XSB0010 : PREDICTED: poly (ADP-ribose) polymerase family, member 1 isoform 3 [Macaca mulatta]
[ CaMP Format ]
This entry is computationally expanded from SB0065
* Basic Information
| Organism | Macaca mulatta (rhesus monkey) |
| Protein Names | poly (ADP-ribose) polymerase family; member 1 isoform 3 |
| Gene Names | PARP1; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 13 mRNAs, 552 ESTs, 12 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001090871 | XM_001090871 | 698806 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 929 aa
Average Mass: 103.568 kDa
Monoisotopic Mass: 103.502 kDa
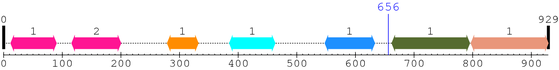
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| zf-PARP 1. | 12 | 90 | 151.0 | 3.5e-42 |
| zf-PARP 2. | 116 | 200 | 157.3 | 4.5e-44 |
| PADR1 1. | 279 | 332 | 124.2 | 4.3e-34 |
| BRCT 1. | 385 | 463 | 50.0 | 9.1e-12 |
| WGR 1. | 548 | 633 | 144.5 | 3.4e-40 |
| --- cleavage 656 --- | ||||
| PARP_reg 1. | 662 | 795 | 284.4 | 2.5e-82 |
| PARP 1. | 797 | 929 | 210.9 | 3.5e-60 |
3. Sequence Information
Fasta Sequence: XSB0010.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] QDEEAVKKLT656-VNPGTKSKLP
Thr656  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gln647 | Asp648 | Glu649 | Glu650 | Ala651 | Val652 | Lys653 | Lys654 | Leu655 | Thr656 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val657 | Asn658 | Pro659 | Gly660 | Thr661 | Lys662 | Ser663 | Lys664 | Leu665 | Pro666 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| HSKNFTKYPKKFYPLEIDYGQDEEAVKKLTVNPGTKSKLPKPVQDLIKLIFDVESMKKAM |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 125.00 | 5 | PREDICTED: poly (ADP-ribose) polymerase family, me |
| 2 | Macaca fascicularis | 125.00 | 1 | unnamed protein product |
| 3 | ADP-ribose | 124.00 | 19 | PARP1_HUMAN Poly |
| 4 | N/A | 124.00 | 9 | - |
| 5 | Homo sapiens | 124.00 | 8 | poly (ADP-ribose) polymerase family, member 1, iso |
| 6 | synthetic construct | 124.00 | 1 | poly (ADP-ribose) polymerase family, member 1 |
| 7 | Bos taurus | 120.00 | 2 | ADP-ribosyltransferase (NAD+) poly (ADP-ribose) po |
| 8 | cattle, Peptide Partial, 607 aa | 120.00 | 1 | NAD+:protein(ADP-ribosyl)-transferase, ADPRT |
| 9 | Mus musculus | 116.00 | 11 | unnamed protein product |
| 10 | mice, Peptide Partial, 606 aa | 115.00 | 1 | NAD+:protein(ADP-ribosyl)-transferase, ADPRT |
| 11 | Rattus norvegicus | 114.00 | 2 | ADP-ribosyltransferase 1 |
| 12 | chickens, Peptide Partial, 607 aa | 114.00 | 1 | NAD+:protein(ADP-ribosyl)-transferase, ADPRT |
| 13 | Gallus gallus | 114.00 | 1 | poly (ADP-ribose) polymerase family, member 1 |
| 14 | Monodelphis domestica | 112.00 | 2 | PREDICTED: similar to poly(ADP-ribose) synthetase |
| 15 | Xenopus laevis | 107.00 | 3 | NAD(+) ADP-ribosyltransferase |
| 16 | Xenopus, Peptide Partial, 607 aa | 107.00 | 1 | NAD+:protein(ADP-ribosyl)-transferase, ADPRT |
| 17 | Tetraodon nigroviridis | 105.00 | 1 | unnamed protein product |
| 18 | Danio rerio | 102.00 | 2 | Zgc:110092 |
| 19 | Anopheles gambiae str. PEST | 60.10 | 1 | ENSANGP00000014723 |
| 20 | adp-ribose | 57.00 | 1 | poly |
| 21 | Arabidopsis thaliana | 49.30 | 2 | poly (ADP-ribose) polymerase, putative / NAD(+) AD |
| 22 | Vitis vinifera | 49.30 | 1 | hypothetical protein |
| 23 | Caenorhabditis elegans | 47.80 | 2 | AF499444_1 poly ADP-ribose metabolism enzyme-1 |
| 24 | Drosophila melanogaster | 47.40 | 3 | CG40411-PC.3 |
| 25 | Drosophila, Peptide Partial, 593 aa | 47.40 | 1 | NAD+:protein(ADP-ribosyl)-transferase, ADPRT |
| 26 | Trypanosoma brucei brucei | 46.60 | 1 | poly(ADP-ribose) polymerase |
| 27 | Caenorhabditis briggsae | 46.60 | 1 | Hypothetical protein CBG04221 |
| 28 | Trypanosoma brucei TREU927 | 46.60 | 1 | poly(ADP-ribose) polymerase, putative |
| 29 | Oryza sativa (japonica cultivar-group) | 45.80 | 5 | hypothetical protein OsJ_022996 |
| 30 | Oryza sativa (indica cultivar-group) | 45.80 | 3 | hypothetical protein OsI_024833 |
| 31 | Zea mays | 45.10 | 1 | poly(ADP-ribose) polymerase |
| 32 | Dictyostelium discoideum AX4 | 42.70 | 3 | poly(ADP-ribosyl)transferase |
| 33 | Dictyostelium discoideum | 42.70 | 2 | NAD(+) ADP-ribosyltransferase-1A |
| 34 | Schistosoma japonicum | 40.80 | 1 | SJCHGC03992 protein |
| 35 | Pan troglodytes | 38.10 | 3 | PREDICTED: poly (ADP-ribose) polymerase family, me |
| 36 | Dictyostelium discoideum, Peptide Partial, 612 aa | 37.00 | 1 | NAD+:protein(ADP-ribosyl)-transferase, ADPRT {EC 2 |
| 37 | Entamoeba histolytica HM-1:IMSS | 35.80 | 1 | poly(ADP-ribose) polymerase |
| 38 | Theileria parva strain Muguga | 33.90 | 1 | hypothetical protein TP01_0611 |
| 39 | Paramecium tetraurelia | 32.70 | 1 | hypothetical protein |
| 40 | Tetrahymena thermophila SB210 | 32.30 | 1 | WGR domain containing protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 |
| Macaca fascicularis | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 |
| ADP-ribose | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 ||||||||||||||||||||||||||||||#||||||||||||||||||+||||||||||| Sbjct 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKMIFDVESMKKAM 686 |
| N/A | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 ||||||||||||||||||||||||||||||#||||||||||||||||||+||||||||||| Sbjct 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKMIFDVESMKKAM 686 |
| Homo sapiens | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 ||||||||||||||||||||||||||||||#||||||||||||||||||+||||||||||| Sbjct 625 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKMIFDVESMKKAM 684 |
| synthetic construct | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 ||||||||||||||||||||||||||||||#||||||||||||||||||+||||||||||| Sbjct 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKMIFDVESMKKAM 686 |
| Bos taurus | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 |||||||+||||||||||||||||||||||#||||||||||||||+|||+||||||||||| Sbjct 629 HSKNFTKHPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQNLIKMIFDVESMKKAM 688 |
| cattle, Peptide Partial, 607 aa | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 |||||||+||||||||||||||||||||||#||||||||||||||+|||+||||||||||| Sbjct 220 HSKNFTKHPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQNLIKMIFDVESMKKAM 279 |
| Mus musculus | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 ||||||||||||||||||||||||||||||#| ||||||||||||+|+ +||||||||||+ Sbjct 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VKPGTKSKLPKPVQELVGMIFDVESMKKAL 686 |
| mice, Peptide Partial, 606 aa | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 ||||||||||||||||||||||||||||||#| ||||||||||||+|+ +||||+|||||+ Sbjct 219 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VKPGTKSKLPKPVQELVGMIFDVDSMKKAL 278 |
| Rattus norvegicus | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 ||||||||||||||||||||||||||||| #| ||||||||||||+|+ +||||||||||+ Sbjct 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLA#VKPGTKSKLPKPVQELVGMIFDVESMKKAL 686 |
| chickens, Peptide Partial, 607 aa | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 ||||||||||||||||||||||||||+|||#|+ |||||| ||+|||||+||||||||||| Sbjct 220 HSKNFTKYPKKFYPLEIDYGQDEEAVRKLT#VSAGTKSKLAKPIQDLIKMIFDVESMKKAM 279 |
| Gallus gallus | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 ||||||||||||||||||||||||||+|||#|+ |||||| ||+|||||+||||||||||| Sbjct 624 HSKNFTKYPKKFYPLEIDYGQDEEAVRKLT#VSAGTKSKLAKPIQDLIKMIFDVESMKKAM 683 |
| Monodelphis domestica | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 || |||||||||||||||||||||||||||#|+ ||||||| |||+|||+||||||||||| Sbjct 624 HSTNFTKYPKKFYPLEIDYGQDEEAVKKLT#VSAGTKSKLPIPVQNLIKMIFDVESMKKAM 683 |
| Xenopus laevis | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 || ||||||||||||||||||+|+ ||||+#| |||||| ||||+||||||||||||||| Sbjct 623 HSPNFTKYPKKFYPLEIDYGQEEDVVKKLS#VGAGTKSKLAKPVQELIKLIFDVESMKKAM 682 |
| Xenopus, Peptide Partial, 607 aa | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 || ||||||||||||||||||+|+ ||||+#| |||||| ||||+||||||||||||||| Sbjct 219 HSPNFTKYPKKFYPLEIDYGQEEDVVKKLS#VGAGTKSKLAKPVQELIKLIFDVESMKKAM 278 |
| Tetraodon nigroviridis | Query 628 SKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 | |||||| ||||||||||||||||||||# |||||| ||||+|||+||||||||||| Sbjct 370 SSNFTKYPNKFYPLEIDYGQDEEAVKKLT#ATAGTKSKLAKPVQELIKMIFDVESMKKAM 428 |
| Danio rerio | Query 628 SKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 | |||||| ||||||||||||||||||||# + | ||+| |||||||++||||||||||| Sbjct 625 SSNFTKYPNKFYPLEIDYGQDEEAVKKLT#QSAGAKSQLEKPVQDLIRMIFDVESMKKAM 683 |
| Anopheles gambiae str. PEST | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 | ++| | |||+|||| | |+| # | | |||| ||||++++|||++| + | Sbjct 610 HDSVYSKMPGAFYPIEIDYS--ETKTKRLA#ENSGIKSKLAPAVQDLVRMLFDVDAMNRVM 667 |
| adp-ribose | Query 631 FTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 | |+| +||++||| +| | | # | |||| ||||++++|||++||| | Sbjct 620 FHKHPGAYYPVDIDYNDEE--TKTLA#ENNDIKSKLEPAVQDLVRMLFDVDTMKKVM 673 |
| Arabidopsis thaliana | Query 630 NFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 || | | || ||+|||| +++ || # | | | + +|+|++||||+ + || Sbjct 603 NFQKQPGKFLPLDIDYGVNKQVAKKEP#FQ--TSSNLAPSLIELMKMLFDVETYRSAM 657 |
| Vitis vinifera | Query 629 KNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 +|| | | +|+||+|||| +++ || #++ |+| | +|+|++|+||+ + || Sbjct 626 QNFQKQPGRFFPLDIDYGVNKQVSKKNN#LS-NVNSQLAPQVVELMKMLFNVETYRSAM 682 |
| Caenorhabditis elegans | Query 627 HSKNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 + |+| | | | +| || + + + #+ ||+|+ ||| |++++ |||||+|| |+ Sbjct 552 YRKHFRKMPGMFSYVETDYSEFAQ-ITDTE#ITPGSKTLLPKSVKEVVMSIFDVENMKSAL 610 |
| Drosophila melanogaster | Query 630 NFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 || | + ||+|| | |++ || # + ||| ||+|||||||++|| | + Sbjct 614 NFVKRTGRMYPIEIQYDDDQKLVKH--#ESHFFTSKLEISVQNLIKLIFDIDSMNKTL 668 |
| Drosophila, Peptide Partial, 593 aa | Query 630 NFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 || | + ||+|| | |++ || # + ||| ||+|||||||++|| | + Sbjct 213 NFVKRTGRMYPIEIQYDDDQKLVKH--#ESHFFTSKLEISVQNLIKLIFDIDSMNKTL 267 |
| Trypanosoma brucei brucei | Query 631 FTKYPKKFYPLEIDYGQDEE--------AVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESM 682 | | | |+ +|+|||+|++ || # | +||||| ||||++|| ||| Sbjct 190 FKKVPGKYEYMEVDYGEDDDDDAGGEKPGKKKKA#EKPVRESKLPKEVQDLMRLIGCRESM 249 Query 683 KKAM 686 + + Sbjct 250 SQVL 253 |
| Caenorhabditis briggsae | Query 629 KNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 |+| | | | +| || + ++ +|+ #+ ||+++ ||+ |++++ |||+|+|| |+ Sbjct 545 KHFRKMPGAFSFVETDYSEFDQ-LKEQD#IIPGSRTLLPRAVKEIVMSIFDIENMKSAL 601 |
| Trypanosoma brucei TREU927 | Query 631 FTKYPKKFYPLEIDYGQDEE--------AVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESM 682 | | | |+ +|+|||+|++ || # | +||||| ||||++|| ||| Sbjct 190 FKKVPGKYEYMEVDYGEDDDDDAGGEKPGKKKKA#EKPVRESKLPKEVQDLMRLIGCRESM 249 Query 683 KKAM 686 + + Sbjct 250 SQVL 253 |
| Oryza sativa (japonica cultivar-group) | Query 630 NFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 || | | |||||+|||| + |+ #++ || || + +|+ ++|++|+ + || Sbjct 560 NFQKQPGKFYPLDIDYGV-RQGPKRKD#IDK-MKSSLPPQLLELMNMLFNIETYRAAM 614 |
| Oryza sativa (indica cultivar-group) | Query 630 NFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 || | | |||||+|||| + |+ #++ || || + +|+ ++|++|+ + || Sbjct 579 NFQKQPGKFYPLDIDYGV-RQGPKRKD#IDK-MKSSLPPQLLELMNMLFNIETYRAAM 633 |
| Zea mays | Query 630 NFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 || | | +||||++||| ++| |+ #++ || | + +|+|++|+||+ + || Sbjct 588 NFRKQPGRFYPLDVDYGV-KKAPKRKD#ISE-MKSSLAPQLLELMKMLFNVETYRAAM 642 |
| Dictyostelium discoideum AX4 | Query 631 FTKYPKKFYPLEIDYGQDEEAVKKL-----------T#VNPGT-------KSKLPKPVQDL 672 | | |+|+| + +| | ||| + | |# | | ||+ | || Sbjct 541 FKKLPRKYYMISLDDGNDEEEEENLVETVLKRKKSET#TTTSTSQEIEPKKEGLPQRVLDL 600 Query 673 IKLIFDVESMKKAM 686 +||+|| | ||| + Sbjct 601 VKLMFDPEMMKKQL 614 |
| Dictyostelium discoideum | Query 631 FTKYPKKFYPLEIDYGQDEEAVKKL-----------T#VNPGT-------KSKLPKPVQDL 672 | | |+|+| + +| | ||| + | |# | | ||+ | || Sbjct 541 FKKLPRKYYMISLDDGNDEEEEENLVETVLKRKKSET#TTTSTSQEIEPKKEGLPQRVLDL 600 Query 673 IKLIFDVESMKKAM 686 +||+|| | ||| + Sbjct 601 VKLMFDPEMMKKQL 614 |
| Schistosoma japonicum | Query 630 NFTKYPKKFYPLEIDYG-QDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 +| |+ |+ +||||| ||| + # ||+|| || |++|| |+++|++ | Sbjct 133 HFIKHKGKYDLVEIDYGVQDENVTIETK#KTEEIKSRLPVAVQVLMQLICDMDNMEQVM 190 |
| Pan troglodytes | Query 629 KNFTKYPKKFYPLEIDYG---QDEEAVKK---LT#VNPGTKSKLPKPVQDLIKLIFDVESM 682 + | | | |+ |++|| |||| || | # +|+| ||+||||| +|++| Sbjct 192 EKFEKVPGKYDMLQMDYATNTQDEEETKKEESLK#SPLKPESQLDLRVQELIKLICNVQAM 251 Query 683 KKAM 686 ++ | Sbjct 252 EEMM 255 |
| Dictyostelium discoideum, Peptide Partial, 612 aa | Query 630 NFTKYPKKFYPLEIDYGQDEE-----------AVKKLT#VNPGTKSKLPKPVQDLIKLIFD 678 || | |+ +|+|| | + | #| + | + ||+|+||||| Sbjct 228 NFKKVAGKYDMIELDYSTDSKPKNGASTTATTTTTKKV#VEHKKECSLDERVQELVKLIFD 287 Query 679 VESMKKAM 686 |+ |++ | Sbjct 288 VKMMERTM 295 |
| Entamoeba histolytica HM-1:IMSS | Query 631 FTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKP----------------VQDLIK 674 | | |++ +| |+|| ||| #+| || |+ + | |||| Sbjct 467 FKKVGGKYFMQALDTGKDETERKKL-#INEETKKKMEEKRQQLKEKAKENYLDPRVSDLIK 525 Query 675 LIFDVESMKKAM 686 +||| + || + Sbjct 526 MIFDTDMMKNTL 537 |
| Theileria parva strain Muguga | Query 629 KNFTKYPKKFYPLEIDYGQDEEAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKK 684 + ++| ||| + | |+ |+ |#| | +| |+| +| | ||| || Sbjct 140 EKYSKRPKKPEKRKGDTDDTEKTKKRST#VEPQPQSDQPEPEVTQVKSIEDVEREKK 195 |
| Paramecium tetraurelia | Query 633 KYPKKFYPLEIDYGQDEEAVKKLT#VNPGT----------KSKLPKPVQDLIKLIFDVESM 682 +| | | | | + ++| #+| + |||| | ++||++||+||+ | Sbjct 157 EYENKIYEKTIT--GDYKILEKFL#INQNSELSNDLAHYEKSKLQKEIKDLLQLIYDVKMM 214 Query 683 KKAM 686 | | Sbjct 215 DKQM 218 |
| Tetrahymena thermophila SB210 | Query 641 LEIDYGQDE--EAVKKLT#VNPGTKSKLPKPVQDLIKLIFDVESMKKAM 686 |++||||| + + + # |||| | || ||||++ + | Sbjct 240 LDMDYGQDATLDELDDVM#KKDAETCKLPKEVISLISLIFDMKMINNQM 287 |
* References
None.