCHGC06221 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRKAGL 802 ||||||||||||||||||||||| ||||||#|||||| Sbjct 60 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 95 |
| Homo sapiens | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRKAGL 802 ||||||||||||||||||||||| ||||||#|||||| Sbjct 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 |
| Mus musculus | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRKAGL 802 ||||||||||||||||||||||| ||||||#|||||| Sbjct 44 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 79 |
| Canis familiaris | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRKAGL 802 ||||||||||||||||||||||| ||||||#|||||| Sbjct 789 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 824 |
| Bos taurus | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRKAGL 802 ||||||||||||||||||||||| ||||||#|||||| Sbjct 766 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 801 |
| Gallus gallus | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRKAGL 802 ||||||||||||||||||||||| ||||||#|||||| Sbjct 23 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 58 |
| Monodelphis domestica | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRKAGL 802 ||||||||||||||||||||||| ||||||#|||||| Sbjct 767 FEKEKMNAKWDTQENPIYKSPINNFKNPNY#GRKAGL 802 |
| Drosophila melanogaster | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 ||||+||||||| |||||| + |||| |# | Sbjct 814 FEKERMNAKWDTGENPIYKQATSTFKNPMY#AGK 846 |
| Biomphalaria glabrata | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#G 797 ||||+ |||||| |||||| + |||| |#| Sbjct 756 FEKERQNAKWDTGENPIYKQATSTFKNPTY#G 786 |
| Pan troglodytes | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||||||||||| ||||||| + || |# | Sbjct 878 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 910 |
| Rattus norvegicus | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||||||||||| ||||||| + || |# | Sbjct 767 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 799 |
| Tetraodon nigroviridis | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||||||||||| ||||||| + || |# | Sbjct 759 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 791 |
| Sus scrofa | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 798 |
| Xenopus laevis | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 798 |
| synthetic construct | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 798 |
| Oryctolagus cuniculus | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||||||||||| ||||||| + || |# | Sbjct 31 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 63 |
| Pongo pygmaeus | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 798 |
| Felis catus | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 798 |
| Xenopus tropicalis | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EGK 798 |
| Danio rerio | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||||||||||| ||||||| + || |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVINPKY#EGK 798 |
| Ostrinia furnacalis | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 ||||+| ||||| |||||| + |||| |# | Sbjct 799 FEKERMMAKWDTGENPIYKQATSTFKNPTY#AGK 831 |
| Pseudoplusia includens | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 ||||+| ||||| |||||| + |||| |# | Sbjct 805 FEKERMMAKWDTGENPIYKQATSTFKNPTY#AGK 837 |
| Anopheles gambiae str. PEST | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 ||||+| ||||| |||||| |||| |# | Sbjct 805 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AGK 837 |
| Anopheles gambiae | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 ||||+| ||||| |||||| |||| |# | Sbjct 805 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AGK 837 |
| Aedes aegypti | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 ||||+| ||||| |||||| |||| |# | Sbjct 803 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AGK 835 |
| Ictalurus punctatus | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 ||||||||||| ||||||| + || |# | Sbjct 775 FEKEKMNAKWDAGENPIYKSAVTTVVNPKY#EGK 807 |
| Tribolium castaneum | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRKA 800 |||| | |+||| |||||| + |||| |# |+ Sbjct 797 FEKEAMMARWDTGENPIYKQATSTFKNPTY#AGKS 830 |
| Podocoryne carnea | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 |||++ | |||+ |||||| + |+|| |#| | Sbjct 749 FEKDRQNPKWDSGENPIYKKATSTFQNPMY#GNK 781 |
| Crassostrea gigas | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY# 796 |||| |||+||| |||||| + | || |# Sbjct 768 FEKEAMNARWDTGENPIYKQATSTFVNPTY# 797 |
| Caenorhabditis briggsae | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRKA 800 | |++ ||||| |||||| |||| |# || Sbjct 776 FNNERLMAKWDTNENPIYKQATTTFKNPVY#AGKA 809 |
| Caenorhabditis elegans | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRKA 800 | |++ ||||| |||||| |||| |# || Sbjct 775 FNNERLMAKWDTNENPIYKQATTTFKNPVY#AGKA 808 |
| Strongylocentrotus purpuratus | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 ||||+ || |+ |||||| + |||| |# | Sbjct 774 FEKERANATWEGGENPIYKPSTSVFKNPTY#NIK 806 |
| Lytechinus variegatus | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 ||||+ || |+ |||||| + |||| |# | Sbjct 774 FEKERANATWEGGENPIYKPSTSVFKNPTY#NIK 806 |
| Acropora millepora | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY# 796 ||+|+|++|| ++||+|++ |+|| |# Sbjct 758 FERERMHSKWTREKNPLYQAAKTTFENPTY# 787 |
| Macaca mulatta | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY# 796 ||+|+ ||||| ||+|| + | | |# Sbjct 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 |
| Equus caballus | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY# 796 ||+|+ ||||| ||+|| + | | |# Sbjct 752 FEEERARAKWDTANNPLYKEATSTFTNITY# 781 |
| rats, Peptide Partial, 723 aa | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY# 796 ||+|+ ||||| ||+|| + | | |# Sbjct 691 FEEERARAKWDTANNPLYKEATSTFTNITY# 720 |
| Canis lupus familiaris | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY# 796 ||+|+ ||||| ||+|| + | | |# Sbjct 752 FEEERARAKWDTANNPLYKEATSTFTNITY# 781 |
| Ophlitaspongia tenuis | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GR 798 ||+| |||+ |||+|+| ++|| |#|+ Sbjct 807 FEREIKNAKYTKNENPLYRSATKDYQNPLY#GK 838 |
| Halocynthia roretzi | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#G 797 ||+| ++| ||++ +| ||+||+|#| Sbjct 819 FEREMKQSRWTKDINPVFVNPSAKFENPSY#G 849 |
| Ovis aries | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GRK 799 || |+ ||| | ||+|+ + ||| |# | Sbjct 744 FEAERSKAKWQTGTNPLYRGSTSTFKNVTY#KHK 776 |
| mice, Peptide Partial, 680 aa | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKN 793 ||+|+ ||||| ||+|| + | | Sbjct 652 FEEERARAKWDTANNPLYKEATSTFTN 678 |
| Apis mellifera | Query 767 FEKEKMNA-KWDTQENPIYKSPINKFKNPNY# 796 ||+|++ | ||+ ++||+|| + |||| +# Sbjct 783 FERERILANKWNRRDNPLYKEATSTFKNPTF# 813 |
| Suberites domuncula | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GR 798 |||| |+ +||+|+| ++|| |#|+ Sbjct 733 FEKELAKTKYPKHQNPLYRSAAKDYQNPIY#GQ 764 |
| Mus sp. | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY# 796 ||||+ | ||+||| | || +# Sbjct 760 FEKEQQQLNWKQDNNPLYKSAITTTVNPRF# 789 |
| Geodia cydonium | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY#GR 798 || | |+ |||+| | + ||| |#|+ Sbjct 847 FENELAKVKYTKNENPLYHSATTEHKNPIY#GQ 878 |
| Schistosoma japonicum | Query 767 FEKEKMNAKWDTQENPIYKSPINKFKNPNY# 796 |+++ | +|+ ||||++|| || +# Sbjct 254 FKQQGENMRWEMAENPIFESPTTNVLNPTF# 283 |
[Site 2] ENPIYKSPIN789-KFKNPNYGRK
Asn789  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu780 | Asn781 | Pro782 | Ile783 | Tyr784 | Lys785 | Ser786 | Pro787 | Ile788 | Asn789 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys790 | Phe791 | Lys792 | Asn793 | Pro794 | Asn795 | Tyr796 | Gly797 | Arg798 | Lys799 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DRREFAKFEKEKMNAKWDTQENPIYKSPINKFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 93.60 | 31 | S |
| 2 | Homo sapiens | 93.60 | 25 | integrin beta 1 isoform 1D precursor |
| 3 | Mus musculus | 93.60 | 14 | beta 1 integrin |
| 4 | Bos taurus | 93.60 | 13 | ITGB1 protein |
| 5 | Canis familiaris | 93.60 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 6 | Gallus gallus | 93.60 | 7 | integrin beta1D |
| 7 | Monodelphis domestica | 93.60 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 8 | Drosophila melanogaster | 66.20 | 2 | myospheroid CG1560-PA |
| 9 | Pan troglodytes | 65.90 | 14 | PREDICTED: hypothetical protein |
| 10 | Rattus norvegicus | 65.90 | 10 | integrin beta 1 |
| 11 | Sus scrofa | 65.90 | 6 | integrin beta-1 subunit |
| 12 | Xenopus laevis | 65.90 | 5 | integrin beta-1 subunit |
| 13 | Tetraodon nigroviridis | 65.90 | 5 | unnamed protein product |
| 14 | synthetic construct | 65.90 | 5 | hypothetical protein |
| 15 | Pongo pygmaeus | 65.90 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 16 | Oryctolagus cuniculus | 65.90 | 2 | integrin beta 1 |
| 17 | Xenopus tropicalis | 65.90 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 18 | Danio rerio | 65.50 | 10 | integrin beta1 subunit-like protein 1 |
| 19 | Biomphalaria glabrata | 65.10 | 1 | beta integrin subunit |
| 20 | Ictalurus punctatus | 63.90 | 2 | beta-1 integrin |
| 21 | Pseudoplusia includens | 63.90 | 1 | integrin beta 1 |
| 22 | Ostrinia furnacalis | 63.90 | 1 | integrin beta 1 precursor |
| 23 | Anopheles gambiae | 63.50 | 2 | integrin beta subunit |
| 24 | Anopheles gambiae str. PEST | 63.50 | 2 | ENSANGP00000021373 |
| 25 | Felis catus | 63.50 | 1 | integrin beta 1 |
| 26 | Aedes aegypti | 63.50 | 1 | integrin beta subunit |
| 27 | Tribolium castaneum | 63.20 | 3 | PREDICTED: similar to CG1560-PA |
| 28 | Podocoryne carnea | 62.40 | 1 | AF308652_1 integrin beta chain |
| 29 | Crassostrea gigas | 57.40 | 1 | integrin beta cgh |
| 30 | Caenorhabditis briggsae | 57.40 | 1 | Hypothetical protein CBG03601 |
| 31 | Caenorhabditis elegans | 55.10 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Strongylocentrotus purpuratus | 54.30 | 5 | integrin beta-C subunit |
| 33 | Lytechinus variegatus | 54.30 | 1 | beta-C integrin subunit |
| 34 | Macaca mulatta | 51.20 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 35 | Equus caballus | 51.20 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 36 | rats, Peptide Partial, 723 aa | 51.20 | 1 | beta 3 integrin, GPIIIA |
| 37 | Canis lupus familiaris | 51.20 | 1 | integrin beta chain, beta 3 |
| 38 | mice, Peptide Partial, 680 aa | 49.70 | 1 | beta 3 integrin, GPIIIA |
| 39 | Ovis aries | 47.40 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 40 | Acropora millepora | 47.00 | 1 | integrin subunit betaCn1 |
| 41 | Apis mellifera | 45.80 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 42 | Halocynthia roretzi | 44.30 | 2 | integrin beta Hr2 precursor |
| 43 | Mus sp. | 43.10 | 1 | beta 7 integrin |
| 44 | Pacifastacus leniusculus | 42.70 | 1 | integrin |
| 45 | Manduca sexta | 42.40 | 1 | plasmatocyte-specific integrin beta 1 |
| 46 | Schistosoma japonicum | 42.40 | 1 | SJCHGC06221 protein |
| 47 | Ophlitaspongia tenuis | 41.60 | 1 | integrin subunit betaPo1 |
| 48 | Spodoptera frugiperda | 38.90 | 1 | integrin beta 1 |
| 49 | nu | 38.50 | 1 | beta |
| 50 | Ovis canadensis | 38.10 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 51 | Capra hircus | 38.10 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 52 | Geodia cydonium | 38.10 | 1 | integrin beta subunit |
| 53 | Bubalus bubalis | 38.10 | 1 | integrin beta-2 |
| 54 | Suberites domuncula | 37.70 | 1 | integrin beta subunit |
| 55 | Sigmodon hispidus | 37.40 | 1 | AF445415_1 integrin beta-2 precursor |
| 56 | Drosophila pseudoobscura | 37.00 | 1 | GA14581-PA |
| 57 | Oncorhynchus mykiss | 34.30 | 1 | CD18 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||# |||||||||||| Sbjct 53 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 95 |
| Homo sapiens | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||# |||||||||||| Sbjct 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 |
| Mus musculus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||# |||||||||||| Sbjct 37 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 79 |
| Bos taurus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||# |||||||||||| Sbjct 759 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 801 |
| Canis familiaris | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||# |||||||||||| Sbjct 782 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 824 |
| Gallus gallus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||# |||||||||||| Sbjct 16 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 58 |
| Monodelphis domestica | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||# |||||||||||| Sbjct 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#NFKNPNYGRKAGL 802 |
| Drosophila melanogaster | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||+||||+||||||| |||||| +# |||| | | Sbjct 807 DRREFARFEKERMNAKWDTGENPIYKQATS#TFKNPMYAGK 846 |
| Pan troglodytes | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||||||||||||||| ||||||| + # || | | Sbjct 871 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 910 |
| Rattus norvegicus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||||||||||||||| ||||||| + # || | | Sbjct 760 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 799 |
| Sus scrofa | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||||||||||||||| ||||||| + # || | | Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 798 |
| Xenopus laevis | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||||||||||||||| ||||||| + # || | | Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 798 |
| Tetraodon nigroviridis | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||||||||||||||| ||||||| + # || | | Sbjct 752 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 791 |
| synthetic construct | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||||||||||||||| ||||||| + # || | | Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 798 |
| Pongo pygmaeus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||||||||||||||| ||||||| + # || | | Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 798 |
| Oryctolagus cuniculus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||||||||||||||| ||||||| + # || | | Sbjct 24 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 63 |
| Xenopus tropicalis | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||||||||||||||| ||||||| + # || | | Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 798 |
| Danio rerio | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||||||||||||||| ||||||| + # || | | Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVT#TVINPKYEGK 798 |
| Biomphalaria glabrata | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYG 797 | |||||||||+ |||||| |||||| +# |||| || Sbjct 749 DTREFAKFEKERQNAKWDTGENPIYKQATS#TFKNPTYG 786 |
| Ictalurus punctatus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 768 DRREFAKFEKEKMNAKWDAGENPIYKSAVT#TVVNPKYEGK 807 |
| Pseudoplusia includens | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||+||||+| ||||| |||||| +# |||| | | Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATS#TFKNPTYAGK 837 |
| Ostrinia furnacalis | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||+||||+| ||||| |||||| +# |||| | | Sbjct 792 DRREFARFEKERMMAKWDTGENPIYKQATS#TFKNPTYAGK 831 |
| Anopheles gambiae | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||+||||+| ||||| |||||| # |||| | | Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATT#TFKNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||+||||+| ||||| |||||| # |||| | | Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATT#TFKNPTYAGK 837 |
| Felis catus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 | ||||||||||||||||| ||||||| + # || | | Sbjct 759 DTREFAKFEKEKMNAKWDTGENPIYKSAVT#TVVNPKYEGK 798 |
| Aedes aegypti | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||+||||+| ||||| |||||| # |||| | | Sbjct 796 DRREFARFEKERMMAKWDTGENPIYKQATT#TFKNPTYAGK 835 |
| Tribolium castaneum | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRKA 800 ||||||||||| | |+||| |||||| +# |||| | |+ Sbjct 790 DRREFAKFEKEAMMARWDTGENPIYKQATS#TFKNPTYAGKS 830 |
| Podocoryne carnea | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||||||||++ | |||+ |||||| +# |+|| || | Sbjct 742 DRREFAKFEKDRQNPKWDSGENPIYKKATS#TFQNPMYGNK 781 |
| Crassostrea gigas | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 |+|| |+|||| |||+||| |||||| +# | || | Sbjct 761 DKRELARFEKEAMNARWDTGENPIYKQATS#TFVNPTY 797 |
| Caenorhabditis briggsae | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRKA 800 || |+||| |++ ||||| |||||| # |||| | || Sbjct 769 DRAEYAKFNNERLMAKWDTNENPIYKQATT#TFKNPVYAGKA 809 |
| Caenorhabditis elegans | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRKA 800 || |+| | |++ ||||| |||||| # |||| | || Sbjct 768 DRSEYATFNNERLMAKWDTNENPIYKQATT#TFKNPVYAGKA 808 |
| Strongylocentrotus purpuratus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||| ||||+ || |+ |||||| +# |||| | | Sbjct 767 DRREFQNFEKERANATWEGGENPIYKPSTS#VFKNPTYNIK 806 |
| Lytechinus variegatus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||||| ||||+ || |+ |||||| +# |||| | | Sbjct 767 DRREFQNFEKERANATWEGGENPIYKPSTS#VFKNPTYNIK 806 |
| Macaca mulatta | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 ||+||||||+|+ ||||| ||+|| +# | | | Sbjct 749 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTNITY 785 |
| Equus caballus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 ||+||||||+|+ ||||| ||+|| +# | | | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTNITY 781 |
| rats, Peptide Partial, 723 aa | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 ||+||||||+|+ ||||| ||+|| +# | | | Sbjct 684 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTNITY 720 |
| Canis lupus familiaris | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 ||+||||||+|+ ||||| ||+|| +# | | | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTNITY 781 |
| mice, Peptide Partial, 680 aa | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKN 793 ||+||||||+|+ ||||| ||+|| +# | | Sbjct 645 DRKEFAKFEEERARAKWDTANNPLYKEATS#TFTN 678 |
| Ovis aries | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 ||+| |||| |+ ||| | ||+|+ +# ||| | | Sbjct 737 DRKEVAKFEAERSKAKWQTGTNPLYRGSTS#TFKNVTYKHK 776 |
| Acropora millepora | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 || |+ |||+|+|++|| ++||+|++ # |+|| | Sbjct 751 DRIEYQKFERERMHSKWTREKNPLYQAAKT#TFENPTY 787 |
| Apis mellifera | Query 760 DRREFAKFEKEKMNA-KWDTQENPIYKSPIN#KFKNPNY 796 | ||| |||+|++ | ||+ ++||+|| +# |||| + Sbjct 776 DNREFKKFERERILANKWNRRDNPLYKEATS#TFKNPTF 813 |
| Halocynthia roretzi | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYG 797 |+||+ ||+||+ || |||+| + +#|| || +| Sbjct 795 DKREYEKFKKEEDLRKWTKGENPVYVNASS#KFDNPMFG 832 |
| Mus sp. | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 ||||+ +||||+ | ||+||| | # || + Sbjct 753 DRREYRRFEKEQQQLNWKQDNNPLYKSAIT#TTVNPRF 789 |
| Pacifastacus leniusculus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGR 798 ||||+||||||+ |||+||| # |+|| + + Sbjct 761 DRREYAKFEKERKLPSGKRAENPLYKSAKT#TFQNPAFAQ 799 |
| Manduca sexta | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRKA 800 |+||+||||+| + +| ||+|+ | # | || | | Sbjct 725 DKREYAKFEEESRSRGFDVSLNPLYQEPEI#NFSNPVYNANA 765 |
| Schistosoma japonicum | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 |||| | |+++ | +|+ ||||++|| # || + Sbjct 247 DRRELANFKQQGENMRWEMAENPIFESPTT#NVLNPTF 283 |
| Ophlitaspongia tenuis | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGR 798 | |||+| |||+ |||+|+| # ++|| ||+ Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATK#DYQNPLYGK 838 |
| Spodoptera frugiperda | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGRK 799 |++|+ ||+ | + | +| ||+|+ | # | || | + Sbjct 740 DKKEYKKFQDEALAAGYDVSLNPLYQDPSI#NFSNPVYNNQ 779 |
| nu | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNP 794 | ||+||||+++ |+ ||||||+ |+ #+++ | Sbjct 753 DAREYAKFEEDQKNSV--RQENPIYRDPVG#RYEVP 785 |
| Ovis canadensis | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 | ||+ +|||||+ ++|+ +||++|| # || + Sbjct 732 DLREYHRFEKEKLKSQWN-NDNPLFKSATT#TVMNPKF 767 |
| Capra hircus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 | ||+ +|||||+ ++|+ +||++|| # || + Sbjct 732 DLREYHRFEKEKLKSQWN-NDNPLFKSATT#TVMNPKF 767 |
| Geodia cydonium | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGR 798 | | ||| | |+ |||+| | #+ ||| ||+ Sbjct 840 DYTELKKFENELAKVKYTKNENPLYHSATT#EHKNPIYGQ 878 |
| Bubalus bubalis | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 | ||+ +|||||+ ++|+ +||++|| # || + Sbjct 731 DLREYHRFEKEKLKSQWN-NDNPLFKSATT#TVMNPKF 766 |
| Suberites domuncula | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNYGR 798 | ||||| |+ +||+|+| # ++|| ||+ Sbjct 729 EVKKFEKELAKTKYPKHQNPLYRSAAK#DYQNPIYGQ 764 |
| Sigmodon hispidus | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 | ||+ |||||+ ++|+ +||++|| # || + Sbjct 731 DLREYRHFEKEKLKSQWN-NDNPLFKSATT#TVMNPKF 766 |
| Drosophila pseudoobscura | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNP 794 | ||+|+||+|+ || +||||+|+ |+ #++ | Sbjct 726 DAREYARFEEEQ--AKRVSQENPLYRDPVG#RYVVP 758 |
| Oncorhynchus mykiss | Query 760 DRREFAKFEKEKMNAKWDTQENPIYKSPIN#KFKNPNY 796 | +|| +|| |+ || +||++ + # || + Sbjct 738 DVKEFRRFENEQKKGKWSPGDNPLFMNATT#TVANPTF 774 |
[Site 3] NPIYKSPINK790-FKNPNYGRKA
Lys790  Phe
Phe
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn781 | Pro782 | Ile783 | Tyr784 | Lys785 | Ser786 | Pro787 | Ile788 | Asn789 | Lys790 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Phe791 | Lys792 | Asn793 | Pro794 | Asn795 | Tyr796 | Gly797 | Arg798 | Lys799 | Ala800 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RREFAKFEKEKMNAKWDTQENPIYKSPINKFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 91.30 | 30 | S |
| 2 | Homo sapiens | 91.30 | 25 | integrin beta 1 isoform 1D precursor |
| 3 | Mus musculus | 91.30 | 14 | beta 1 integrin |
| 4 | Bos taurus | 91.30 | 13 | ITGB1 protein |
| 5 | Canis familiaris | 91.30 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 6 | Gallus gallus | 91.30 | 7 | integrin beta1D |
| 7 | Monodelphis domestica | 91.30 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 8 | Drosophila melanogaster | 63.90 | 2 | myospheroid CG1560-PA |
| 9 | Pan troglodytes | 63.50 | 14 | PREDICTED: hypothetical protein |
| 10 | Rattus norvegicus | 63.50 | 10 | integrin beta 1 |
| 11 | Sus scrofa | 63.50 | 6 | integrin beta-1 subunit |
| 12 | Xenopus laevis | 63.50 | 5 | integrin beta 1 |
| 13 | Tetraodon nigroviridis | 63.50 | 5 | unnamed protein product |
| 14 | synthetic construct | 63.50 | 5 | hypothetical protein |
| 15 | Pongo pygmaeus | 63.50 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 16 | Oryctolagus cuniculus | 63.50 | 2 | integrin beta 1 |
| 17 | Xenopus tropicalis | 63.50 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 18 | Danio rerio | 63.20 | 10 | integrin beta1 subunit-like protein 1 |
| 19 | Biomphalaria glabrata | 63.20 | 1 | beta integrin subunit |
| 20 | Felis catus | 61.60 | 1 | integrin beta 1 |
| 21 | Pseudoplusia includens | 61.60 | 1 | integrin beta 1 |
| 22 | Ictalurus punctatus | 61.60 | 1 | beta-1 integrin |
| 23 | Ostrinia furnacalis | 61.60 | 1 | integrin beta 1 precursor |
| 24 | Anopheles gambiae str. PEST | 61.20 | 2 | ENSANGP00000021373 |
| 25 | Anopheles gambiae | 61.20 | 2 | integrin beta subunit |
| 26 | Aedes aegypti | 61.20 | 1 | integrin beta subunit |
| 27 | Tribolium castaneum | 60.80 | 3 | PREDICTED: similar to CG1560-PA |
| 28 | Podocoryne carnea | 60.10 | 1 | AF308652_1 integrin beta chain |
| 29 | Crassostrea gigas | 55.10 | 1 | integrin beta cgh |
| 30 | Caenorhabditis briggsae | 55.10 | 1 | Hypothetical protein CBG03601 |
| 31 | Caenorhabditis elegans | 52.80 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Strongylocentrotus purpuratus | 52.00 | 5 | integrin beta-C subunit |
| 33 | Lytechinus variegatus | 52.00 | 1 | beta-C integrin subunit |
| 34 | Macaca mulatta | 48.90 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 35 | Equus caballus | 48.90 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 36 | rats, Peptide Partial, 723 aa | 48.90 | 1 | beta 3 integrin, GPIIIA |
| 37 | Canis lupus familiaris | 48.90 | 1 | integrin beta chain, beta 3 |
| 38 | mice, Peptide Partial, 680 aa | 47.40 | 1 | beta 3 integrin, GPIIIA |
| 39 | Ovis aries | 45.10 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 40 | Acropora millepora | 44.70 | 1 | integrin subunit betaCn1 |
| 41 | Apis mellifera | 43.50 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 42 | Halocynthia roretzi | 42.00 | 2 | integrin beta Hr2 precursor |
| 43 | Ophlitaspongia tenuis | 41.60 | 1 | integrin subunit betaPo1 |
| 44 | Mus sp. | 40.80 | 1 | beta 7 integrin |
| 45 | Pacifastacus leniusculus | 40.40 | 1 | integrin |
| 46 | Schistosoma japonicum | 40.00 | 1 | SJCHGC06221 protein |
| 47 | Manduca sexta | 40.00 | 1 | plasmatocyte-specific integrin beta 1 |
| 48 | Suberites domuncula | 37.70 | 1 | integrin beta subunit |
| 49 | Geodia cydonium | 37.00 | 1 | integrin beta subunit |
| 50 | Capra hircus | 36.60 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 51 | Spodoptera frugiperda | 36.60 | 1 | integrin beta 1 |
| 52 | nu | 36.60 | 1 | beta |
| 53 | Bubalus bubalis | 36.60 | 1 | integrin beta-2 |
| 54 | Ovis canadensis | 36.60 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 55 | Sigmodon hispidus | 35.80 | 1 | AF445415_1 integrin beta-2 precursor |
| 56 | Drosophila pseudoobscura | 35.00 | 1 | GA14581-PA |
| 57 | Oncorhynchus mykiss | 33.10 | 1 | CD18 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRKAGL 802 ||||||||||||||||||||||||||||| #|||||||||||| Sbjct 54 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 95 |
| Homo sapiens | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRKAGL 802 ||||||||||||||||||||||||||||| #|||||||||||| Sbjct 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 |
| Mus musculus | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRKAGL 802 ||||||||||||||||||||||||||||| #|||||||||||| Sbjct 38 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 79 |
| Bos taurus | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRKAGL 802 ||||||||||||||||||||||||||||| #|||||||||||| Sbjct 760 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 801 |
| Canis familiaris | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRKAGL 802 ||||||||||||||||||||||||||||| #|||||||||||| Sbjct 783 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 824 |
| Gallus gallus | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRKAGL 802 ||||||||||||||||||||||||||||| #|||||||||||| Sbjct 17 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 58 |
| Monodelphis domestica | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRKAGL 802 ||||||||||||||||||||||||||||| #|||||||||||| Sbjct 761 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRKAGL 802 |
| Drosophila melanogaster | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||+||||+||||||| |||||| + #|||| | | Sbjct 808 RREFARFEKERMNAKWDTGENPIYKQATST#FKNPMYAGK 846 |
| Pan troglodytes | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 872 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 910 |
| Rattus norvegicus | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 799 |
| Sus scrofa | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Xenopus laevis | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Tetraodon nigroviridis | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 753 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 791 |
| synthetic construct | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Pongo pygmaeus | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Oryctolagus cuniculus | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 25 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 63 |
| Xenopus tropicalis | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Danio rerio | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||||||||||||||| ||||||| + # || | | Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VINPKYEGK 798 |
| Biomphalaria glabrata | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYG 797 |||||||||+ |||||| |||||| + #|||| || Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATST#FKNPTYG 786 |
| Felis catus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Pseudoplusia includens | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||+||||+| ||||| |||||| + #|||| | | Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYAGK 837 |
| Ictalurus punctatus | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 769 RREFAKFEKEKMNAKWDAGENPIYKSAVTT#VVNPKYEGK 807 |
| Ostrinia furnacalis | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||+||||+| ||||| |||||| + #|||| | | Sbjct 793 RREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYAGK 831 |
| Anopheles gambiae str. PEST | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||+||||+| ||||| |||||| #|||| | | Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYAGK 837 |
| Anopheles gambiae | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||+||||+| ||||| |||||| #|||| | | Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYAGK 837 |
| Aedes aegypti | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||+||||+| ||||| |||||| #|||| | | Sbjct 797 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYAGK 835 |
| Tribolium castaneum | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRKA 800 |||||||||| | |+||| |||||| + #|||| | |+ Sbjct 791 RREFAKFEKEAMMARWDTGENPIYKQATST#FKNPTYAGKS 830 |
| Podocoryne carnea | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||||||||++ | |||+ |||||| + #|+|| || | Sbjct 743 RREFAKFEKDRQNPKWDSGENPIYKKATST#FQNPMYGNK 781 |
| Crassostrea gigas | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 +|| |+|||| |||+||| |||||| + #| || | Sbjct 762 KRELARFEKEAMNARWDTGENPIYKQATST#FVNPTY 797 |
| Caenorhabditis briggsae | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRKA 800 | |+||| |++ ||||| |||||| #|||| | || Sbjct 770 RAEYAKFNNERLMAKWDTNENPIYKQATTT#FKNPVYAGKA 809 |
| Caenorhabditis elegans | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRKA 800 | |+| | |++ ||||| |||||| #|||| | || Sbjct 769 RSEYATFNNERLMAKWDTNENPIYKQATTT#FKNPVYAGKA 808 |
| Strongylocentrotus purpuratus | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||| ||||+ || |+ |||||| + #|||| | | Sbjct 768 RREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTYNIK 806 |
| Lytechinus variegatus | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |||| ||||+ || |+ |||||| + #|||| | | Sbjct 768 RREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTYNIK 806 |
| Macaca mulatta | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 |+||||||+|+ ||||| ||+|| + #| | | Sbjct 750 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITY 785 |
| Equus caballus | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 |+||||||+|+ ||||| ||+|| + #| | | Sbjct 746 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITY 781 |
| rats, Peptide Partial, 723 aa | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 |+||||||+|+ ||||| ||+|| + #| | | Sbjct 685 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITY 720 |
| Canis lupus familiaris | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 |+||||||+|+ ||||| ||+|| + #| | | Sbjct 746 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITY 781 |
| mice, Peptide Partial, 680 aa | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKN 793 |+||||||+|+ ||||| ||+|| + #| | Sbjct 646 RKEFAKFEEERARAKWDTANNPLYKEATST#FTN 678 |
| Ovis aries | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 |+| |||| |+ ||| | ||+|+ + #||| | | Sbjct 738 RKEVAKFEAERSKAKWQTGTNPLYRGSTST#FKNVTYKHK 776 |
| Acropora millepora | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 | |+ |||+|+|++|| ++||+|++ #|+|| | Sbjct 752 RIEYQKFERERMHSKWTREKNPLYQAAKTT#FENPTY 787 |
| Apis mellifera | Query 762 REFAKFEKEKMNA-KWDTQENPIYKSPINK#FKNPNY 796 ||| |||+|++ | ||+ ++||+|| + #|||| + Sbjct 778 REFKKFERERILANKWNRRDNPLYKEATST#FKNPTF 813 |
| Halocynthia roretzi | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYG 797 +||+ ||+||+ || |||+| + +|#| || +| Sbjct 796 KREYEKFKKEEDLRKWTKGENPVYVNASSK#FDNPMFG 832 |
| Ophlitaspongia tenuis | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGR 798 | |||+| |||+ |||+|+| #++|| ||+ Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATKD#YQNPLYGK 838 |
| Mus sp. | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 |||+ +||||+ | ||+||| | # || + Sbjct 754 RREYRRFEKEQQQLNWKQDNNPLYKSAITT#TVNPRF 789 |
| Pacifastacus leniusculus | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGR 798 |||+||||||+ |||+||| #|+|| + + Sbjct 762 RREYAKFEKERKLPSGKRAENPLYKSAKTT#FQNPAFAQ 799 |
| Schistosoma japonicum | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 ||| | |+++ | +|+ ||||++|| # || + Sbjct 248 RRELANFKQQGENMRWEMAENPIFESPTTN#VLNPTF 283 |
| Manduca sexta | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRKA 800 +||+||||+| + +| ||+|+ | #| || | | Sbjct 726 KREYAKFEEESRSRGFDVSLNPLYQEPEIN#FSNPVYNANA 765 |
| Suberites domuncula | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGR 798 | ||||| |+ +||+|+| #++|| ||+ Sbjct 729 EVKKFEKELAKTKYPKHQNPLYRSAAKD#YQNPIYGQ 764 |
| Geodia cydonium | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGR 798 | ||| | |+ |||+| | +# ||| ||+ Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTE#HKNPIYGQ 878 |
| Capra hircus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 ||+ +|||||+ ++|+ +||++|| # || + Sbjct 734 REYHRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 767 |
| Spodoptera frugiperda | Query 761 RREFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNYGRK 799 ++|+ ||+ | + | +| ||+|+ | #| || | + Sbjct 741 KKEYKKFQDEALAAGYDVSLNPLYQDPSIN#FSNPVYNNQ 779 |
| nu | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINK#FKNP 794 ||+||||+++ |+ ||||||+ |+ +#++ | Sbjct 755 REYAKFEEDQKNSV--RQENPIYRDPVGR#YEVP 785 |
| Bubalus bubalis | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 ||+ +|||||+ ++|+ +||++|| # || + Sbjct 733 REYHRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 766 |
| Ovis canadensis | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 ||+ +|||||+ ++|+ +||++|| # || + Sbjct 734 REYHRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 767 |
| Sigmodon hispidus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 ||+ |||||+ ++|+ +||++|| # || + Sbjct 733 REYRHFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 766 |
| Drosophila pseudoobscura | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINK#FKNP 794 ||+|+||+|+ || +||||+|+ |+ +#+ | Sbjct 728 REYARFEEEQ--AKRVSQENPLYRDPVGR#YVVP 758 |
| Oncorhynchus mykiss | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINK#FKNPNY 796 +|| +|| |+ || +||++ + # || + Sbjct 740 KEFRRFENEQKKGKWSPGDNPLFMNATTT#VANPTF 774 |
[Site 4] PIYKSPINKF791-KNPNYGRKAG
Phe791  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro782 | Ile783 | Tyr784 | Lys785 | Ser786 | Pro787 | Ile788 | Asn789 | Lys790 | Phe791 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys792 | Asn793 | Pro794 | Asn795 | Tyr796 | Gly797 | Arg798 | Lys799 | Ala800 | Gly801 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| REFAKFEKEKMNAKWDTQENPIYKSPINKFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 89.40 | 28 | S |
| 2 | Homo sapiens | 89.40 | 23 | integrin beta 1 isoform 1D precursor |
| 3 | Mus musculus | 89.40 | 14 | beta 1 integrin |
| 4 | Bos taurus | 89.40 | 13 | ITGB1 protein |
| 5 | Canis familiaris | 89.40 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 6 | Gallus gallus | 89.40 | 7 | integrin beta1D |
| 7 | Monodelphis domestica | 89.40 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 8 | Biomphalaria glabrata | 63.20 | 1 | beta integrin subunit |
| 9 | Drosophila melanogaster | 62.00 | 2 | myospheroid CG1560-PA |
| 10 | Pan troglodytes | 61.60 | 12 | PREDICTED: hypothetical protein |
| 11 | Rattus norvegicus | 61.60 | 10 | integrin beta 1 |
| 12 | Sus scrofa | 61.60 | 6 | integrin beta-1 subunit |
| 13 | Tetraodon nigroviridis | 61.60 | 5 | unnamed protein product |
| 14 | Xenopus laevis | 61.60 | 5 | integrin beta 1 |
| 15 | synthetic construct | 61.60 | 5 | hypothetical protein |
| 16 | Oryctolagus cuniculus | 61.60 | 2 | integrin beta 1 |
| 17 | Pongo pygmaeus | 61.60 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 18 | Xenopus tropicalis | 61.60 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 19 | Felis catus | 61.60 | 1 | integrin beta 1 |
| 20 | Danio rerio | 61.20 | 8 | integrin beta1 subunit-like protein 1 |
| 21 | Ostrinia furnacalis | 59.70 | 1 | integrin beta 1 precursor |
| 22 | Ictalurus punctatus | 59.70 | 1 | beta-1 integrin |
| 23 | Pseudoplusia includens | 59.70 | 1 | integrin beta 1 |
| 24 | Anopheles gambiae str. PEST | 59.30 | 2 | ENSANGP00000021373 |
| 25 | Anopheles gambiae | 59.30 | 2 | integrin beta subunit |
| 26 | Aedes aegypti | 59.30 | 1 | integrin beta subunit |
| 27 | Tribolium castaneum | 58.90 | 3 | PREDICTED: similar to CG1560-PA |
| 28 | Podocoryne carnea | 58.20 | 1 | AF308652_1 integrin beta chain |
| 29 | Crassostrea gigas | 54.30 | 1 | integrin beta cgh |
| 30 | Caenorhabditis briggsae | 53.50 | 1 | Hypothetical protein CBG03601 |
| 31 | Caenorhabditis elegans | 51.20 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Strongylocentrotus purpuratus | 50.10 | 5 | integrin beta-C subunit |
| 33 | Lytechinus variegatus | 50.10 | 1 | beta-C integrin subunit |
| 34 | Macaca mulatta | 47.00 | 12 | PREDICTED: similar to integrin beta chain, beta 3 |
| 35 | rats, Peptide Partial, 723 aa | 47.00 | 1 | beta 3 integrin, GPIIIA |
| 36 | Canis lupus familiaris | 47.00 | 1 | integrin beta chain, beta 3 |
| 37 | Equus caballus | 47.00 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 38 | mice, Peptide Partial, 680 aa | 45.40 | 1 | beta 3 integrin, GPIIIA |
| 39 | Acropora millepora | 43.90 | 1 | integrin subunit betaCn1 |
| 40 | Apis mellifera | 43.50 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 41 | Ovis aries | 43.10 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 42 | Ophlitaspongia tenuis | 41.60 | 1 | integrin subunit betaPo1 |
| 43 | Halocynthia roretzi | 41.20 | 2 | integrin beta Hr2 precursor |
| 44 | Manduca sexta | 39.30 | 1 | plasmatocyte-specific integrin beta 1 |
| 45 | Mus sp. | 38.90 | 1 | beta 7 integrin |
| 46 | Pacifastacus leniusculus | 38.50 | 1 | integrin |
| 47 | Schistosoma japonicum | 38.10 | 1 | SJCHGC06221 protein |
| 48 | Suberites domuncula | 37.70 | 1 | integrin beta subunit |
| 49 | Geodia cydonium | 37.00 | 1 | integrin beta subunit |
| 50 | nu | 36.60 | 1 | beta |
| 51 | Capra hircus | 36.60 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 52 | Bubalus bubalis | 36.60 | 1 | integrin beta-2 |
| 53 | Ovis canadensis | 36.60 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 54 | Sigmodon hispidus | 35.80 | 1 | AF445415_1 integrin beta-2 precursor |
| 55 | Spodoptera frugiperda | 35.80 | 1 | integrin beta 1 |
| 56 | Drosophila pseudoobscura | 35.00 | 1 | GA14581-PA |
| 57 | Oncorhynchus mykiss | 33.10 | 1 | CD18 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRKAGL 802 |||||||||||||||||||||||||||| |#||||||||||| Sbjct 55 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 95 |
| Homo sapiens | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRKAGL 802 |||||||||||||||||||||||||||| |#||||||||||| Sbjct 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 |
| Mus musculus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRKAGL 802 |||||||||||||||||||||||||||| |#||||||||||| Sbjct 39 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 79 |
| Bos taurus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRKAGL 802 |||||||||||||||||||||||||||| |#||||||||||| Sbjct 761 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 801 |
| Canis familiaris | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRKAGL 802 |||||||||||||||||||||||||||| |#||||||||||| Sbjct 784 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 824 |
| Gallus gallus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRKAGL 802 |||||||||||||||||||||||||||| |#||||||||||| Sbjct 18 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 58 |
| Monodelphis domestica | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRKAGL 802 |||||||||||||||||||||||||||| |#||||||||||| Sbjct 762 REFAKFEKEKMNAKWDTQENPIYKSPINNF#KNPNYGRKAGL 802 |
| Biomphalaria glabrata | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYG 797 |||||||||+ |||||| |||||| + |#||| || Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATSTF#KNPTYG 786 |
| Drosophila melanogaster | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||+||||+||||||| |||||| + |#||| | | Sbjct 809 REFARFEKERMNAKWDTGENPIYKQATSTF#KNPMYAGK 846 |
| Pan troglodytes | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 873 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 910 |
| Rattus norvegicus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 762 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 799 |
| Sus scrofa | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 798 |
| Tetraodon nigroviridis | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 754 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 791 |
| Xenopus laevis | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 798 |
| synthetic construct | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 798 |
| Oryctolagus cuniculus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 26 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 63 |
| Pongo pygmaeus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 798 |
| Xenopus tropicalis | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 798 |
| Felis catus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYEGK 798 |
| Danio rerio | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||||||||||| ||||||| + # || | | Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#INPKYEGK 798 |
| Ostrinia furnacalis | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||+||||+| ||||| |||||| + |#||| | | Sbjct 794 REFARFEKERMMAKWDTGENPIYKQATSTF#KNPTYAGK 831 |
| Ictalurus punctatus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 |||||||||||||||| ||||||| + # || | | Sbjct 770 REFAKFEKEKMNAKWDAGENPIYKSAVTTV#VNPKYEGK 807 |
| Pseudoplusia includens | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||+||||+| ||||| |||||| + |#||| | | Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATSTF#KNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||+||||+| ||||| |||||| |#||| | | Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTF#KNPTYAGK 837 |
| Anopheles gambiae | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||+||||+| ||||| |||||| |#||| | | Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTF#KNPTYAGK 837 |
| Aedes aegypti | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||+||||+| ||||| |||||| |#||| | | Sbjct 798 REFARFEKERMMAKWDTGENPIYKQATTTF#KNPTYAGK 835 |
| Tribolium castaneum | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRKA 800 ||||||||| | |+||| |||||| + |#||| | |+ Sbjct 792 REFAKFEKEAMMARWDTGENPIYKQATSTF#KNPTYAGKS 830 |
| Podocoryne carnea | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||||||||++ | |||+ |||||| + |#+|| || | Sbjct 744 REFAKFEKDRQNPKWDSGENPIYKKATSTF#QNPMYGNK 781 |
| Crassostrea gigas | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 || |+|||| |||+||| |||||| + |# || | Sbjct 763 RELARFEKEAMNARWDTGENPIYKQATSTF#VNPTY 797 |
| Caenorhabditis briggsae | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRKA 800 |+||| |++ ||||| |||||| |#||| | || Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTF#KNPVYAGKA 809 |
| Caenorhabditis elegans | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRKA 800 |+| | |++ ||||| |||||| |#||| | || Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTF#KNPVYAGKA 808 |
| Strongylocentrotus purpuratus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||| ||||+ || |+ |||||| + |#||| | | Sbjct 769 REFQNFEKERANATWEGGENPIYKPSTSVF#KNPTYNIK 806 |
| Lytechinus variegatus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 ||| ||||+ || |+ |||||| + |#||| | | Sbjct 769 REFQNFEKERANATWEGGENPIYKPSTSVF#KNPTYNIK 806 |
| Macaca mulatta | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 +||||||+|+ ||||| ||+|| + |# | | Sbjct 751 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 785 |
| rats, Peptide Partial, 723 aa | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 +||||||+|+ ||||| ||+|| + |# | | Sbjct 686 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 720 |
| Canis lupus familiaris | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 +||||||+|+ ||||| ||+|| + |# | | Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 781 |
| Equus caballus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 +||||||+|+ ||||| ||+|| + |# | | Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTF#TNITY 781 |
| mice, Peptide Partial, 680 aa | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KN 793 +||||||+|+ ||||| ||+|| + |# | Sbjct 647 KEFAKFEEERARAKWDTANNPLYKEATSTF#TN 678 |
| Acropora millepora | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 |+ |||+|+|++|| ++||+|++ |#+|| | Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTF#ENPTY 787 |
| Apis mellifera | Query 762 REFAKFEKEKMNA-KWDTQENPIYKSPINKF#KNPNY 796 ||| |||+|++ | ||+ ++||+|| + |#||| + Sbjct 778 REFKKFERERILANKWNRRDNPLYKEATSTF#KNPTF 813 |
| Ovis aries | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 +| |||| |+ ||| | ||+|+ + |#|| | | Sbjct 739 KEVAKFEAERSKAKWQTGTNPLYRGSTSTF#KNVTYKHK 776 |
| Ophlitaspongia tenuis | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGR 798 | |||+| |||+ |||+|+| +#+|| ||+ Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATKDY#QNPLYGK 838 |
| Halocynthia roretzi | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYG 797 ||+ ||+||+ || |||+| + +||# || +| Sbjct 797 REYEKFKKEEDLRKWTKGENPVYVNASSKF#DNPMFG 832 |
| Manduca sexta | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRKA 800 ||+||||+| + +| ||+|+ | |# || | | Sbjct 727 REYAKFEEESRSRGFDVSLNPLYQEPEINF#SNPVYNANA 765 |
| Mus sp. | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 ||+ +||||+ | ||+||| | # || + Sbjct 755 REYRRFEKEQQQLNWKQDNNPLYKSAITTT#VNPRF 789 |
| Pacifastacus leniusculus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGR 798 ||+||||||+ |||+||| |#+|| + + Sbjct 763 REYAKFEKERKLPSGKRAENPLYKSAKTTF#QNPAFAQ 799 |
| Schistosoma japonicum | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 || | |+++ | +|+ ||||++|| # || + Sbjct 249 RELANFKQQGENMRWEMAENPIFESPTTNV#LNPTF 283 |
| Suberites domuncula | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGR 798 | ||||| |+ +||+|+| +#+|| ||+ Sbjct 729 EVKKFEKELAKTKYPKHQNPLYRSAAKDY#QNPIYGQ 764 |
| Geodia cydonium | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGR 798 | ||| | |+ |||+| | + #||| ||+ Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTEH#KNPIYGQ 878 |
| nu | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNP 794 ||+||||+++ |+ ||||||+ |+ ++#+ | Sbjct 755 REYAKFEEDQKNSV--RQENPIYRDPVGRY#EVP 785 |
| Capra hircus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 ||+ +|||||+ ++|+ +||++|| # || + Sbjct 734 REYHRFEKEKLKSQWN-NDNPLFKSATTTV#MNPKF 767 |
| Bubalus bubalis | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 ||+ +|||||+ ++|+ +||++|| # || + Sbjct 733 REYHRFEKEKLKSQWN-NDNPLFKSATTTV#MNPKF 766 |
| Ovis canadensis | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 ||+ +|||||+ ++|+ +||++|| # || + Sbjct 734 REYHRFEKEKLKSQWN-NDNPLFKSATTTV#MNPKF 767 |
| Sigmodon hispidus | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 ||+ |||||+ ++|+ +||++|| # || + Sbjct 733 REYRHFEKEKLKSQWN-NDNPLFKSATTTV#MNPKF 766 |
| Spodoptera frugiperda | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNYGRK 799 +|+ ||+ | + | +| ||+|+ | |# || | + Sbjct 742 KEYKKFQDEALAAGYDVSLNPLYQDPSINF#SNPVYNNQ 779 |
| Drosophila pseudoobscura | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNP 794 ||+|+||+|+ || +||||+|+ |+ ++# | Sbjct 728 REYARFEEEQ--AKRVSQENPLYRDPVGRY#VVP 758 |
| Oncorhynchus mykiss | Query 762 REFAKFEKEKMNAKWDTQENPIYKSPINKF#KNPNY 796 +|| +|| |+ || +||++ + # || + Sbjct 740 KEFRRFENEQKKGKWSPGDNPLFMNATTTV#ANPTF 774 |
[Site 5] IYKSPINKFK792-NPNYGRKAGL
Lys792  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile783 | Tyr784 | Lys785 | Ser786 | Pro787 | Ile788 | Asn789 | Lys790 | Phe791 | Lys792 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn793 | Pro794 | Asn795 | Tyr796 | Gly797 | Arg798 | Lys799 | Ala800 | Gly801 | Leu802 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EFAKFEKEKMNAKWDTQENPIYKSPINKFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 87.40 | 28 | S |
| 2 | Homo sapiens | 87.40 | 23 | integrin beta 1 isoform 1D precursor |
| 3 | Mus musculus | 87.40 | 14 | beta 1 integrin |
| 4 | Bos taurus | 87.40 | 13 | ITGB1 protein |
| 5 | Canis familiaris | 87.40 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 6 | Gallus gallus | 87.40 | 6 | integrin beta1D |
| 7 | Monodelphis domestica | 87.40 | 5 | PREDICTED: similar to integrin beta-1 subunit isof |
| 8 | Biomphalaria glabrata | 61.20 | 1 | beta integrin subunit |
| 9 | Drosophila melanogaster | 60.10 | 2 | myospheroid CG1560-PA |
| 10 | Pan troglodytes | 59.70 | 12 | PREDICTED: hypothetical protein |
| 11 | Rattus norvegicus | 59.70 | 10 | integrin beta 1 |
| 12 | Sus scrofa | 59.70 | 6 | integrin beta-1 subunit |
| 13 | Tetraodon nigroviridis | 59.70 | 5 | unnamed protein product |
| 14 | Xenopus laevis | 59.70 | 5 | integrin beta 1 |
| 15 | synthetic construct | 59.70 | 5 | hypothetical protein |
| 16 | Oryctolagus cuniculus | 59.70 | 2 | integrin beta 1 |
| 17 | Pongo pygmaeus | 59.70 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 18 | Xenopus tropicalis | 59.70 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 19 | Felis catus | 59.70 | 1 | integrin beta 1 |
| 20 | Danio rerio | 59.30 | 8 | integrin beta1 subunit-like protein 1 |
| 21 | Ostrinia furnacalis | 57.80 | 1 | integrin beta 1 precursor |
| 22 | Ictalurus punctatus | 57.80 | 1 | beta-1 integrin |
| 23 | Pseudoplusia includens | 57.80 | 1 | integrin beta 1 |
| 24 | Anopheles gambiae str. PEST | 57.40 | 2 | ENSANGP00000021373 |
| 25 | Anopheles gambiae | 57.40 | 2 | integrin beta subunit |
| 26 | Aedes aegypti | 57.40 | 1 | integrin beta subunit |
| 27 | Tribolium castaneum | 57.00 | 2 | PREDICTED: similar to CG1560-PA |
| 28 | Podocoryne carnea | 56.20 | 1 | AF308652_1 integrin beta chain |
| 29 | Caenorhabditis briggsae | 53.50 | 1 | Hypothetical protein CBG03601 |
| 30 | Crassostrea gigas | 52.40 | 1 | integrin beta cgh |
| 31 | Caenorhabditis elegans | 51.20 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Strongylocentrotus purpuratus | 48.10 | 5 | integrin beta-C subunit |
| 33 | Lytechinus variegatus | 48.10 | 1 | beta-C integrin subunit |
| 34 | Macaca mulatta | 46.20 | 12 | PREDICTED: similar to integrin beta chain, beta 3 |
| 35 | rats, Peptide Partial, 723 aa | 46.20 | 1 | beta 3 integrin, GPIIIA |
| 36 | Canis lupus familiaris | 46.20 | 1 | integrin beta chain, beta 3 |
| 37 | Equus caballus | 46.20 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 38 | mice, Peptide Partial, 680 aa | 44.70 | 1 | beta 3 integrin, GPIIIA |
| 39 | Acropora millepora | 43.90 | 1 | integrin subunit betaCn1 |
| 40 | Ovis aries | 42.40 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 41 | Apis mellifera | 41.60 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 42 | Ophlitaspongia tenuis | 41.60 | 1 | integrin subunit betaPo1 |
| 43 | Halocynthia roretzi | 39.30 | 2 | integrin beta Hr2 precursor |
| 44 | Suberites domuncula | 37.70 | 1 | integrin beta subunit |
| 45 | Manduca sexta | 37.40 | 1 | plasmatocyte-specific integrin beta 1 |
| 46 | Mus sp. | 37.00 | 1 | beta 7 integrin |
| 47 | Geodia cydonium | 37.00 | 1 | integrin beta subunit |
| 48 | Pacifastacus leniusculus | 36.60 | 1 | integrin |
| 49 | Schistosoma japonicum | 36.20 | 1 | SJCHGC06221 protein |
| 50 | Spodoptera frugiperda | 35.00 | 1 | integrin beta 1 |
| 51 | Ovis canadensis | 34.70 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 52 | Capra hircus | 34.70 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 53 | Bubalus bubalis | 34.70 | 1 | integrin beta-2 |
| 54 | nu | 34.70 | 1 | beta |
| 55 | Sigmodon hispidus | 33.90 | 1 | AF445415_1 integrin beta-2 precursor |
| 56 | Drosophila pseudoobscura | 33.10 | 1 | GA14581-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRKAGL 802 ||||||||||||||||||||||||||| ||#|||||||||| Sbjct 56 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 95 |
| Homo sapiens | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRKAGL 802 ||||||||||||||||||||||||||| ||#|||||||||| Sbjct 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 |
| Mus musculus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRKAGL 802 ||||||||||||||||||||||||||| ||#|||||||||| Sbjct 40 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 79 |
| Bos taurus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRKAGL 802 ||||||||||||||||||||||||||| ||#|||||||||| Sbjct 762 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 801 |
| Canis familiaris | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRKAGL 802 ||||||||||||||||||||||||||| ||#|||||||||| Sbjct 785 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 824 |
| Gallus gallus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRKAGL 802 ||||||||||||||||||||||||||| ||#|||||||||| Sbjct 19 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 58 |
| Monodelphis domestica | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRKAGL 802 ||||||||||||||||||||||||||| ||#|||||||||| Sbjct 763 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRKAGL 802 |
| Biomphalaria glabrata | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYG 797 ||||||||+ |||||| |||||| + ||#|| || Sbjct 752 EFAKFEKERQNAKWDTGENPIYKQATSTFK#NPTYG 786 |
| Drosophila melanogaster | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||+||||+||||||| |||||| + ||#|| | | Sbjct 810 EFARFEKERMNAKWDTGENPIYKQATSTFK#NPMYAGK 846 |
| Pan troglodytes | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 874 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 910 |
| Rattus norvegicus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 763 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 799 |
| Sus scrofa | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Tetraodon nigroviridis | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 755 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 791 |
| Xenopus laevis | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| synthetic construct | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Oryctolagus cuniculus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 27 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 63 |
| Pongo pygmaeus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Xenopus tropicalis | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Felis catus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Danio rerio | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||||||||||| ||||||| + #|| | | Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVI#NPKYEGK 798 |
| Ostrinia furnacalis | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||+||||+| ||||| |||||| + ||#|| | | Sbjct 795 EFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYAGK 831 |
| Ictalurus punctatus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 ||||||||||||||| ||||||| + #|| | | Sbjct 771 EFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKYEGK 807 |
| Pseudoplusia includens | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||+||||+| ||||| |||||| + ||#|| | | Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||+||||+| ||||| |||||| ||#|| | | Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYAGK 837 |
| Anopheles gambiae | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||+||||+| ||||| |||||| ||#|| | | Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYAGK 837 |
| Aedes aegypti | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||+||||+| ||||| |||||| ||#|| | | Sbjct 799 EFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYAGK 835 |
| Tribolium castaneum | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRKA 800 |||||||| | |+||| |||||| + ||#|| | |+ Sbjct 793 EFAKFEKEAMMARWDTGENPIYKQATSTFK#NPTYAGKS 830 |
| Podocoryne carnea | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |||||||++ | |||+ |||||| + |+#|| || | Sbjct 745 EFAKFEKDRQNPKWDSGENPIYKKATSTFQ#NPMYGNK 781 |
| Caenorhabditis briggsae | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRKA 800 |+||| |++ ||||| |||||| ||#|| | || Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTFK#NPVYAGKA 809 |
| Crassostrea gigas | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 | |+|||| |||+||| |||||| + | #|| | Sbjct 764 ELARFEKEAMNARWDTGENPIYKQATSTFV#NPTY 797 |
| Caenorhabditis elegans | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRKA 800 |+| | |++ ||||| |||||| ||#|| | || Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTFK#NPVYAGKA 808 |
| Strongylocentrotus purpuratus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 || ||||+ || |+ |||||| + ||#|| | | Sbjct 770 EFQNFEKERANATWEGGENPIYKPSTSVFK#NPTYNIK 806 |
| Lytechinus variegatus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 || ||||+ || |+ |||||| + ||#|| | | Sbjct 770 EFQNFEKERANATWEGGENPIYKPSTSVFK#NPTYNIK 806 |
| Macaca mulatta | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 ||||||+|+ ||||| ||+|| + | #| | Sbjct 752 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITY 785 |
| rats, Peptide Partial, 723 aa | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 ||||||+|+ ||||| ||+|| + | #| | Sbjct 687 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITY 720 |
| Canis lupus familiaris | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 ||||||+|+ ||||| ||+|| + | #| | Sbjct 748 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITY 781 |
| Equus caballus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 ||||||+|+ ||||| ||+|| + | #| | Sbjct 748 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITY 781 |
| mice, Peptide Partial, 680 aa | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#N 793 ||||||+|+ ||||| ||+|| + | #| Sbjct 648 EFAKFEEERARAKWDTANNPLYKEATSTFT#N 678 |
| Acropora millepora | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 |+ |||+|+|++|| ++||+|++ |+#|| | Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTFE#NPTY 787 |
| Ovis aries | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 | |||| |+ ||| | ||+|+ + ||#| | | Sbjct 740 EVAKFEAERSKAKWQTGTNPLYRGSTSTFK#NVTYKHK 776 |
| Apis mellifera | Query 763 EFAKFEKEKMNA-KWDTQENPIYKSPINKFK#NPNY 796 || |||+|++ | ||+ ++||+|| + ||#|| + Sbjct 779 EFKKFERERILANKWNRRDNPLYKEATSTFK#NPTF 813 |
| Ophlitaspongia tenuis | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGR 798 | |||+| |||+ |||+|+| ++#|| ||+ Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATKDYQ#NPLYGK 838 |
| Halocynthia roretzi | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYG 797 |+ ||+||+ || |||+| + +|| #|| +| Sbjct 798 EYEKFKKEEDLRKWTKGENPVYVNASSKFD#NPMFG 832 |
| Suberites domuncula | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGR 798 | ||||| |+ +||+|+| ++#|| ||+ Sbjct 729 EVKKFEKELAKTKYPKHQNPLYRSAAKDYQ#NPIYGQ 764 |
| Manduca sexta | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRKA 800 |+||||+| + +| ||+|+ | | #|| | | Sbjct 728 EYAKFEEESRSRGFDVSLNPLYQEPEINFS#NPVYNANA 765 |
| Mus sp. | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 |+ +||||+ | ||+||| | #|| + Sbjct 756 EYRRFEKEQQQLNWKQDNNPLYKSAITTTV#NPRF 789 |
| Geodia cydonium | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGR 798 | ||| | |+ |||+| | + |#|| ||+ Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTEHK#NPIYGQ 878 |
| Pacifastacus leniusculus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGR 798 |+||||||+ |||+||| |+#|| + + Sbjct 764 EYAKFEKERKLPSGKRAENPLYKSAKTTFQ#NPAFAQ 799 |
| Schistosoma japonicum | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 | | |+++ | +|+ ||||++|| #|| + Sbjct 250 ELANFKQQGENMRWEMAENPIFESPTTNVL#NPTF 283 |
| Spodoptera frugiperda | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNYGRK 799 |+ ||+ | + | +| ||+|+ | | #|| | + Sbjct 743 EYKKFQDEALAAGYDVSLNPLYQDPSINFS#NPVYNNQ 779 |
| Ovis canadensis | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 |+ +|||||+ ++|+ +||++|| #|| + Sbjct 735 EYHRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 767 |
| Capra hircus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 |+ +|||||+ ++|+ +||++|| #|| + Sbjct 735 EYHRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 767 |
| Bubalus bubalis | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 |+ +|||||+ ++|+ +||++|| #|| + Sbjct 734 EYHRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 766 |
| nu | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NP 794 |+||||+++ |+ ||||||+ |+ +++# | Sbjct 756 EYAKFEEDQKNSV--RQENPIYRDPVGRYE#VP 785 |
| Sigmodon hispidus | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NPNY 796 |+ |||||+ ++|+ +||++|| #|| + Sbjct 734 EYRHFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 766 |
| Drosophila pseudoobscura | Query 763 EFAKFEKEKMNAKWDTQENPIYKSPINKFK#NP 794 |+|+||+|+ || +||||+|+ |+ ++ # | Sbjct 729 EYARFEEEQ--AKRVSQENPLYRDPVGRYV#VP 758 |
[Site 6] KWDTQENPIY784-KSPINKFKNP
Tyr784  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys775 | Trp776 | Asp777 | Thr778 | Gln779 | Glu780 | Asn781 | Pro782 | Ile783 | Tyr784 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys785 | Ser786 | Pro787 | Ile788 | Asn789 | Lys790 | Phe791 | Lys792 | Asn793 | Pro794 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LMIIHDRREFAKFEKEKMNAKWDTQENPIYKSPINKFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 103.00 | 32 | S |
| 2 | Homo sapiens | 103.00 | 25 | integrin beta 1 isoform 1D precursor |
| 3 | Mus musculus | 103.00 | 16 | beta 1 integrin |
| 4 | Bos taurus | 103.00 | 13 | ITGB1 protein |
| 5 | Canis familiaris | 103.00 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 6 | Gallus gallus | 103.00 | 7 | integrin beta1D |
| 7 | Monodelphis domestica | 103.00 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 8 | Pan troglodytes | 75.50 | 14 | PREDICTED: hypothetical protein |
| 9 | Rattus norvegicus | 75.50 | 10 | integrin beta 1 |
| 10 | Sus scrofa | 75.50 | 6 | integrin beta-1 subunit |
| 11 | Xenopus laevis | 75.50 | 6 | integrin beta-1 subunit |
| 12 | Tetraodon nigroviridis | 75.50 | 5 | unnamed protein product |
| 13 | synthetic construct | 75.50 | 5 | hypothetical protein |
| 14 | Pongo pygmaeus | 75.50 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Oryctolagus cuniculus | 75.50 | 2 | integrin beta 1 |
| 16 | Xenopus tropicalis | 75.50 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 17 | Danio rerio | 75.10 | 10 | integrin beta1 subunit-like protein 1 |
| 18 | Ictalurus punctatus | 73.60 | 2 | beta-1 integrin |
| 19 | Felis catus | 73.20 | 1 | integrin beta 1 |
| 20 | Drosophila melanogaster | 71.60 | 2 | myospheroid CG1560-PA |
| 21 | Biomphalaria glabrata | 70.90 | 1 | beta integrin subunit |
| 22 | Aedes aegypti | 68.90 | 1 | integrin beta subunit |
| 23 | Anopheles gambiae str. PEST | 68.60 | 2 | ENSANGP00000021373 |
| 24 | Anopheles gambiae | 68.60 | 2 | integrin beta subunit |
| 25 | Ostrinia furnacalis | 68.60 | 1 | integrin beta 1 precursor |
| 26 | Tribolium castaneum | 67.80 | 3 | PREDICTED: similar to CG1560-PA |
| 27 | Pseudoplusia includens | 67.00 | 1 | integrin beta 1 |
| 28 | Podocoryne carnea | 64.70 | 1 | AF308652_1 integrin beta chain |
| 29 | Caenorhabditis briggsae | 63.50 | 1 | Hypothetical protein CBG03601 |
| 30 | Caenorhabditis elegans | 61.20 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 31 | Strongylocentrotus purpuratus | 59.70 | 5 | integrin beta-C subunit |
| 32 | Lytechinus variegatus | 59.70 | 1 | beta-C integrin subunit |
| 33 | Crassostrea gigas | 58.50 | 1 | integrin beta cgh |
| 34 | Macaca mulatta | 57.40 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 35 | rats, Peptide Partial, 723 aa | 57.40 | 1 | beta 3 integrin, GPIIIA |
| 36 | Equus caballus | 57.40 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 37 | Canis lupus familiaris | 57.40 | 1 | integrin beta chain, beta 3 |
| 38 | mice, Peptide Partial, 680 aa | 55.80 | 1 | beta 3 integrin, GPIIIA |
| 39 | Ovis aries | 51.60 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 40 | Apis mellifera | 48.90 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 41 | Pacifastacus leniusculus | 47.40 | 1 | integrin |
| 42 | Acropora millepora | 47.40 | 1 | integrin subunit betaCn1 |
| 43 | Manduca sexta | 47.00 | 1 | plasmatocyte-specific integrin beta 1 |
| 44 | Halocynthia roretzi | 45.40 | 2 | integrin beta Hr2 precursor |
| 45 | Mus sp. | 45.40 | 1 | beta 7 integrin |
| 46 | Ophlitaspongia tenuis | 44.70 | 1 | integrin subunit betaPo1 |
| 47 | Schistosoma japonicum | 43.90 | 1 | SJCHGC06221 protein |
| 48 | Spodoptera frugiperda | 43.10 | 1 | integrin beta 1 |
| 49 | Geodia cydonium | 40.80 | 1 | integrin beta subunit |
| 50 | Suberites domuncula | 38.90 | 1 | integrin beta subunit |
| 51 | Capra hircus | 38.50 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 52 | nu | 38.50 | 1 | beta |
| 53 | Bubalus bubalis | 38.50 | 1 | integrin beta-2 |
| 54 | Ovis canadensis | 38.50 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 55 | Sigmodon hispidus | 37.40 | 1 | AF445415_1 integrin beta-2 precursor |
| 56 | Drosophila pseudoobscura | 37.00 | 1 | GA14581-PA |
| 57 | Oncorhynchus mykiss | 35.80 | 1 | CD18 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||| |||||||||||| Sbjct 48 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 95 |
| Homo sapiens | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||| |||||||||||| Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 |
| Mus musculus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||| |||||||||||| Sbjct 32 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 79 |
| Bos taurus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||| |||||||||||| Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 801 |
| Canis familiaris | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||| |||||||||||| Sbjct 777 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 824 |
| Gallus gallus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||| |||||||||||| Sbjct 11 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 58 |
| Monodelphis domestica | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||| |||||||||||| Sbjct 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINNFKNPNYGRKAGL 802 |
| Pan troglodytes | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||||||||||||||||||||| |||||#|| + || | | Sbjct 866 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 910 |
| Rattus norvegicus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||||||||||||||||||||| |||||#|| + || | | Sbjct 755 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 799 |
| Sus scrofa | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||||||||||||||||||||| |||||#|| + || | | Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 798 |
| Xenopus laevis | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||||||||||||||||||||| |||||#|| + || | | Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 798 |
| Tetraodon nigroviridis | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||||||||||||||||||||| |||||#|| + || | | Sbjct 747 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 791 |
| synthetic construct | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||||||||||||||||||||| |||||#|| + || | | Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 798 |
| Pongo pygmaeus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||||||||||||||||||||| |||||#|| + || | | Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 798 |
| Oryctolagus cuniculus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||||||||||||||||||||| |||||#|| + || | | Sbjct 19 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 63 |
| Xenopus tropicalis | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||||||||||||||||||||| |||||#|| + || | | Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 798 |
| Danio rerio | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||||||||||||||||||||| |||||#|| + || | | Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVINPKYEGK 798 |
| Ictalurus punctatus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 ||||||||||||||||||||||| |||||#|| + || | | Sbjct 763 LMIIHDRREFAKFEKEKMNAKWDAGENPIY#KSAVTTVVNPKYEGK 807 |
| Felis catus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||| ||||||||||||||||| |||||#|| + || | | Sbjct 754 LMIIHDTREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEGK 798 |
| Drosophila melanogaster | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 | ||||||||+||||+||||||| |||||#| + |||| | | Sbjct 802 LTTIHDRREFARFEKERMNAKWDTGENPIY#KQATSTFKNPMYAGK 846 |
| Biomphalaria glabrata | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYG 797 | ||| |||||||||+ |||||| |||||#| + |||| || Sbjct 744 LTFIHDTREFAKFEKERQNAKWDTGENPIY#KQATSTFKNPTYG 786 |
| Aedes aegypti | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 | ||||||||+||||+| ||||| |||||#| |||| | | Sbjct 791 LTTIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAGK 835 |
| Anopheles gambiae str. PEST | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 | ||||||||+||||+| ||||| |||||#| |||| | | Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAGK 837 |
| Anopheles gambiae | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 | ||||||||+||||+| ||||| |||||#| |||| | | Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAGK 837 |
| Ostrinia furnacalis | Query 758 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 ||||||||+||||+| ||||| |||||#| + |||| | | Sbjct 790 IHDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYAGK 831 |
| Tribolium castaneum | Query 758 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRKA 800 ||||||||||||| | |+||| |||||#| + |||| | |+ Sbjct 788 IHDRREFAKFEKEAMMARWDTGENPIY#KQATSTFKNPTYAGKS 830 |
| Pseudoplusia includens | Query 759 HDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |||||||+||||+| ||||| |||||#| + |||| | | Sbjct 797 HDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYAGK 837 |
| Podocoryne carnea | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 | | ||||||||||++ | |||+ |||||#| + |+|| || | Sbjct 737 LATIQDRREFAKFEKDRQNPKWDSGENPIY#KKATSTFQNPMYGNK 781 |
| Caenorhabditis briggsae | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRKA 800 | ++||| |+||| |++ ||||| |||||#| |||| | || Sbjct 764 LTVLHDRAEYAKFNNERLMAKWDTNENPIY#KQATTTFKNPVYAGKA 809 |
| Caenorhabditis elegans | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRKA 800 | ++||| |+| | |++ ||||| |||||#| |||| | || Sbjct 763 LTVLHDRSEYATFNNERLMAKWDTNENPIY#KQATTTFKNPVYAGKA 808 |
| Strongylocentrotus purpuratus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 | ||||||| ||||+ || |+ |||||#| + |||| | | Sbjct 762 LTYIHDRREFQNFEKERANATWEGGENPIY#KPSTSVFKNPTYNIK 806 |
| Lytechinus variegatus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 | ||||||| ||||+ || |+ |||||#| + |||| | | Sbjct 762 LTYIHDRREFQNFEKERANATWEGGENPIY#KPSTSVFKNPTYNIK 806 |
| Crassostrea gigas | Query 758 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 | |+|| |+|||| |||+||| |||||#| + | || | Sbjct 759 ISDKRELARFEKEAMNARWDTGENPIY#KQATSTFVNPTY 797 |
| Macaca mulatta | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 |+ ||||+||||||+|+ ||||| ||+|#| + | | | Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 785 |
| rats, Peptide Partial, 723 aa | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 |+ ||||+||||||+|+ ||||| ||+|#| + | | | Sbjct 679 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 720 |
| Equus caballus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 |+ ||||+||||||+|+ ||||| ||+|#| + | | | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 781 |
| Canis lupus familiaris | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 |+ ||||+||||||+|+ ||||| ||+|#| + | | | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITY 781 |
| mice, Peptide Partial, 680 aa | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKN 793 |+ ||||+||||||+|+ ||||| ||+|#| + | | Sbjct 640 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 678 |
| Ovis aries | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |+ |||+| |||| |+ ||| | ||+|#+ + ||| | | Sbjct 732 LVSFHDRKEVAKFEAERSKAKWQTGTNPLY#RGSTSTFKNVTYKHK 776 |
| Apis mellifera | Query 755 LMIIHDRREFAKFEKEKMNA-KWDTQENPIY#KSPINKFKNPNY 796 | +| | ||| |||+|++ | ||+ ++||+|#| + |||| + Sbjct 771 LTMIRDNREFKKFERERILANKWNRRDNPLY#KEATSTFKNPTF 813 |
| Pacifastacus leniusculus | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGR 798 | +|||||+||||||+ |||+|#|| |+|| + + Sbjct 756 LTTLHDRREYAKFEKERKLPSGKRAENPLY#KSAKTTFQNPAFAQ 799 |
| Acropora millepora | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 | + || |+ |||+|+|++|| ++||+|#++ |+|| | Sbjct 746 LFTMVDRIEYQKFERERMHSKWTREKNPLY#QAAKTTFENPTY 787 |
| Manduca sexta | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRKA 800 |+ +||+||+||||+| + +| ||+|#+ | | || | | Sbjct 720 LVDLHDKREYAKFEEESRSRGFDVSLNPLY#QEPEINFSNPVYNANA 765 |
| Halocynthia roretzi | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYG 797 + + |+||+ ||+||+ || |||+|# + +|| || +| Sbjct 790 IQTLRDKREYEKFKKEEDLRKWTKGENPVY#VNASSKFDNPMFG 832 |
| Mus sp. | Query 758 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 |+||||+ +||||+ | ||+|#|| | || + Sbjct 751 IYDRREYRRFEKEQQQLNWKQDNNPLY#KSAITTTVNPRF 789 |
| Ophlitaspongia tenuis | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGR 798 |+++ | | |||+| |||+ |||+|#+| ++|| ||+ Sbjct 795 LLLLWDVVEVRKFEREIKNAKYTKNENPLY#RSATKDYQNPLYGK 838 |
| Schistosoma japonicum | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 ++ | |||| | |+++ | +|+ ||||+#+|| || + Sbjct 242 VITIDDRRELANFKQQGENMRWEMAENPIF#ESPTTNVLNPTF 283 |
| Spodoptera frugiperda | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGRK 799 |+ +||++|+ ||+ | + | +| ||+|#+ | | || | + Sbjct 735 LVDMHDKKEYKKFQDEALAAGYDVSLNPLY#QDPSINFSNPVYNNQ 779 |
| Geodia cydonium | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGR 798 |+++ | | ||| | |+ |||+|# | + ||| ||+ Sbjct 835 LLMVLDYTELKKFENELAKVKYTKNENPLY#HSATTEHKNPIYGQ 878 |
| Suberites domuncula | Query 756 MIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNYGR 798 + | | | ||||| |+ +||+|#+| ++|| ||+ Sbjct 722 LFIIDVIEVKKFEKELAKTKYPKHQNPLY#RSAAKDYQNPIYGQ 764 |
| Capra hircus | Query 758 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 + | ||+ +|||||+ ++|+ +||++#|| || + Sbjct 730 LSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 767 |
| nu | Query 760 DRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNP 794 | ||+||||+++ |+ ||||||#+ |+ +++ | Sbjct 753 DAREYAKFEEDQKNSV--RQENPIY#RDPVGRYEVP 785 |
| Bubalus bubalis | Query 758 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 + | ||+ +|||||+ ++|+ +||++#|| || + Sbjct 729 LSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 766 |
| Ovis canadensis | Query 758 IHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 + | ||+ +|||||+ ++|+ +||++#|| || + Sbjct 730 LSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 767 |
| Sigmodon hispidus | Query 760 DRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 | ||+ |||||+ ++|+ +||++#|| || + Sbjct 731 DLREYRHFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 766 |
| Drosophila pseudoobscura | Query 760 DRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNP 794 | ||+|+||+|+ || +||||+|#+ |+ ++ | Sbjct 726 DAREYARFEEEQ--AKRVSQENPLY#RDPVGRYVVP 758 |
| Oncorhynchus mykiss | Query 755 LMIIHDRREFAKFEKEKMNAKWDTQENPIY#KSPINKFKNPNY 796 ++ + | +|| +|| |+ || +||++# + || + Sbjct 733 ILYMRDVKEFRRFENEQKKGKWSPGDNPLF#MNATTTVANPTF 774 |
[Site 7] KEKMNAKWDT778-QENPIYKSPI
Thr778  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys769 | Glu770 | Lys771 | Met772 | Asn773 | Ala774 | Lys775 | Trp776 | Asp777 | Thr778 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln779 | Glu780 | Asn781 | Pro782 | Ile783 | Tyr784 | Lys785 | Ser786 | Pro787 | Ile788 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LLIWKLLMIIHDRREFAKFEKEKMNAKWDTQENPIYKSPINKFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 115.00 | 32 | S |
| 2 | Homo sapiens | 115.00 | 25 | integrin beta 1 isoform 1D precursor |
| 3 | Mus musculus | 115.00 | 18 | beta 1 integrin |
| 4 | Bos taurus | 115.00 | 13 | ITGB1 protein |
| 5 | Canis familiaris | 115.00 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 6 | Gallus gallus | 115.00 | 7 | integrin beta1D |
| 7 | Monodelphis domestica | 115.00 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 8 | Pan troglodytes | 87.80 | 14 | PREDICTED: hypothetical protein |
| 9 | Rattus norvegicus | 87.80 | 10 | integrin beta 1 |
| 10 | Tetraodon nigroviridis | 87.80 | 8 | unnamed protein product |
| 11 | Sus scrofa | 87.80 | 6 | integrin beta-1 subunit |
| 12 | Xenopus laevis | 87.80 | 6 | integrin beta 1 |
| 13 | synthetic construct | 87.80 | 5 | hypothetical protein |
| 14 | Pongo pygmaeus | 87.80 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Oryctolagus cuniculus | 87.80 | 2 | integrin beta 1 |
| 16 | Xenopus tropicalis | 87.80 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 17 | Danio rerio | 87.40 | 13 | integrin beta1 subunit-like protein 1 |
| 18 | Felis catus | 85.50 | 1 | integrin beta 1 |
| 19 | Ictalurus punctatus | 84.30 | 2 | beta-1 integrin |
| 20 | Drosophila melanogaster | 83.20 | 2 | myospheroid CG1560-PA |
| 21 | Biomphalaria glabrata | 82.40 | 1 | beta integrin subunit |
| 22 | Aedes aegypti | 80.50 | 2 | integrin beta subunit |
| 23 | Anopheles gambiae str. PEST | 79.00 | 2 | ENSANGP00000021373 |
| 24 | Anopheles gambiae | 79.00 | 2 | integrin beta subunit |
| 25 | Tribolium castaneum | 78.60 | 3 | PREDICTED: similar to CG1560-PA |
| 26 | Ostrinia furnacalis | 78.20 | 1 | integrin beta 1 precursor |
| 27 | Podocoryne carnea | 77.00 | 1 | AF308652_1 integrin beta chain |
| 28 | Pseudoplusia includens | 75.10 | 1 | integrin beta 1 |
| 29 | Caenorhabditis briggsae | 75.10 | 1 | Hypothetical protein CBG03601 |
| 30 | Caenorhabditis elegans | 72.80 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 31 | Strongylocentrotus purpuratus | 70.90 | 5 | integrin beta-C subunit |
| 32 | Lytechinus variegatus | 70.90 | 1 | beta-C integrin subunit |
| 33 | Crassostrea gigas | 70.10 | 1 | integrin beta cgh |
| 34 | Macaca mulatta | 69.70 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 35 | rats, Peptide Partial, 723 aa | 69.70 | 1 | beta 3 integrin, GPIIIA |
| 36 | Equus caballus | 69.70 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 37 | Canis lupus familiaris | 69.70 | 1 | integrin beta chain, beta 3 |
| 38 | mice, Peptide Partial, 680 aa | 68.20 | 1 | beta 3 integrin, GPIIIA |
| 39 | Ovis aries | 62.00 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 40 | Pacifastacus leniusculus | 59.70 | 1 | integrin |
| 41 | Apis mellifera | 58.50 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 42 | Halocynthia roretzi | 55.80 | 2 | integrin beta Hr2 precursor |
| 43 | Manduca sexta | 55.80 | 1 | plasmatocyte-specific integrin beta 1 |
| 44 | Schistosoma japonicum | 52.80 | 1 | SJCHGC06221 protein |
| 45 | Spodoptera frugiperda | 52.80 | 1 | integrin beta 1 |
| 46 | Acropora millepora | 50.10 | 1 | integrin subunit betaCn1 |
| 47 | Mus sp. | 48.50 | 1 | beta 7 integrin |
| 48 | Ophlitaspongia tenuis | 48.10 | 1 | integrin subunit betaPo1 |
| 49 | Capra hircus | 47.80 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 50 | Bubalus bubalis | 47.80 | 1 | integrin beta-2 |
| 51 | Ovis canadensis | 47.00 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 52 | Sigmodon hispidus | 46.60 | 1 | AF445415_1 integrin beta-2 precursor |
| 53 | Geodia cydonium | 45.80 | 1 | integrin beta subunit |
| 54 | Suberites domuncula | 45.40 | 1 | integrin beta subunit |
| 55 | Oncorhynchus mykiss | 40.80 | 1 | CD18 protein |
| 56 | nu | 39.70 | 1 | beta |
| 57 | Drosophila pseudoobscura | 37.70 | 1 | GA14581-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||| Sbjct 42 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 95 |
| Homo sapiens | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||| Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 |
| Mus musculus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||| Sbjct 26 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 79 |
| Bos taurus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||| Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 801 |
| Canis familiaris | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||| Sbjct 771 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 824 |
| Gallus gallus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||| Sbjct 5 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 58 |
| Monodelphis domestica | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||| Sbjct 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRKAGL 802 |
| Pan troglodytes | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 860 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 910 |
| Rattus norvegicus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 799 |
| Tetraodon nigroviridis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 741 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 791 |
| Sus scrofa | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Xenopus laevis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| synthetic construct | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Pongo pygmaeus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Oryctolagus cuniculus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 13 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 63 |
| Xenopus tropicalis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Danio rerio | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVINPKYEGK 798 |
| Felis catus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 |||||||||||| |||||||||||||||||# ||||||| + || | | Sbjct 748 LLIWKLLMIIHDTREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Ictalurus punctatus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 | ||||||||||||||||||||||||||| # ||||||| + || | | Sbjct 757 LFIWKLLMIIHDRREFAKFEKEKMNAKWDA#GENPIYKSAVTTVVNPKYEGK 807 |
| Drosophila melanogaster | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||+|||| ||||||||+||||+|||||||# |||||| + |||| | | Sbjct 796 LLLWKLLTTIHDRREFARFEKERMNAKWDT#GENPIYKQATSTFKNPMYAGK 846 |
| Biomphalaria glabrata | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYG 797 ||+|||| ||| |||||||||+ ||||||# |||||| + |||| || Sbjct 738 LLLWKLLTFIHDTREFAKFEKERQNAKWDT#GENPIYKQATSTFKNPTYG 786 |
| Aedes aegypti | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||+|||| ||||||||+||||+| |||||# |||||| |||| | | Sbjct 785 LLLWKLLTTIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAGK 835 |
| Anopheles gambiae str. PEST | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||+||+| ||||||||+||||+| |||||# |||||| |||| | | Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAGK 837 |
| Anopheles gambiae | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||+||+| ||||||||+||||+| |||||# |||||| |||| | | Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAGK 837 |
| Tribolium castaneum | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRKA 800 ||+||| ||||||||||||| | |+|||# |||||| + |||| | |+ Sbjct 779 LLLWKLFTTIHDRREFAKFEKEAMMARWDT#GENPIYKQATSTFKNPTYAGKS 830 |
| Ostrinia furnacalis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 |++||++ ||||||||+||||+| |||||# |||||| + |||| | | Sbjct 781 LMLWKMVTTIHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAGK 831 |
| Podocoryne carnea | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||||| | ||||||||||++ | |||+# |||||| + |+|| || | Sbjct 731 LLIWKLLATIQDRREFAKFEKDRQNPKWDS#GENPIYKKATSTFQNPMYGNK 781 |
| Pseudoplusia includens | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 |++||+ |||||||+||||+| |||||# |||||| + |||| | | Sbjct 787 LMLWKMATTSHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAGK 837 |
| Caenorhabditis briggsae | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRKA 800 ||+|||| ++||| |+||| |++ |||||# |||||| |||| | || Sbjct 758 LLLWKLLTVLHDRAEYAKFNNERLMAKWDT#NENPIYKQATTTFKNPVYAGKA 809 |
| Caenorhabditis elegans | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRKA 800 ||+|||| ++||| |+| | |++ |||||# |||||| |||| | || Sbjct 757 LLLWKLLTVLHDRSEYATFNNERLMAKWDT#NENPIYKQATTTFKNPVYAGKA 808 |
| Strongylocentrotus purpuratus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||+|| ||||||| ||||+ || |+ # |||||| + |||| | | Sbjct 756 LLIWRLLTYIHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTYNIK 806 |
| Lytechinus variegatus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ||||+|| ||||||| ||||+ || |+ # |||||| + |||| | | Sbjct 756 LLIWRLLTYIHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTYNIK 806 |
| Crassostrea gigas | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 |||||| | |+|| |+|||| |||+|||# |||||| + | || | Sbjct 750 LLIWKLFTTISDKRELARFEKEAMNARWDT#GENPIYKQATSTFVNPTY 797 |
| Macaca mulatta | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | | Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 785 |
| rats, Peptide Partial, 723 aa | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | | Sbjct 673 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 720 |
| Equus caballus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | | Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 781 |
| Canis lupus familiaris | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | | Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITY 781 |
| mice, Peptide Partial, 680 aa | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKN 793 |||||||+ ||||+||||||+|+ |||||# ||+|| + | | Sbjct 634 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTN 678 |
| Ovis aries | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 | |||||+ |||+| |||| |+ ||| |# ||+|+ + ||| | | Sbjct 726 LCIWKLLVSFHDRKEVAKFEAERSKAKWQT#GTNPLYRGSTSTFKNVTYKHK 776 |
| Pacifastacus leniusculus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGR 798 ||||||| +|||||+||||||+ # |||+||| |+|| + + Sbjct 750 LLIWKLLTTLHDRREYAKFEKERKLPSGKR#AENPLYKSAKTTFQNPAFAQ 799 |
| Apis mellifera | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNA-KWDT#QENPIYKSPINKFKNPNY 796 || ||+| +| | ||| |||+|++ | ||+ #++||+|| + |||| + Sbjct 765 LLTWKILTMIRDNREFKKFERERILANKWNR#RDNPLYKEATSTFKNPTF 813 |
| Halocynthia roretzi | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYG 797 |+|||++ + |+||+ ||+||+ || # |||+| + +|| || +| Sbjct 784 LIIWKVIQTLRDKREYEKFKKEEDLRKWTK#GENPVYVNASSKFDNPMFG 832 |
| Manduca sexta | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRKA 800 |+ ||+|+ +||+||+||||+| + +| # ||+|+ | | || | | Sbjct 714 LIAWKILVDLHDKREYAKFEEESRSRGFDV#SLNPLYQEPEINFSNPVYNANA 765 |
| Schistosoma japonicum | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 |||+||++ | |||| | |+++ | +|+ # ||||++|| || + Sbjct 236 LLIYKLVITIDDRRELANFKQQGENMRWEM#AENPIFESPTTNVLNPTF 283 |
| Spodoptera frugiperda | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGRK 799 ++|||+|+ +||++|+ ||+ | + | +| # ||+|+ | | || | + Sbjct 729 VIIWKILVDMHDKKEYKKFQDEALAAGYDV#SLNPLYQDPSINFSNPVYNNQ 779 |
| Acropora millepora | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 |++ | | + || |+ |||+|+|++|| #++||+|++ |+|| | Sbjct 740 LIVIKGLFTMVDRIEYQKFERERMHSKWTR#EKNPLYQAAKTTFENPTY 787 |
| Mus sp. | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 +| ++| + |+||||+ +||||+ | # ||+||| | || + Sbjct 742 VLAYRLSVEIYDRREYRRFEKEQQQLNWKQ#DNNPLYKSAITTTVNPRF 789 |
| Ophlitaspongia tenuis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGR 798 ||+ | |+++ | | |||+| |||+ # |||+|+| ++|| ||+ Sbjct 789 LLLLKGLLLLWDVVEVRKFEREIKNAKYTK#NENPLYRSATKDYQNPLYGK 838 |
| Capra hircus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 |+||| | + | ||+ +|||||+ ++|+ # +||++|| || + Sbjct 721 LVIWKALTHLSDLREYHRFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 767 |
| Bubalus bubalis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 |+||| | + | ||+ +|||||+ ++|+ # +||++|| || + Sbjct 720 LVIWKALTHLSDLREYHRFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 766 |
| Ovis canadensis | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 | ||| | + | ||+ +|||||+ ++|+ # +||++|| || + Sbjct 721 LAIWKALTHLSDLREYHRFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 767 |
| Sigmodon hispidus | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 |+||| | + | ||+ |||||+ ++|+ # +||++|| || + Sbjct 720 LVIWKALTHLTDLREYRHFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 766 |
| Geodia cydonium | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGR 798 |++ |+|+++ | | ||| | |+ # |||+| | + ||| ||+ Sbjct 829 LILLKILLMVLDYTELKKFENELAKVKYTK#NENPLYHSATTEHKNPIYGQ 878 |
| Suberites domuncula | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNYGR 798 ||+ || + | | | ||||| |+ # +||+|+| ++|| ||+ Sbjct 715 LLLLKLFLFIIDVIEVKKFEKELAKTKYPK#HQNPLYRSAAKDYQNPIYGQ 764 |
| Oncorhynchus mykiss | Query 749 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNPNY 796 ||| | ++ + | +|| +|| |+ || # +||++ + || + Sbjct 727 LLIIKAILYMRDVKEFRRFENEQKKGKWSP#GDNPLFMNATTTVANPTF 774 |
| nu | Query 749 LLIWKLLMIIH--DRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNP 794 |+|+ +| | | ||+||||+++ |+ #||||||+ |+ +++ | Sbjct 740 LIIFIILWYIRAKDAREYAKFEEDQKNSV--R#QENPIYRDPVGRYEVP 785 |
| Drosophila pseudoobscura | Query 749 LLIWKLLMIIH--DRREFAKFEKEKMNAKWDT#QENPIYKSPINKFKNP 794 |||+ + | | ||+|+||+|+ || +#||||+|+ |+ ++ | Sbjct 713 LLIFLFIWCIRRKDAREYARFEEEQ--AKRVS#QENPLYRDPVGRYVVP 758 |
[Site 8] EKMNAKWDTQ779-ENPIYKSPIN
Gln779  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu770 | Lys771 | Met772 | Asn773 | Ala774 | Lys775 | Trp776 | Asp777 | Thr778 | Gln779 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu780 | Asn781 | Pro782 | Ile783 | Tyr784 | Lys785 | Ser786 | Pro787 | Ile788 | Asn789 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LIWKLLMIIHDRREFAKFEKEKMNAKWDTQENPIYKSPINKFKNPNYGRKAGL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 114.00 | 32 | S |
| 2 | Homo sapiens | 114.00 | 25 | integrin beta 1 isoform 1D precursor |
| 3 | Mus musculus | 114.00 | 18 | beta 1 integrin |
| 4 | Bos taurus | 114.00 | 13 | ITGB1 protein |
| 5 | Canis familiaris | 114.00 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 6 | Gallus gallus | 114.00 | 7 | integrin beta1D |
| 7 | Monodelphis domestica | 114.00 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 8 | Pan troglodytes | 86.30 | 14 | PREDICTED: hypothetical protein |
| 9 | Rattus norvegicus | 86.30 | 10 | integrin beta 1 |
| 10 | Tetraodon nigroviridis | 86.30 | 8 | unnamed protein product |
| 11 | Sus scrofa | 86.30 | 6 | integrin beta-1 subunit |
| 12 | Xenopus laevis | 86.30 | 6 | integrin beta-1 subunit |
| 13 | synthetic construct | 86.30 | 5 | hypothetical protein |
| 14 | Pongo pygmaeus | 86.30 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Oryctolagus cuniculus | 86.30 | 2 | integrin beta 1 |
| 16 | Xenopus tropicalis | 86.30 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 17 | Danio rerio | 85.90 | 13 | integrin beta1 subunit-like protein 1 |
| 18 | Felis catus | 84.00 | 1 | integrin beta 1 |
| 19 | Ictalurus punctatus | 82.80 | 2 | beta-1 integrin |
| 20 | Drosophila melanogaster | 81.60 | 2 | myospheroid CG1560-PA |
| 21 | Biomphalaria glabrata | 80.90 | 1 | beta integrin subunit |
| 22 | Aedes aegypti | 79.00 | 2 | integrin beta subunit |
| 23 | Anopheles gambiae str. PEST | 77.40 | 2 | ENSANGP00000021373 |
| 24 | Anopheles gambiae | 77.40 | 2 | integrin beta subunit |
| 25 | Tribolium castaneum | 77.00 | 3 | PREDICTED: similar to CG1560-PA |
| 26 | Ostrinia furnacalis | 76.60 | 1 | integrin beta 1 precursor |
| 27 | Podocoryne carnea | 75.50 | 1 | AF308652_1 integrin beta chain |
| 28 | Pseudoplusia includens | 73.60 | 1 | integrin beta 1 |
| 29 | Caenorhabditis briggsae | 73.60 | 1 | Hypothetical protein CBG03601 |
| 30 | Caenorhabditis elegans | 71.20 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 31 | Strongylocentrotus purpuratus | 69.30 | 5 | integrin beta-C subunit |
| 32 | Lytechinus variegatus | 69.30 | 1 | beta-C integrin subunit |
| 33 | Crassostrea gigas | 68.60 | 1 | integrin beta cgh |
| 34 | Macaca mulatta | 68.20 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 35 | rats, Peptide Partial, 723 aa | 68.20 | 1 | beta 3 integrin, GPIIIA |
| 36 | Equus caballus | 68.20 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 37 | Canis lupus familiaris | 68.20 | 1 | integrin beta chain, beta 3 |
| 38 | mice, Peptide Partial, 680 aa | 66.60 | 1 | beta 3 integrin, GPIIIA |
| 39 | Ovis aries | 60.80 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 40 | Pacifastacus leniusculus | 58.20 | 1 | integrin |
| 41 | Apis mellifera | 57.00 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 42 | Halocynthia roretzi | 54.30 | 2 | integrin beta Hr2 precursor |
| 43 | Manduca sexta | 54.30 | 1 | plasmatocyte-specific integrin beta 1 |
| 44 | Spodoptera frugiperda | 52.40 | 1 | integrin beta 1 |
| 45 | Schistosoma japonicum | 51.20 | 1 | SJCHGC06221 protein |
| 46 | Acropora millepora | 48.50 | 1 | integrin subunit betaCn1 |
| 47 | Mus sp. | 48.10 | 1 | beta 7 integrin |
| 48 | Ophlitaspongia tenuis | 46.60 | 1 | integrin subunit betaPo1 |
| 49 | Capra hircus | 46.20 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 50 | Bubalus bubalis | 46.20 | 1 | integrin beta-2 |
| 51 | Ovis canadensis | 45.80 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 52 | Sigmodon hispidus | 45.10 | 1 | AF445415_1 integrin beta-2 precursor |
| 53 | Geodia cydonium | 44.30 | 1 | integrin beta subunit |
| 54 | Suberites domuncula | 43.90 | 1 | integrin beta subunit |
| 55 | Oncorhynchus mykiss | 39.30 | 1 | CD18 protein |
| 56 | nu | 38.50 | 1 | beta |
| 57 | Drosophila pseudoobscura | 37.00 | 1 | GA14581-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#|||||||||| |||||||||||| Sbjct 43 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 95 |
| Homo sapiens | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#|||||||||| |||||||||||| Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 |
| Mus musculus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#|||||||||| |||||||||||| Sbjct 27 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 79 |
| Bos taurus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#|||||||||| |||||||||||| Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 801 |
| Canis familiaris | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#|||||||||| |||||||||||| Sbjct 772 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 824 |
| Gallus gallus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#|||||||||| |||||||||||| Sbjct 6 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 58 |
| Monodelphis domestica | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRKAGL 802 ||||||||||||||||||||||||||||||#|||||||||| |||||||||||| Sbjct 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRKAGL 802 |
| Pan troglodytes | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 861 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 910 |
| Rattus norvegicus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 799 |
| Tetraodon nigroviridis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 742 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 791 |
| Sus scrofa | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Xenopus laevis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| synthetic construct | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Pongo pygmaeus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Oryctolagus cuniculus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 14 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 63 |
| Xenopus tropicalis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Danio rerio | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVINPKYEGK 798 |
| Felis catus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||| ||||||||||||||||| #||||||| + || | | Sbjct 749 LIWKLLMIIHDTREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Ictalurus punctatus | Query 751 IWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ||||||||||||||||||||||||||| #||||||| + || | | Sbjct 759 IWKLLMIIHDRREFAKFEKEKMNAKWDAG#ENPIYKSAVTTVVNPKYEGK 807 |
| Drosophila melanogaster | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 |+|||| ||||||||+||||+||||||| #|||||| + |||| | | Sbjct 797 LLWKLLTTIHDRREFARFEKERMNAKWDTG#ENPIYKQATSTFKNPMYAGK 846 |
| Biomphalaria glabrata | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYG 797 |+|||| ||| |||||||||+ |||||| #|||||| + |||| || Sbjct 739 LLWKLLTFIHDTREFAKFEKERQNAKWDTG#ENPIYKQATSTFKNPTYG 786 |
| Aedes aegypti | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 |+|||| ||||||||+||||+| ||||| #|||||| |||| | | Sbjct 786 LLWKLLTTIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTYAGK 835 |
| Anopheles gambiae str. PEST | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 |+||+| ||||||||+||||+| ||||| #|||||| |||| | | Sbjct 788 LLWKVLTSIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTYAGK 837 |
| Anopheles gambiae | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 |+||+| ||||||||+||||+| ||||| #|||||| |||| | | Sbjct 788 LLWKVLTSIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTYAGK 837 |
| Tribolium castaneum | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRKA 800 |+||| ||||||||||||| | |+||| #|||||| + |||| | |+ Sbjct 780 LLWKLFTTIHDRREFAKFEKEAMMARWDTG#ENPIYKQATSTFKNPTYAGKS 830 |
| Ostrinia furnacalis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ++||++ ||||||||+||||+| ||||| #|||||| + |||| | | Sbjct 782 MLWKMVTTIHDRREFARFEKERMMAKWDTG#ENPIYKQATSTFKNPTYAGK 831 |
| Podocoryne carnea | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 |||||| | ||||||||||++ | |||+ #|||||| + |+|| || | Sbjct 732 LIWKLLATIQDRREFAKFEKDRQNPKWDSG#ENPIYKKATSTFQNPMYGNK 781 |
| Pseudoplusia includens | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 ++||+ |||||||+||||+| ||||| #|||||| + |||| | | Sbjct 788 MLWKMATTSHDRREFARFEKERMMAKWDTG#ENPIYKQATSTFKNPTYAGK 837 |
| Caenorhabditis briggsae | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRKA 800 |+|||| ++||| |+||| |++ ||||| #|||||| |||| | || Sbjct 759 LLWKLLTVLHDRAEYAKFNNERLMAKWDTN#ENPIYKQATTTFKNPVYAGKA 809 |
| Caenorhabditis elegans | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRKA 800 |+|||| ++||| |+| | |++ ||||| #|||||| |||| | || Sbjct 758 LLWKLLTVLHDRSEYATFNNERLMAKWDTN#ENPIYKQATTTFKNPVYAGKA 808 |
| Strongylocentrotus purpuratus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 |||+|| ||||||| ||||+ || |+ #|||||| + |||| | | Sbjct 757 LIWRLLTYIHDRREFQNFEKERANATWEGG#ENPIYKPSTSVFKNPTYNIK 806 |
| Lytechinus variegatus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 |||+|| ||||||| ||||+ || |+ #|||||| + |||| | | Sbjct 757 LIWRLLTYIHDRREFQNFEKERANATWEGG#ENPIYKPSTSVFKNPTYNIK 806 |
| Crassostrea gigas | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 ||||| | |+|| |+|||| |||+||| #|||||| + | || | Sbjct 751 LIWKLFTTISDKRELARFEKEAMNARWDTG#ENPIYKQATSTFVNPTY 797 |
| Macaca mulatta | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | | Sbjct 739 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITY 785 |
| rats, Peptide Partial, 723 aa | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | | Sbjct 674 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITY 720 |
| Equus caballus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | | Sbjct 735 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITY 781 |
| Canis lupus familiaris | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | | Sbjct 735 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITY 781 |
| mice, Peptide Partial, 680 aa | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKN 793 ||||||+ ||||+||||||+|+ ||||| # ||+|| + | | Sbjct 635 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTN 678 |
| Ovis aries | Query 751 IWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 |||||+ |||+| |||| |+ ||| | # ||+|+ + ||| | | Sbjct 728 IWKLLVSFHDRKEVAKFEAERSKAKWQTG#TNPLYRGSTSTFKNVTYKHK 776 |
| Pacifastacus leniusculus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGR 798 |||||| +|||||+||||||+ #|||+||| |+|| + + Sbjct 751 LIWKLLTTLHDRREYAKFEKERKLPSGKRA#ENPLYKSAKTTFQNPAFAQ 799 |
| Apis mellifera | Query 750 LIWKLLMIIHDRREFAKFEKEKMNA-KWDTQ#ENPIYKSPINKFKNPNY 796 | ||+| +| | ||| |||+|++ | ||+ +#+||+|| + |||| + Sbjct 766 LTWKILTMIRDNREFKKFERERILANKWNRR#DNPLYKEATSTFKNPTF 813 |
| Halocynthia roretzi | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYG 797 +|||++ + |+||+ ||+||+ || #|||+| + +|| || +| Sbjct 785 IIWKVIQTLRDKREYEKFKKEEDLRKWTKG#ENPVYVNASSKFDNPMFG 832 |
| Manduca sexta | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRKA 800 + ||+|+ +||+||+||||+| + +| # ||+|+ | | || | | Sbjct 715 IAWKILVDLHDKREYAKFEEESRSRGFDVS#LNPLYQEPEINFSNPVYNANA 765 |
| Spodoptera frugiperda | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGRK 799 +|||+|+ +||++|+ ||+ | + | +| # ||+|+ | | || | + Sbjct 730 IIWKILVDMHDKKEYKKFQDEALAAGYDVS#LNPLYQDPSINFSNPVYNNQ 779 |
| Schistosoma japonicum | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 ||+||++ | |||| | |+++ | +|+ #||||++|| || + Sbjct 237 LIYKLVITIDDRRELANFKQQGENMRWEMA#ENPIFESPTTNVLNPTF 283 |
| Acropora millepora | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 ++ | | + || |+ |||+|+|++|| +#+||+|++ |+|| | Sbjct 741 IVIKGLFTMVDRIEYQKFERERMHSKWTRE#KNPLYQAAKTTFENPTY 787 |
| Mus sp. | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 | ++| + |+||||+ +||||+ | # ||+||| | || + Sbjct 743 LAYRLSVEIYDRREYRRFEKEQQQLNWKQD#NNPLYKSAITTTVNPRF 789 |
| Ophlitaspongia tenuis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGR 798 |+ | |+++ | | |||+| |||+ #|||+|+| ++|| ||+ Sbjct 790 LLLKGLLLLWDVVEVRKFEREIKNAKYTKN#ENPLYRSATKDYQNPLYGK 838 |
| Capra hircus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 +||| | + | ||+ +|||||+ ++|+ #+||++|| || + Sbjct 722 VIWKALTHLSDLREYHRFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 767 |
| Bubalus bubalis | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 +||| | + | ||+ +|||||+ ++|+ #+||++|| || + Sbjct 721 VIWKALTHLSDLREYHRFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 766 |
| Ovis canadensis | Query 751 IWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 ||| | + | ||+ +|||||+ ++|+ #+||++|| || + Sbjct 723 IWKALTHLSDLREYHRFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 767 |
| Sigmodon hispidus | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 +||| | + | ||+ |||||+ ++|+ #+||++|| || + Sbjct 721 VIWKALTHLTDLREYRHFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 766 |
| Geodia cydonium | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGR 798 ++ |+|+++ | | ||| | |+ #|||+| | + ||| ||+ Sbjct 830 ILLKILLMVLDYTELKKFENELAKVKYTKN#ENPLYHSATTEHKNPIYGQ 878 |
| Suberites domuncula | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNYGR 798 |+ || + | | | ||||| |+ #+||+|+| ++|| ||+ Sbjct 716 LLLKLFLFIIDVIEVKKFEKELAKTKYPKH#QNPLYRSAAKDYQNPIYGQ 764 |
| Oncorhynchus mykiss | Query 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNPNY 796 || | ++ + | +|| +|| |+ || #+||++ + || + Sbjct 728 LIIKAILYMRDVKEFRRFENEQKKGKWSPG#DNPLFMNATTTVANPTF 774 |
| nu | Query 760 DRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNP 794 | ||+||||+++ |+ |#|||||+ |+ +++ | Sbjct 753 DAREYAKFEEDQKNSV--RQ#ENPIYRDPVGRYEVP 785 |
| Drosophila pseudoobscura | Query 760 DRREFAKFEKEKMNAKWDTQ#ENPIYKSPINKFKNP 794 | ||+|+||+|+ || +|#|||+|+ |+ ++ | Sbjct 726 DAREYARFEEEQ--AKRVSQ#ENPLYRDPVGRYVVP 758 |
* References
None.