CL1430 : putative peptidase C2
[ CaMP Format ]
* Basic Information
| Organism | Bradyrhizobium sp. ORS278 |
| Protein Names | calpain; putative peptidase C2, calpain [Bradyrhizobium sp. ORS278]; Putative uncharacterized protein |
| Gene Names | BRADO2763 |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| YP_001204811 | N/A | 5118052 | A4YRP8 | N/A | N/A | N/A | bra:BRADO2763 |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 567 aa
Average Mass: 57.804 kDa
Monoisotopic Mass: 57.769 kDa
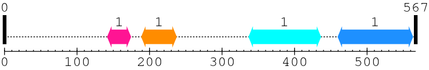
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Flg_bb_rod 1. | 142 | 174 | -5.7 | 8.7 |
| CBM_19 1. | 189 | 237 | -21.5 | 8.8 |
| Fork_head_N 1. | 337 | 436 | -34.0 | 3.3 |
| Nrf1_activ_bdg 1. | 460 | 563 | -20.2 | 8.1 |
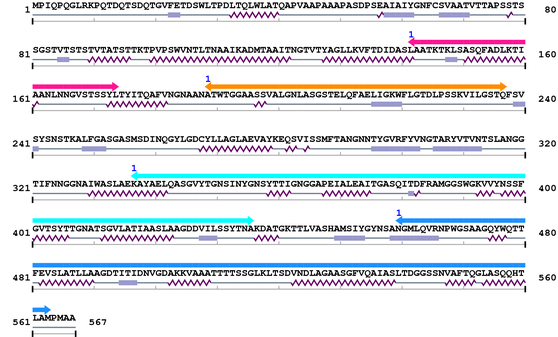
3. Sequence Information
Fasta Sequence: CL1430.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* References
[PubMed ID: 17540897] Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre JC, Jaubert M, Simon D, Cartieaux F, Prin Y, Bena G, Hannibal L, Fardoux J, Kojadinovic M, Vuillet L, Lajus A, Cruveiller S, Rouy Z, Mangenot S, Segurens B, Dossat C, Franck WL, Chang WS, Saunders E, Bruce D, Richardson P, Normand P, Dreyfus B, Pignol D, Stacey G, Emerich D, Vermeglio A, Medigue C, Sadowsky M, Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science. 2007 Jun 1;316(5829):1307-12.