CL0010 : Chain A, Crystal Structure Of A Mu-Like Calpain
[ CaMP Format ]
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | Calpain-2 catalytic subunit; 3.4.22.53; Calpain-2 large subunit; Calcium-activated neutral proteinase 2; CANP 2; Calpain M-type; M-calpain; Millimolar-calpain |
| Gene Names | Capn2 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 900 aa
Average Mass: 103.383 kDa
Monoisotopic Mass: 103.318 kDa
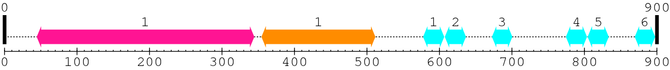
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Peptidase_C2 1. | 45 | 344 | 754.2 | 9.3e-224 |
| Calpain_III 1. | 355 | 511 | 365.0 | 1.4e-106 |
| efhand 1. | 578 | 606 | 16.6 | 0.11 |
| efhand 2. | 608 | 636 | 18.5 | 0.027 |
| efhand 3. | 673 | 700 | 3.2 | 9.5 |
| efhand 4. | 775 | 803 | 18.4 | 0.03 |
| efhand 5. | 805 | 833 | 7.9 | 2.9 |
| efhand 6. | 870 | 897 | 7.5 | 3.2 |
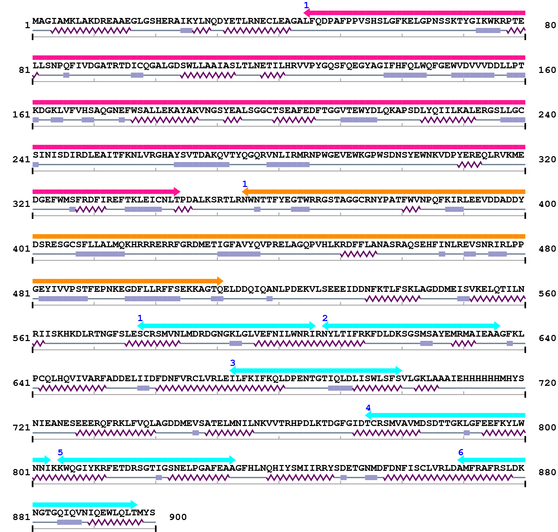
3. Sequence Information
Fasta Sequence: CL0010.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* References
[PubMed ID: 14656436] Pal GP, De Veyra T, Elce JS, Jia Z, Crystal structure of a micro-like calpain reveals a partially activated conformation with low Ca2+ requirement. Structure. 2003 Dec;11(12):1521-6.