CL0007 : Chain B, Crystal Structure Of Calcium-Bound Protease Core Of Calpain I
[ CaMP Format ]
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | Calpain-1 catalytic subunit; 3.4.22.52; Calpain-1 large subunit; Calcium-activated neutral proteinase 1; CANP 1; Calpain mu-type; muCANP; Micromolar-calpain |
| Gene Names | Cls1 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 339 aa
Average Mass: 38.744 kDa
Monoisotopic Mass: 38.719 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
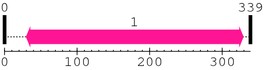
| Peptidase_C2 1. | 30 | 329 | 752.3 | 3.5e-223 |
3. Sequence Information
Fasta Sequence: CL0007.fasta
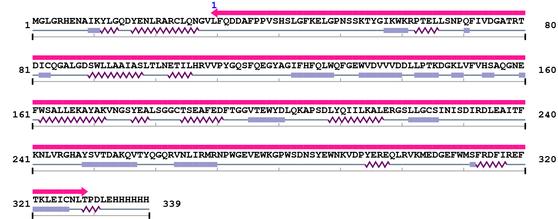
Amino Acid Sequence and Secondary Structures (PsiPred):
* References
[PubMed ID: 11893336] Moldoveanu T, Hosfield CM, Lim D, Elce JS, Jia Z, Davies PL, A Ca(2+) switch aligns the active site of calpain. Cell. 2002 Mar 8;108(5):649-60.