XSB0011 : PREDICTED: growth associated protein 43 isoform 2 [Macaca mulatta]
[ CaMP Format ]
This entry is computationally expanded from SB0067
* Basic Information
| Organism | Macaca mulatta (rhesus monkey) |
| Protein Names | growth associated protein 43 isoform 2 |
| Gene Names | GAP43; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 11 mRNAs, 230 ESTs, 3 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 238 aa
Average Mass: 24.789 kDa
Monoisotopic Mass: 24.774 kDa
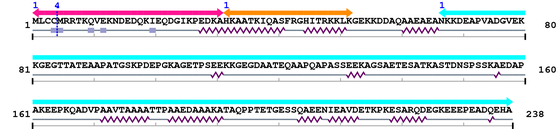
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
| Neuromodulin_N 1. | 1 | 31 | 65.8 | 1.6e-16 |
| --- cleavage 4 (inside Neuromodulin_N 1..31) --- |
| IQ 1. | 32 | 52 | 32.4 | 1.9e-06 |
| Neuromodulin 1. | 67 | 238 | 358.4 | 1.4e-104 |
3. Sequence Information
Fasta Sequence: XSB0011.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] MLCC4-MRRTKQVEKN
Cys4  Met
Met
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| - | - | - | - | - | - | Met1 | Leu2 | Cys3 | Cys4 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Met5 | Arg6 | Arg7 | Thr8 | Lys9 | Gln10 | Val11 | Glu12 | Lys13 | Asn14 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
MLCCMRRTKQVEKNDEDQKIEQDGIKPEDKAHKA
|
Summary
| # |
organism |
max score |
hits |
top seq |
| 1 |
N/A |
75.10 |
4 |
N |
| 2 |
Bos taurus |
75.10 |
2 |
growth associated protein 43 |
| 3 |
Macaca mulatta |
75.10 |
2 |
PREDICTED: growth associated protein 43 isoform 2 |
| 4 |
Canis familiaris |
75.10 |
1 |
PREDICTED: similar to Neuromodulin (Axonal membran |
| 5 |
Felis catus |
75.10 |
1 |
growth associated protein 43 |
| 6 |
Rattus norvegicus |
74.70 |
1 |
growth associated protein 43 |
| 7 |
Mus musculus |
74.70 |
1 |
growth associated protein 43 |
| 8 |
synthetic construct |
73.90 |
3 |
growth associated protein 43 |
| 9 |
Pongo pygmaeus |
73.90 |
1 |
NEUM_PONPY Neuromodulin (Axonal membrane protein G |
| 10 |
Homo sapiens |
73.90 |
1 |
growth associated protein 43 |
| 11 |
Lonchura striata |
67.80 |
1 |
growth-associated protein 43 |
| 12 |
Xenopus tropicalis |
65.50 |
1 |
hypothetical protein LOC779658 |
| 13 |
Xenopus laevis |
63.90 |
2 |
growth associated protein 43 |
| 14 |
Saimiri boliviensis |
62.80 |
1 |
growth associated protein 43 |
| 15 |
Danio rerio |
58.50 |
2 |
Gap43 protein |
| 16 |
Capra hircus |
57.00 |
1 |
growth associated protein 43 |
| 17 |
Tetraodon nigroviridis |
56.20 |
1 |
unnamed protein product |
| 18 |
Serinus canaria |
51.60 |
1 |
NEUM_SERCA Neuromodulin (Axonal membrane protein G |
| 19 |
Monodelphis domestica |
47.40 |
1 |
PREDICTED: similar to neuron growth-associated pro |
| 20 |
Gallus gallus |
45.80 |
1 |
PREDICTED: similar to growth-associated polypeptid |
Top-ranked sequences
| organism | matching |
|---|
| N/A |
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
||||#||||||||||||||||||||||||||||||
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
|
| Bos taurus |
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
||||#||||||||||||||||||||||||||||||
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
|
| Macaca mulatta |
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
||||#||||||||||||||||||||||||||||||
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
|
| Canis familiaris |
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
||||#||||||||||||||||||||||||||||||
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
|
| Felis catus |
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
||||#||||||||||||||||||||||||||||||
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
|
| Rattus norvegicus |
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
||||#||||||||||||||||||||+|||||||||
Sbjct 1 MLCC#MRRTKQVEKNDEDQKIEQDGVKPEDKAHKA 34
|
| Mus musculus |
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
||||#||||||||||||||||||||+|||||||||
Sbjct 1 MLCC#MRRTKQVEKNDEDQKIEQDGVKPEDKAHKA 34
|
| synthetic construct |
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
||||#|||||||||||+||||||||||||||||||
Sbjct 1 MLCC#MRRTKQVEKNDDDQKIEQDGIKPEDKAHKA 34
|
| Pongo pygmaeus |
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
||||#|||||||||||+||||||||||||||||||
Sbjct 1 MLCC#MRRTKQVEKNDDDQKIEQDGIKPEDKAHKA 34
|
| Homo sapiens |
Query 1 MLCC#MRRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
||||#|||||||||||+||||||||||||||||||
Sbjct 1 MLCC#MRRTKQVEKNDDDQKIEQDGIKPEDKAHKA 34
|
| Lonchura striata |
Query 1 MLCC#MRRTKQVEKNDE-DQKIEQDGIKPEDKAHKA 34
||||#||||||||||++ ||||||||||||||||||
Sbjct 1 MLCC#MRRTKQVEKNEDGDQKIEQDGIKPEDKAHKA 35
|
| Xenopus tropicalis |
Query 1 MLCC#MRRTKQVEKN-DEDQKIEQDGIKPEDKAHKA 34
||||#|||||||||| | |||||||| |||||||||
Sbjct 1 MLCC#MRRTKQVEKNEDADQKIEQDGNKPEDKAHKA 35
|
| Xenopus laevis |
Query 1 MLCC#MRRTKQVEKNDE-DQKIEQDGIKPEDKAHKA 34
||||#||||||||||++ ||||+||| |||||||||
Sbjct 1 MLCC#MRRTKQVEKNEDGDQKIDQDGNKPEDKAHKA 35
|
| Saimiri boliviensis |
Query 6 RRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
|||||||||||||||||||||||||||||
Query 6 RRTKQVEKNDEDQKIEQDGIKPEDKAHKA 34
|
| Danio rerio |
Query 1 MLCC#MRRTKQVEKNDE-DQKIEQDGIKPEDKAHKA 34
||||#+|||| ||||+| ||+|+|||||||+ ||||
Sbjct 1 MLCC#IRRTKPVEKNEEADQEIKQDGIKPEENAHKA 35
|
| Capra hircus |
Query 9 KQVEKNDEDQKIEQDGIKPEDKAHKA 34
||||||||||||||||||||||||||
Query 9 KQVEKNDEDQKIEQDGIKPEDKAHKA 34
|
| Tetraodon nigroviridis |
Query 1 MLCC#MRRTKQVEKNDE-DQKIEQDGI--KPEDKAHKA 34
||||#+|||| ||||+| +||+|||| |||||||||
Sbjct 1 MLCC#IRRTKPVEKNEEAEQKVEQDGTTNKPEDKAHKA 37
|
| Serinus canaria |
Query 8 TKQVEKNDE-DQKIEQDGIKPEDKAHKA 34
|||||||++ ||||||||||||||||||
Sbjct 1 TKQVEKNEDGDQKIEQDGIKPEDKAHKA 28
|
| Monodelphis domestica |
Query 11 VEKNDE-DQKIEQDGIKPEDKAHKA 34
|||||+ ||||||||||||||||||
Sbjct 21 VEKNDDADQKIEQDGIKPEDKAHKA 45
|
| Gallus gallus |
Query 11 VEKNDE-DQKIEQDGIKPEDKAHKA 34
||||++ ||||||||||||||||||
Sbjct 204 VEKNEDGDQKIEQDGIKPEDKAHKA 228
|
 Met
Met

 Sequence conservation (by blast)
Sequence conservation (by blast)