CL1269 : hypothetical protein
[ CaMP Format ]
* Basic Information
| Organism | Kluyveromyces lactis NRRL Y-1140 |
| Protein Names | unnamed protein product [Kluyveromyces lactis]; Calpain-like protease palB/RIM13; 3.4.22.-; Cysteine protease RIM13 |
| Gene Names | KLLA0F07183g |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_455406 | XM_455406 | 2895973 | Q6CKY3 | N/A | N/A | N/A | kla:KLLA0F07183g |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 685 aa
Average Mass: 79.686 kDa
Monoisotopic Mass: 79.635 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| DUF1477 1. | 1 | 85 | -55.5 | 8 |
| CBM_10 1. | 445 | 467 | -2.2 | 4.5 |
| Shugoshin_N 1. | 600 | 643 | 0.4 | 4.6 |
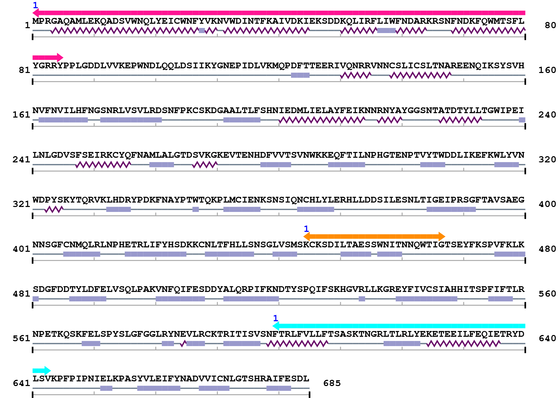
3. Sequence Information
Fasta Sequence: CL1269.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* References
[PubMed ID: 15229592] Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisrame A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wesolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL, Genome evolution in yeasts. Nature. 2004 Jul 1;430(6995):35-44.