| # |
organism |
max score |
hits |
top seq |
| 1 |
Macaca mulatta |
103.00 |
2 |
PREDICTED: similar to Beta crystallin A4 (Beta-A4- |
| 2 |
Homo sapiens |
100.00 |
8 |
CRYBA4 |
| 3 |
Bos taurus |
86.30 |
6 |
crystallin, beta A4 |
| 4 |
Mus musculus |
78.60 |
8 |
unnamed protein product |
| 5 |
Canis lupus familiaris |
78.20 |
2 |
crystallin, beta A4 |
| 6 |
Pan troglodytes |
77.40 |
6 |
PREDICTED: crystallin, beta A4 |
| 7 |
Rattus norvegicus |
77.00 |
7 |
crystallin, beta A4 |
| 8 |
Monodelphis domestica |
72.00 |
3 |
PREDICTED: similar to Beta crystallin A4 (Beta-A4- |
| 9 |
Xenopus tropicalis |
63.90 |
2 |
Unknown (protein for MGC:121449) |
| 10 |
Tetraodon nigroviridis |
58.50 |
2 |
unnamed protein product |
| 11 |
Danio rerio |
57.00 |
5 |
hypothetical protein LOC553178 |
| 12 |
rats, Sprague-Dawley, lens, Peptide Partial, 24 aa |
54.30 |
1 |
beta-crystallin isoform A4 {N-terminal} |
| 13 |
N/A |
47.00 |
8 |
J |
| 14 |
Gallus gallus |
47.00 |
3 |
crystallin, beta A4 |
| 15 |
Xenopus laevis |
45.40 |
3 |
hypothetical protein LOC495288 |
| 16 |
Contains: Beta crystallin A3, isoform A1, Delta4 form; Beta crystallin A3, isoform A1, Delta7 form; Beta crystallin A3, isoform A1, Delta8 form |
41.20 |
1 |
CRBA1_BOVIN Beta crystallin A3 |
| 17 |
Oryctolagus cuniculus |
40.80 |
2 |
beta A3-crystallin |
| 18 |
Canis familiaris |
39.70 |
1 |
PREDICTED: similar to crystallin, beta B1 |
| 19 |
Cavia porcellus |
38.10 |
2 |
beta A3-crystallin |
| 20 |
Iguana iguana |
37.70 |
1 |
betaA2-crystallin |
| 21 |
Rana catesbeiana |
37.40 |
4 |
CRB32_RANCA Beta crystallin A3-2 emb |
| 22 |
Rana temporaria |
37.40 |
2 |
unnamed protein product |
| 23 |
Ovis aries |
35.40 |
1 |
crystallin, beta B3 |
| 24 |
Contains: Beta crystallin B1B |
35.40 |
1 |
CRBB1_RAT Beta crystallin B1 |
| 25 |
synthetic construct |
35.40 |
1 |
crystallin beta B1 |
| 26 |
Sus scrofa |
35.00 |
1 |
beta crystallin subunit beta B1 |
| organism | matching |
|---|
| Macaca mulatta |
Query 1 MFPGPISEGATMTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAECP 45
|||||||||||||||#||||||||||||||||||||||||||||||
Query 1 MFPGPISEGATMTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAECP 45
|
| Homo sapiens |
Query 1 MFPGPISEGATMTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAECP 45
|||||||||||||||#|||||| |||||||||||||||||||||||
Sbjct 1 MFPGPISEGATMTLQ#CTKSAGPWKMVVWDEDGFQGRRHEFTAECP 45
|
| Bos taurus |
Query 1 MFPGPISEGATMTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAECP 45
|| | ||| + |+||#|||||||||+|||||+||||||||||||||
Sbjct 4 MFSGSISETSGMSLQ#CTKSAGHWKIVVWDEEGFQGRRHEFTAECP 48
|
| Mus musculus |
Query 12 MTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAECP 45
||||#||||||||+||||||+||||||||||||||
Sbjct 1 MTLQ#CTKSAGHWRMVVWDEEGFQGRRHEFTAECP 34
|
| Canis lupus familiaris |
Query 12 MTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAECP 45
||||#|||||||||+|||||+||||||||||||||
Sbjct 1 MTLQ#CTKSAGHWKIVVWDEEGFQGRRHEFTAECP 34
|
| Pan troglodytes |
Query 12 MTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAECP 45
||||#|||||| |||||||||||||||||||||||
Sbjct 1 MTLQ#CTKSAGPWKMVVWDEDGFQGRRHEFTAECP 34
|
| Rattus norvegicus |
Query 12 MTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAECP 45
||||#||||||||++|||||+||||||||||||||
Sbjct 1 MTLQ#CTKSAGHWRVVVWDEEGFQGRRHEFTAECP 34
|
| Monodelphis domestica |
Query 12 MTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAECP 45
|+ |#||||+||||+||||++||||||||||||||
Sbjct 1 MSQQ#CTKSSGHWKIVVWDQEGFQGRRHEFTAECP 34
|
| Xenopus tropicalis |
Query 12 MTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAEC 44
|| #||| +||||++||||+ ||||||||||||
Sbjct 1 MTQH#CTKFSGHWKIIVWDEECFQGRRHEFTAEC 33
|
| Tetraodon nigroviridis |
Query 12 MTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAEC 44
|| #||| +||||++|+||+ |||||||||+||
Sbjct 1 MTHH#CTKFSGHWKIIVFDEECFQGRRHEFTSEC 33
|
| Danio rerio |
Query 12 MTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAEC 44
|| #||| +||||++|+||+ |||| ||||+||
Sbjct 1 MTHH#CTKFSGHWKIIVYDEECFQGRHHEFTSEC 33
|
| rats, Sprague-Dawley, lens, Peptide Partial, 24 aa |
Query 12 MTLQ#CTKSAGHWKMVVWDEDGFQG 35
||||#||||||||++|||||+||||
Sbjct 1 MTLQ#CTKSAGHWRVVVWDEEGFQG 24
|
| N/A |
Query 19 SAGHWKMVVWDEDGFQGRRHEFTAEC 44
|+| ||+||||| |||+||||| +|
Sbjct 8 SSGLWKIVVWDEPFFQGKRHEFTTDC 33
|
| Gallus gallus |
Query 19 SAGHWKMVVWDEDGFQGRRHEFTAEC 44
|+| ||+||||| |||+||||| +|
Sbjct 8 SSGLWKIVVWDEPFFQGKRHEFTTDC 33
|
| Xenopus laevis |
Query 12 MTLQ#CTKSAGHWKMVVWDEDGFQGRRHEFTAECP 45
|| # ++ | ||| ||+|+ |||+| || |||
Sbjct 1 MTSH#PMETQGQWKMTVWEEENFQGKRCEFHLECP 34
|
| Contains: Beta crystallin A3, isoform A1, Delta4 form; Beta crystallin A3, isoform A1, Delta7 form; Beta crystallin A3, isoform A1, Delta8 form |
Query 19 SAGHWKMVVWDEDGFQGRRHEFTAECP 45
| | ||+ ++|++ |||+| |||+ ||
Sbjct 27 SVGPWKITIYDQENFQGKRMEFTSSCP 53
|
| Oryctolagus cuniculus |
Query 19 SAGHWKMVVWDEDGFQGRRHEFTAECP 45
| | ||+ ++|++ |||+| |||+ ||
Sbjct 27 SLGPWKITIYDQENFQGKRMEFTSSCP 53
|
| Canis familiaris |
Query 3 PGPISEGATMTLQ#CTKSA----GHWKMVVWDEDGFQGRRHEFTAEC 44
||| | |+ # ||+ | +|+||++++ ||||| ||+ ||
Sbjct 31 PGPGPALAPATVP#TTKAGDLPPGSYKLVVFEQENFQGRRVEFSGEC 76
|
| Cavia porcellus |
Query 19 SAGHWKMVVWDEDGFQGRRHEFTAEC 44
| | ||+ ++|++ |||+| |||+ |
Sbjct 27 SLGPWKITIYDQENFQGKRMEFTSSC 52
|
| Iguana iguana |
Query 21 GHWKMVVWDEDGFQGRRHEFTAECP 45
| +|+ ||+|+ ||| | || |||
Sbjct 3 GQFKITVWEEENFQGERCEFLMECP 27
|
| Rana catesbeiana |
Query 21 GHWKMVVWDEDGFQGRRHEFTAEC 44
| ||+ |+|++ |||+| |||+ |
Sbjct 29 GPWKITVYDQENFQGKRMEFTSSC 52
|
| Rana temporaria |
Query 21 GHWKMVVWDEDGFQGRRHEFTAEC 44
| ||+ |+|++ |||+| |||+ |
Sbjct 12 GPWKITVYDQENFQGKRMEFTSSC 35
|
| Ovis aries |
Query 21 GHWKMVVWDEDGFQGRRHEFTAECP 45
| +|++|++ + |||+| | |||||
Sbjct 22 GSYKVIVYEMENFQGKRCELTAECP 46
|
| Contains: Beta crystallin B1B |
Query 3 PGPISEGATMTLQ#CTKSA----GHWKMVVWDEDGFQGRRHEFTAEC 44
||| |++ # | | +++||++++ ||||| ||+ ||
Sbjct 33 PGPTPVPASVPRP#AAKVGELPPGSYRLVVFEQENFQGRRVEFSGEC 78
|
| synthetic construct |
Query 3 PGPISEGATMTLQ#CTKSA----GHWKMVVWDEDGFQGRRHEFTAEC 44
|| |+ + # |+| |++++||++ + ||||| ||+ ||
Sbjct 35 PGTTLAPTTVPIT#SAKAAELPPGNYRLVVFELENFQGRRAEFSGEC 80
|
| Sus scrofa |
Query 21 GHWKMVVWDEDGFQGRRHEFTAEC 44
| +|+||++++ ||||| ||+ ||
Sbjct 54 GSYKLVVFEQENFQGRRVEFSGEC 77
|
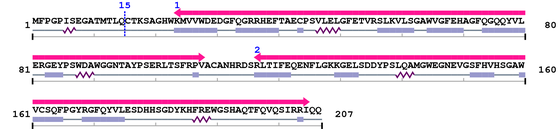
 Cys
Cys

 Sequence conservation (by blast)
Sequence conservation (by blast)