CL0004 : Chain L, Crystal Structure Of Human M-Calpain Form Ii
[ CaMP Format ]
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Calpain-2 catalytic subunit; 3.4.22.53; Calpain-2 large subunit; Calcium-activated neutral proteinase 2; CANP 2; Calpain M-type; M-calpain; Millimolar-calpain; Calpain large polypeptide L2 |
| Gene Names | CANPL2 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 699 aa
Average Mass: 79.875 kDa
Monoisotopic Mass: 79.825 kDa
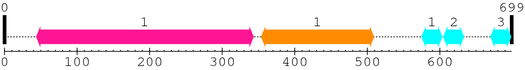
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Peptidase_C2 1. | 44 | 343 | 808.0 | 6.2e-240 |
| Calpain_III 1. | 354 | 509 | 393.6 | 3.4e-115 |
| efhand 1. | 575 | 603 | 20.4 | 0.0074 |
| efhand 2. | 605 | 633 | 19.4 | 0.015 |
| efhand 3. | 670 | 698 | 4.0 | 7.6 |
3. Sequence Information
Fasta Sequence: CL0004.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* References
[PubMed ID: 10666632] Masumoto H, Nakagawa K, Irie S, Sorimachi H, Suzuki K, Bourenkov GP, Bartunik H, Fernandez-Catalan C, Bode W, Strobl S, Crystallization and preliminary X-ray analysis of recombinant full-length human m-calpain. Acta Crystallogr D Biol Crystallogr. 2000 Jan;56(Pt 1):73-5.