XSB0021 : PREDICTED: integrin, beta 7 [Macaca mulatta]
[ CaMP Format ]
This entry is computationally expanded from SB0062
* Basic Information
| Organism | Macaca mulatta (rhesus monkey) |
| Protein Names | integrin; beta 7 |
| Gene Names | ITGB7; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 1 mRNA, 6 ESTs, 24 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001087864 | XM_001087864 | 699405 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 674 aa
Average Mass: 72.550 kDa
Monoisotopic Mass: 72.502 kDa
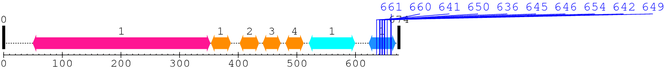
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Integrin_beta 1. | 50 | 352 | 489.0 | 6.6e-144 |
| EGF_2 1. | 354 | 387 | 11.6 | 0.14 |
| EGF_2 2. | 403 | 435 | 14.7 | 0.059 |
| EGF_2 3. | 442 | 472 | 20.2 | 0.0088 |
| EGF_2 4. | 481 | 511 | 31.5 | 3.4e-06 |
| Integrin_B_tail 1. | 521 | 599 | 86.5 | 9.3e-23 |
| Integrin_b_cyt 1. | 623 | 668 | 81.9 | 2.3e-21 |
| --- cleavage 661 (inside Integrin_b_cyt 623..668) --- | ||||
| --- cleavage 660 (inside Integrin_b_cyt 623..668) --- | ||||
| --- cleavage 641 (inside Integrin_b_cyt 623..668) --- | ||||
| --- cleavage 650 (inside Integrin_b_cyt 623..668) --- | ||||
| --- cleavage 636 (inside Integrin_b_cyt 623..668) --- | ||||
| --- cleavage 645 (inside Integrin_b_cyt 623..668) --- | ||||
| --- cleavage 646 (inside Integrin_b_cyt 623..668) --- | ||||
| --- cleavage 654 (inside Integrin_b_cyt 623..668) --- | ||||
| --- cleavage 642 (inside Integrin_b_cyt 623..668) --- | ||||
| --- cleavage 649 (inside Integrin_b_cyt 623..668) --- | ||||
3. Sequence Information
Fasta Sequence: XSB0021.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
10 [sites]
Cleavage sites (±10aa)
[Site 1] PLYKSAITTT661-INPRFQEADS
Thr661  Ile
Ile
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro652 | Leu653 | Tyr654 | Lys655 | Ser656 | Ala657 | Ile658 | Thr659 | Thr660 | Thr661 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ile662 | Asn663 | Pro664 | Arg665 | Phe666 | Gln667 | Glu668 | Ala669 | Asp670 | Ser671 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| REYSRFEKEQQQLNWKQDSNPLYKSAITTTINPRFQEADSPIL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 91.70 | 2 | PREDICTED: integrin, beta 7 |
| 2 | Homo sapiens | 89.70 | 14 | integrin, beta 7 |
| 3 | Pan troglodytes | 89.70 | 7 | PREDICTED: integrin, beta 7 isoform 2 |
| 4 | Bos taurus | 80.50 | 6 | PREDICTED: similar to Integrin beta-7 |
| 5 | Mus musculus | 75.10 | 10 | integrin beta 7 |
| 6 | Mus sp. | 75.10 | 1 | beta 7 integrin |
| 7 | Rattus norvegicus | 74.30 | 8 | integrin beta-7 subunit |
| 8 | Canis familiaris | 70.90 | 4 | PREDICTED: similar to Integrin beta-7 precursor |
| 9 | N/A | 65.90 | 21 | C |
| 10 | Gallus gallus | 51.20 | 6 | integrin, beta 2 |
| 11 | Danio rerio | 50.10 | 10 | integrin beta1 subunit-like protein 3 |
| 12 | Xenopus laevis | 49.30 | 6 | integrin beta-1 subunit |
| 13 | Tetraodon nigroviridis | 49.30 | 5 | unnamed protein product |
| 14 | Monodelphis domestica | 49.30 | 5 | PREDICTED: similar to integrin beta-1 subunit isof |
| 15 | Sus scrofa | 49.30 | 4 | integrin beta-1 subunit |
| 16 | synthetic construct | 49.30 | 3 | hypothetical protein |
| 17 | Ictalurus punctatus | 49.30 | 2 | beta-1 integrin |
| 18 | Oryctolagus cuniculus | 49.30 | 2 | integrin beta 1 |
| 19 | Felis catus | 49.30 | 1 | integrin beta 1 |
| 20 | Xenopus tropicalis | 49.30 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 21 | Pongo pygmaeus | 49.30 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 22 | Capra hircus | 48.50 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 23 | Ovis canadensis | 48.50 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 24 | Ovis aries | 48.50 | 1 | antigen CD18 |
| 25 | Bubalus bubalis | 48.50 | 1 | integrin beta-2 |
| 26 | Sigmodon hispidus | 46.60 | 1 | AF445415_1 integrin beta-2 precursor |
| 27 | Pacifastacus leniusculus | 43.10 | 1 | integrin |
| 28 | Anopheles gambiae str. PEST | 42.40 | 1 | ENSANGP00000021373 |
| 29 | Anopheles gambiae | 42.40 | 1 | integrin beta subunit |
| 30 | Aedes aegypti | 42.40 | 1 | integrin beta subunit |
| 31 | Tribolium castaneum | 42.00 | 2 | PREDICTED: similar to CG1560-PA |
| 32 | Crassostrea gigas | 42.00 | 1 | integrin beta cgh |
| 33 | Biomphalaria glabrata | 41.60 | 1 | beta integrin subunit |
| 34 | Strongylocentrotus purpuratus | 41.20 | 5 | integrin beta G subunit |
| 35 | Drosophila melanogaster | 40.80 | 1 | myospheroid CG1560-PA |
| 36 | Ostrinia furnacalis | 40.80 | 1 | integrin beta 1 precursor |
| 37 | Pseudoplusia includens | 40.80 | 1 | integrin beta 1 |
| 38 | Podocoryne carnea | 40.00 | 1 | AF308652_1 integrin beta chain |
| 39 | Oncorhynchus mykiss | 40.00 | 1 | CD18 protein |
| 40 | Acropora millepora | 39.70 | 1 | integrin subunit betaCn1 |
| 41 | Schistosoma japonicum | 37.40 | 1 | SJCHGC06221 protein |
| 42 | Apis mellifera | 36.60 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 43 | Caenorhabditis briggsae | 36.20 | 1 | Hypothetical protein CBG03601 |
| 44 | Canis lupus familiaris | 35.00 | 1 | integrin beta chain, beta 3 |
| 45 | Lytechinus variegatus | 35.00 | 1 | beta-C integrin subunit |
| 46 | Caenorhabditis elegans | 35.00 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 47 | rats, Peptide Partial, 723 aa | 35.00 | 1 | beta 3 integrin, GPIIIA |
| 48 | Halocynthia roretzi | 35.00 | 1 | integrin beta Hr2 precursor |
| 49 | mice, Peptide Partial, 680 aa | 35.00 | 1 | beta 3 integrin, GPIIIA |
| 50 | Equus caballus | 35.00 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#||||||||||||| Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEADSPIL 674 |
| Homo sapiens | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#||||||||||| | Sbjct 756 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEADSPTL 798 |
| Pan troglodytes | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#||||||||||| | Sbjct 634 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEADSPTL 676 |
| Bos taurus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEADSPIL 674 ||+ |||||+| |||||| ||||+||||||#+||||||||||+| Sbjct 758 REFHRFEKERQHLNWKQDHNPLYQSAITTT#VNPRFQEADSPVL 800 |
| Mus musculus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||| ||||||||||||||+|||||||||||#+||||| Sbjct 754 REYRRFEKEQQQLNWKQDNNPLYKSAITTT#VNPRFQ 789 |
| Mus sp. | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||| ||||||||||||||+|||||||||||#+||||| Sbjct 755 REYRRFEKEQQQLNWKQDNNPLYKSAITTT#VNPRFQ 790 |
| Rattus norvegicus | Query 633 EYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||||||||+||||||||||||||||+|||#+||||| Sbjct 590 EYSRFEKERQQLNWKQDSNPLYKSAVTTT#VNPRFQ 624 |
| Canis familiaris | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEADS 671 ||+|||||||+ |||||++||||+||||||#+||+||| | Sbjct 688 REFSRFEKEQKHLNWKQENNPLYRSAITTT#VNPQFQETGS 727 |
| N/A | Query 644 LNWKQDSNPLYKSAITTT#INPRFQEADSPIL 674 ||||||||||||||||||#||||||||||| | Sbjct 1 LNWKQDSNPLYKSAITTT#INPRFQEADSPTL 31 |
| Gallus gallus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 ||| |||||+ + | + |||+||| || #+|||| Sbjct 735 REYRRFEKEKSKAKWNEADNPLFKSATTTV#MNPRF 769 |
| Danio rerio | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||+ +||||+ | ||+||||+|| #+|||++ Sbjct 750 REFDKFEKEKNNAKWDTGENPIYKSAVTTV#VNPRYE 785 |
| Xenopus laevis | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||+++||||+ | ||+||||+|| #+||+++ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYE 796 |
| Tetraodon nigroviridis | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||+++||||+ | ||+||||+|| #+||+++ Sbjct 754 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYE 789 |
| Monodelphis domestica | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||+++||||+ | ||+||||+|| #+||+++ Sbjct 762 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYE 797 |
| Sus scrofa | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||+++||||+ | ||+||||+|| #+||+++ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYE 796 |
| synthetic construct | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||+++||||+ | ||+||||+|| #+||+++ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYE 796 |
| Ictalurus punctatus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||+++||||+ | ||+||||+|| #+||+++ Sbjct 770 REFAKFEKEKMNAKWDAGENPIYKSAVTTV#VNPKYE 805 |
| Oryctolagus cuniculus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||+++||||+ | ||+||||+|| #+||+++ Sbjct 26 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYE 61 |
| Felis catus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||+++||||+ | ||+||||+|| #+||+++ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYE 796 |
| Xenopus tropicalis | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||+++||||+ | ||+||||+|| #+||+++ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYE 796 |
| Pongo pygmaeus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 ||+++||||+ | ||+||||+|| #+||+++ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTV#VNPKYE 796 |
| Capra hircus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEA 669 ||| |||||+ + | | |||+||| || #+||+| |+ Sbjct 734 REYHRFEKEKLKSQWNND-NPLFKSATTTV#MNPKFAES 770 |
| Ovis canadensis | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEA 669 ||| |||||+ + | | |||+||| || #+||+| |+ Sbjct 734 REYHRFEKEKLKSQWNND-NPLFKSATTTV#MNPKFAES 770 |
| Ovis aries | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEA 669 ||| |||||+ + | | |||+||| || #+||+| |+ Sbjct 734 REYHRFEKEKLKSQWNND-NPLFKSATTTV#MNPKFAES 770 |
| Bubalus bubalis | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEA 669 ||| |||||+ + | | |||+||| || #+||+| |+ Sbjct 733 REYHRFEKEKLKSQWNND-NPLFKSATTTV#MNPKFAES 769 |
| Sigmodon hispidus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEA 669 ||| ||||+ + | | |||+||| || #+||+| |+ Sbjct 733 REYRHFEKEKLKSQWNND-NPLFKSATTTV#MNPKFAES 769 |
| Pacifastacus leniusculus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQEA 669 |||++||||++ + |+ ||||||| || # || | ++ Sbjct 763 REYAKFEKERKLPSGKRAENPLYKSAKTTF#QNPAFAQS 800 |
| Anopheles gambiae str. PEST | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 ||++|||||+ | ||+|| | || # || + Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTF#KNPTY 834 |
| Anopheles gambiae | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 ||++|||||+ | ||+|| | || # || + Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTF#KNPTY 834 |
| Aedes aegypti | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 ||++|||||+ | ||+|| | || # || + Sbjct 798 REFARFEKERMMAKWDTGENPIYKQATTTF#KNPTY 832 |
| Tribolium castaneum | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQ 667 +||++|| |+|++ | ++ ||||+ | +| # || ++ Sbjct 717 QEYAKFENERQKMQWHRNDNPLYRQATSTF#KNPVYR 752 |
| Crassostrea gigas | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQE 668 || +||||| | ||+|| | +| #+|| +++ Sbjct 763 RELARFEKEAMNARWDTGENPIYKQATSTF#VNPTYRQ 799 |
| Biomphalaria glabrata | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 ||+++||||+| | ||+|| | +| # || + Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATSTF#KNPTY 785 |
| Strongylocentrotus purpuratus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQE 668 ||++ ||||+ +| |+ ||+|| + +| # || +|+ Sbjct 747 REFASFEKERAGTHWGQNENPIYKPSTSTF#KNPTYQK 783 |
| Drosophila melanogaster | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 ||++|||||+ | ||+|| | +| # || + Sbjct 809 REFARFEKERMNAKWDTGENPIYKQATSTF#KNPMY 843 |
| Ostrinia furnacalis | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 ||++|||||+ | ||+|| | +| # || + Sbjct 794 REFARFEKERMMAKWDTGENPIYKQATSTF#KNPTY 828 |
| Pseudoplusia includens | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 ||++|||||+ | ||+|| | +| # || + Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATSTF#KNPTY 834 |
| Podocoryne carnea | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 ||+++|||++| | ||+|| | +| # || + Sbjct 744 REFAKFEKDRQNPKWDSGENPIYKKATSTF#QNPMY 778 |
| Oncorhynchus mykiss | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 +|+ ||| ||++ | |||+ +| || # || | Sbjct 740 KEFRRFENEQKKGKWSPGDNPLFMNATTTV#ANPTF 774 |
| Acropora millepora | Query 633 EYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 || +||+|+ | ++ ||||++| || # || + Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTF#ENPTY 787 |
| Schistosoma japonicum | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRFQE 668 || + |+++ + + |+ ||+++| | #+|| |+| Sbjct 249 RELANFKQQGENMRWEMAENPIFESPTTNV#LNPTFEE 285 |
| Apis mellifera | Query 632 REYSRFEKEQQQLN-WKQDSNPLYKSAITTT#INPRF 666 ||+ +||+|+ | | + ||||| | +| # || | Sbjct 778 REFKKFERERILANKWNRRDNPLYKEATSTF#KNPTF 813 |
| Caenorhabditis briggsae | Query 633 EYSRFEKEQQQLNWKQDSNPLYKSAITTT#INP 664 ||++| |+ | + ||+|| | || # || Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTF#KNP 803 |
| Canis lupus familiaris | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#IN 663 +|+++||+|+ + | +||||| | +| # | Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTF#TN 778 |
| Lytechinus variegatus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 ||+ ||||+ |+ ||+|| + + # || + Sbjct 769 REFQNFEKERANATWEGGENPIYKPSTSVF#KNPTY 803 |
| Caenorhabditis elegans | Query 633 EYSRFEKEQQQLNWKQDSNPLYKSAITTT#INP 664 ||+ | |+ | + ||+|| | || # || Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTF#KNP 802 |
| rats, Peptide Partial, 723 aa | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#IN 663 +|+++||+|+ + | +||||| | +| # | Sbjct 686 KEFAKFEEERARAKWDTANNPLYKEATSTF#TN 717 |
| Halocynthia roretzi | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#INPRF 666 ||| +|+||+ | + ||+| +| + # || | Sbjct 797 REYEKFKKEEDLRKWTKGENPVYVNASSKF#DNPMF 831 |
| mice, Peptide Partial, 680 aa | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#IN 663 +|+++||+|+ + | +||||| | +| # | Sbjct 647 KEFAKFEEERARAKWDTANNPLYKEATSTF#TN 678 |
| Equus caballus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTT#IN 663 +|+++||+|+ + | +||||| | +| # | Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTF#TN 778 |
[Site 2] NPLYKSAITT660-TINPRFQEAD
Thr660  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn651 | Pro652 | Leu653 | Tyr654 | Lys655 | Ser656 | Ala657 | Ile658 | Thr659 | Thr660 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr661 | Ile662 | Asn663 | Pro664 | Arg665 | Phe666 | Gln667 | Glu668 | Ala669 | Asp670 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RREYSRFEKEQQQLNWKQDSNPLYKSAITTTINPRFQEADSPIL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 93.60 | 11 | PREDICTED: integrin, beta 7 |
| 2 | Homo sapiens | 91.70 | 19 | integrin, beta 7 |
| 3 | Pan troglodytes | 91.70 | 13 | PREDICTED: integrin, beta 7 isoform 2 |
| 4 | Bos taurus | 82.40 | 13 | PREDICTED: similar to Integrin beta-7 |
| 5 | Mus musculus | 77.00 | 14 | integrin beta 7 |
| 6 | Mus sp. | 77.00 | 1 | beta 7 integrin |
| 7 | Rattus norvegicus | 75.50 | 10 | integrin beta-7 subunit |
| 8 | Canis familiaris | 72.80 | 10 | PREDICTED: similar to Integrin beta-7 precursor |
| 9 | N/A | 65.90 | 25 | C |
| 10 | Gallus gallus | 53.10 | 7 | integrin, beta 2 |
| 11 | Danio rerio | 52.00 | 11 | integrin beta1 subunit-like protein 3 |
| 12 | Xenopus laevis | 51.20 | 6 | integrin beta 1 |
| 13 | Monodelphis domestica | 51.20 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 14 | synthetic construct | 51.20 | 5 | hypothetical protein |
| 15 | Tetraodon nigroviridis | 51.20 | 5 | unnamed protein product |
| 16 | Sus scrofa | 51.20 | 5 | integrin beta-1 subunit |
| 17 | Pongo pygmaeus | 51.20 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 18 | Oryctolagus cuniculus | 51.20 | 2 | integrin beta 1 |
| 19 | Ictalurus punctatus | 51.20 | 2 | beta-1 integrin |
| 20 | Xenopus tropicalis | 51.20 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 21 | Felis catus | 49.30 | 1 | integrin beta 1 |
| 22 | Ovis aries | 48.50 | 2 | antigen CD18 |
| 23 | Bubalus bubalis | 48.50 | 1 | integrin beta-2 |
| 24 | Capra hircus | 48.50 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 25 | Ovis canadensis | 48.50 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 26 | Sigmodon hispidus | 46.60 | 1 | AF445415_1 integrin beta-2 precursor |
| 27 | Pacifastacus leniusculus | 45.10 | 1 | integrin |
| 28 | Anopheles gambiae str. PEST | 44.30 | 1 | ENSANGP00000021373 |
| 29 | Anopheles gambiae | 44.30 | 1 | integrin beta subunit |
| 30 | Aedes aegypti | 44.30 | 1 | integrin beta subunit |
| 31 | Tribolium castaneum | 43.90 | 2 | PREDICTED: similar to CG1560-PA |
| 32 | Pseudoplusia includens | 42.70 | 1 | integrin beta 1 |
| 33 | Crassostrea gigas | 42.70 | 1 | integrin beta cgh |
| 34 | Ostrinia furnacalis | 42.70 | 1 | integrin beta 1 precursor |
| 35 | Drosophila melanogaster | 42.70 | 1 | myospheroid CG1560-PA |
| 36 | Podocoryne carnea | 42.00 | 1 | AF308652_1 integrin beta chain |
| 37 | Biomphalaria glabrata | 41.60 | 1 | beta integrin subunit |
| 38 | Strongylocentrotus purpuratus | 41.20 | 5 | integrin beta G subunit |
| 39 | Acropora millepora | 40.40 | 1 | integrin subunit betaCn1 |
| 40 | Oncorhynchus mykiss | 40.00 | 1 | CD18 protein |
| 41 | Schistosoma japonicum | 39.30 | 1 | SJCHGC06221 protein |
| 42 | Caenorhabditis briggsae | 37.70 | 1 | Hypothetical protein CBG03601 |
| 43 | rats, Peptide Partial, 723 aa | 37.00 | 1 | beta 3 integrin, GPIIIA |
| 44 | Canis lupus familiaris | 37.00 | 1 | integrin beta chain, beta 3 |
| 45 | mice, Peptide Partial, 680 aa | 37.00 | 1 | beta 3 integrin, GPIIIA |
| 46 | Lytechinus variegatus | 37.00 | 1 | beta-C integrin subunit |
| 47 | Equus caballus | 37.00 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 48 | Caenorhabditis elegans | 36.60 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 49 | Apis mellifera | 36.60 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 50 | Halocynthia roretzi | 35.80 | 1 | integrin beta Hr2 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||||| Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEADSPIL 674 |
| Homo sapiens | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||| | Sbjct 755 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEADSPTL 798 |
| Pan troglodytes | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||| | Sbjct 633 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEADSPTL 676 |
| Bos taurus | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEADSPIL 674 |||+ |||||+| |||||| ||||+|||||#|+||||||||||+| Sbjct 757 RREFHRFEKERQHLNWKQDHNPLYQSAITT#TVNPRFQEADSPVL 800 |
| Mus musculus | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||| ||||||||||||||+||||||||||#|+||||| Sbjct 753 RREYRRFEKEQQQLNWKQDNNPLYKSAITT#TVNPRFQ 789 |
| Mus sp. | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||| ||||||||||||||+||||||||||#|+||||| Sbjct 754 RREYRRFEKEQQQLNWKQDNNPLYKSAITT#TVNPRFQ 790 |
| Rattus norvegicus | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 | ||||||||+||||||||||||||||+||#|+||||| Sbjct 588 RLEYSRFEKERQQLNWKQDSNPLYKSAVTT#TVNPRFQ 624 |
| Canis familiaris | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEADS 671 |||+|||||||+ |||||++||||+|||||#|+||+||| | Sbjct 687 RREFSRFEKEQKHLNWKQENNPLYRSAITT#TVNPQFQETGS 727 |
| N/A | Query 644 LNWKQDSNPLYKSAITT#TINPRFQEADSPIL 674 |||||||||||||||||#|||||||||||| | Sbjct 1 LNWKQDSNPLYKSAITT#TINPRFQEADSPTL 31 |
| Gallus gallus | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 |||| |||||+ + | + |||+||| ||# +|||| Sbjct 734 RREYRRFEKEKSKAKWNEADNPLFKSATTT#VMNPRF 769 |
| Danio rerio | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||+ +||||+ | ||+||||+||# +|||++ Sbjct 749 RREFDKFEKEKNNAKWDTGENPIYKSAVTT#VVNPRYE 785 |
| Xenopus laevis | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||+++||||+ | ||+||||+||# +||+++ Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYE 796 |
| Monodelphis domestica | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||+++||||+ | ||+||||+||# +||+++ Sbjct 761 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYE 797 |
| synthetic construct | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||+++||||+ | ||+||||+||# +||+++ Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYE 796 |
| Tetraodon nigroviridis | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||+++||||+ | ||+||||+||# +||+++ Sbjct 753 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYE 789 |
| Sus scrofa | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||+++||||+ | ||+||||+||# +||+++ Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYE 796 |
| Pongo pygmaeus | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||+++||||+ | ||+||||+||# +||+++ Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYE 796 |
| Oryctolagus cuniculus | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||+++||||+ | ||+||||+||# +||+++ Sbjct 25 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYE 61 |
| Ictalurus punctatus | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||+++||||+ | ||+||||+||# +||+++ Sbjct 769 RREFAKFEKEKMNAKWDAGENPIYKSAVTT#VVNPKYE 805 |
| Xenopus tropicalis | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |||+++||||+ | ||+||||+||# +||+++ Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYE 796 |
| Felis catus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 ||+++||||+ | ||+||||+||# +||+++ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYE 796 |
| Ovis aries | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEA 669 ||| |||||+ + | | |||+||| ||# +||+| |+ Sbjct 734 REYHRFEKEKLKSQWNND-NPLFKSATTT#VMNPKFAES 770 |
| Bubalus bubalis | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEA 669 ||| |||||+ + | | |||+||| ||# +||+| |+ Sbjct 733 REYHRFEKEKLKSQWNND-NPLFKSATTT#VMNPKFAES 769 |
| Capra hircus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEA 669 ||| |||||+ + | | |||+||| ||# +||+| |+ Sbjct 734 REYHRFEKEKLKSQWNND-NPLFKSATTT#VMNPKFAES 770 |
| Ovis canadensis | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEA 669 ||| |||||+ + | | |||+||| ||# +||+| |+ Sbjct 734 REYHRFEKEKLKSQWNND-NPLFKSATTT#VMNPKFAES 770 |
| Sigmodon hispidus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEA 669 ||| ||||+ + | | |||+||| ||# +||+| |+ Sbjct 733 REYRHFEKEKLKSQWNND-NPLFKSATTT#VMNPKFAES 769 |
| Pacifastacus leniusculus | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEA 669 ||||++||||++ + |+ ||||||| ||# || | ++ Sbjct 762 RREYAKFEKERKLPSGKRAENPLYKSAKTT#FQNPAFAQS 800 |
| Anopheles gambiae str. PEST | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 |||++|||||+ | ||+|| | ||# || + Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTY 834 |
| Anopheles gambiae | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 |||++|||||+ | ||+|| | ||# || + Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTY 834 |
| Aedes aegypti | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 |||++|||||+ | ||+|| | ||# || + Sbjct 797 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTY 832 |
| Tribolium castaneum | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQ 667 |+||++|| |+|++ | ++ ||||+ | +|# || ++ Sbjct 716 RQEYAKFENERQKMQWHRNDNPLYRQATST#FKNPVYR 752 |
| Pseudoplusia includens | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 |||++|||||+ | ||+|| | +|# || + Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATST#FKNPTY 834 |
| Crassostrea gigas | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQE 668 +|| +||||| | ||+|| | +|# +|| +++ Sbjct 762 KRELARFEKEAMNARWDTGENPIYKQATST#FVNPTYRQ 799 |
| Ostrinia furnacalis | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 |||++|||||+ | ||+|| | +|# || + Sbjct 793 RREFARFEKERMMAKWDTGENPIYKQATST#FKNPTY 828 |
| Drosophila melanogaster | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 |||++|||||+ | ||+|| | +|# || + Sbjct 808 RREFARFEKERMNAKWDTGENPIYKQATST#FKNPMY 843 |
| Podocoryne carnea | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 |||+++|||++| | ||+|| | +|# || + Sbjct 743 RREFAKFEKDRQNPKWDSGENPIYKKATST#FQNPMY 778 |
| Biomphalaria glabrata | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 ||+++||||+| | ||+|| | +|# || + Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATST#FKNPTY 785 |
| Strongylocentrotus purpuratus | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQE 668 ||++ ||||+ +| |+ ||+|| + +|# || +|+ Sbjct 747 REFASFEKERAGTHWGQNENPIYKPSTST#FKNPTYQK 783 |
| Acropora millepora | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 | || +||+|+ | ++ ||||++| ||# || + Sbjct 752 RIEYQKFERERMHSKWTREKNPLYQAAKTT#FENPTY 787 |
| Oncorhynchus mykiss | Query 632 REYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 +|+ ||| ||++ | |||+ +| ||# || | Sbjct 740 KEFRRFENEQKKGKWSPGDNPLFMNATTT#VANPTF 774 |
| Schistosoma japonicum | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQE 668 ||| + |+++ + + |+ ||+++| | # +|| |+| Sbjct 248 RRELANFKQQGENMRWEMAENPIFESPTTN#VLNPTFEE 285 |
| Caenorhabditis briggsae | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINP 664 | ||++| |+ | + ||+|| | ||# || Sbjct 770 RAEYAKFNNERLMAKWDTNENPIYKQATTT#FKNP 803 |
| rats, Peptide Partial, 723 aa | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TIN 663 |+|+++||+|+ + | +||||| | +|# | Sbjct 685 RKEFAKFEEERARAKWDTANNPLYKEATST#FTN 717 |
| Canis lupus familiaris | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TIN 663 |+|+++||+|+ + | +||||| | +|# | Sbjct 746 RKEFAKFEEERARAKWDTANNPLYKEATST#FTN 778 |
| mice, Peptide Partial, 680 aa | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TIN 663 |+|+++||+|+ + | +||||| | +|# | Sbjct 646 RKEFAKFEEERARAKWDTANNPLYKEATST#FTN 678 |
| Lytechinus variegatus | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 |||+ ||||+ |+ ||+|| + + # || + Sbjct 768 RREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTY 803 |
| Equus caballus | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TIN 663 |+|+++||+|+ + | +||||| | +|# | Sbjct 746 RKEFAKFEEERARAKWDTANNPLYKEATST#FTN 778 |
| Caenorhabditis elegans | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINP 664 | ||+ | |+ | + ||+|| | ||# || Sbjct 769 RSEYATFNNERLMAKWDTNENPIYKQATTT#FKNP 802 |
| Apis mellifera | Query 632 REYSRFEKEQQQLN-WKQDSNPLYKSAITT#TINPRF 666 ||+ +||+|+ | | + ||||| | +|# || | Sbjct 778 REFKKFERERILANKWNRRDNPLYKEATST#FKNPTF 813 |
| Halocynthia roretzi | Query 631 RREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRF 666 +||| +|+||+ | + ||+| +| + # || | Sbjct 796 KREYEKFKKEEDLRKWTKGENPVYVNASSK#FDNPMF 831 |
[Site 3] REYSRFEKEQ641-QQLNWKQDSN
Gln641  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg632 | Glu633 | Tyr634 | Ser635 | Arg636 | Phe637 | Glu638 | Lys639 | Glu640 | Gln641 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln642 | Gln643 | Leu644 | Asn645 | Trp646 | Lys647 | Gln648 | Asp649 | Ser650 | Asn651 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| VAVGLGLVLAYRLSVEIYDRREYSRFEKEQQQLNWKQDSNPLYKSAITTTINPRFQEADS |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 122.00 | 25 | integrin, beta 7 |
| 2 | Pan troglodytes | 122.00 | 15 | PREDICTED: integrin, beta 7 isoform 2 |
| 3 | Macaca mulatta | 122.00 | 13 | PREDICTED: integrin, beta 7 |
| 4 | Mus musculus | 111.00 | 14 | integrin beta 7 |
| 5 | Bos taurus | 111.00 | 13 | PREDICTED: similar to Integrin beta-7 |
| 6 | Mus sp. | 111.00 | 1 | beta 7 integrin |
| 7 | Rattus norvegicus | 109.00 | 10 | integrin beta-7 subunit |
| 8 | Canis familiaris | 107.00 | 10 | PREDICTED: similar to Integrin beta-7 precursor |
| 9 | Gallus gallus | 70.50 | 7 | integrin, beta 2 |
| 10 | Danio rerio | 67.80 | 14 | integrin beta1 subunit-like protein 3 |
| 11 | N/A | 66.60 | 30 | - |
| 12 | Sus scrofa | 66.60 | 6 | integrin beta-1 subunit |
| 13 | Monodelphis domestica | 66.60 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 14 | Tetraodon nigroviridis | 66.60 | 6 | unnamed protein product |
| 15 | Xenopus laevis | 66.60 | 6 | integrin beta-1 subunit |
| 16 | synthetic construct | 66.60 | 5 | hypothetical protein |
| 17 | Oryctolagus cuniculus | 66.60 | 2 | integrin beta 1 |
| 18 | Pongo pygmaeus | 66.60 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 19 | Xenopus tropicalis | 66.60 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 20 | Ictalurus punctatus | 65.10 | 2 | beta-1 integrin |
| 21 | Felis catus | 64.30 | 1 | integrin beta 1 |
| 22 | Aedes aegypti | 58.90 | 1 | integrin beta subunit |
| 23 | Pacifastacus leniusculus | 58.50 | 1 | integrin |
| 24 | Drosophila melanogaster | 58.20 | 1 | myospheroid CG1560-PA |
| 25 | Strongylocentrotus purpuratus | 57.80 | 6 | integrin beta G subunit |
| 26 | Anopheles gambiae | 57.40 | 2 | integrin beta subunit |
| 27 | Anopheles gambiae str. PEST | 57.40 | 2 | ENSANGP00000021373 |
| 28 | Biomphalaria glabrata | 57.40 | 1 | beta integrin subunit |
| 29 | Ostrinia furnacalis | 57.40 | 1 | integrin beta 1 precursor |
| 30 | Podocoryne carnea | 57.00 | 1 | AF308652_1 integrin beta chain |
| 31 | Bubalus bubalis | 56.60 | 1 | integrin beta-2 |
| 32 | Capra hircus | 56.60 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 33 | Pseudoplusia includens | 56.20 | 1 | integrin beta 1 |
| 34 | Ovis aries | 55.80 | 2 | antigen CD18 |
| 35 | Ovis canadensis | 55.80 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 36 | Tribolium castaneum | 54.30 | 3 | PREDICTED: similar to CG1560-PA |
| 37 | Sigmodon hispidus | 53.90 | 1 | AF445415_1 integrin beta-2 precursor |
| 38 | Lytechinus variegatus | 53.50 | 1 | beta-C integrin subunit |
| 39 | Crassostrea gigas | 52.80 | 1 | integrin beta cgh |
| 40 | Schistosoma japonicum | 52.40 | 1 | SJCHGC06221 protein |
| 41 | rats, Peptide Partial, 723 aa | 50.80 | 1 | beta 3 integrin, GPIIIA |
| 42 | Canis lupus familiaris | 50.80 | 1 | integrin beta chain, beta 3 |
| 43 | Equus caballus | 50.80 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 44 | mice, Peptide Partial, 680 aa | 50.80 | 1 | beta 3 integrin, GPIIIA |
| 45 | Caenorhabditis briggsae | 49.30 | 1 | Hypothetical protein CBG03601 |
| 46 | Caenorhabditis elegans | 48.10 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 47 | Apis mellifera | 47.80 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 48 | Oncorhynchus mykiss | 45.40 | 1 | CD18 protein |
| 49 | Acropora millepora | 44.70 | 1 | integrin subunit betaCn1 |
| 50 | Manduca sexta | 43.50 | 1 | plasmatocyte-specific integrin beta 1 |
| 51 | Halocynthia roretzi | 42.40 | 2 | integrin beta Hr2 precursor |
| 52 | Cyprinus carpio | 37.00 | 2 | CD18 type 2 |
| 53 | Spodoptera frugiperda | 36.20 | 1 | integrin beta 1 |
| 54 | Ophlitaspongia tenuis | 34.70 | 1 | integrin subunit betaPo1 |
| 55 | Geodia cydonium | 34.30 | 1 | integrin beta subunit |
| 56 | Suberites domuncula | 33.50 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEADS 671 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEADS 671 |
| Pan troglodytes | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEADS 671 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEADS 671 |
| Macaca mulatta | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEADS 671 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEADS 671 |
| Mus musculus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 ||||||||||||||||||||||| ||||||#||||||||+|||||||||||+||||| Sbjct 734 VAVGLGLVLAYRLSVEIYDRREYRRFEKEQ#QQLNWKQDNNPLYKSAITTTVNPRFQ 789 |
| Bos taurus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEADS 671 ||||||||||||||||||||||+ |||||+#| |||||| ||||+||||||+||||||||| Sbjct 738 VAVGLGLVLAYRLSVEIYDRREFHRFEKER#QHLNWKQDHNPLYQSAITTTVNPRFQEADS 797 |
| Mus sp. | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 ||||||||||||||||||||||| ||||||#||||||||+|||||||||||+||||| Sbjct 735 VAVGLGLVLAYRLSVEIYDRREYRRFEKEQ#QQLNWKQDNNPLYKSAITTTVNPRFQ 790 |
| Rattus norvegicus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 ||||||||||||||||+||| ||||||||+#||||||||||||||||+|||+||||| Sbjct 569 VAVGLGLVLAYRLSVEVYDRLEYSRFEKER#QQLNWKQDSNPLYKSAVTTTVNPRFQ 624 |
| Canis familiaris | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEADS 671 ||||||||||||||||||||||+|||||||#+ |||||++||||+||||||+||+||| | Sbjct 668 VAVGLGLVLAYRLSVEIYDRREFSRFEKEQ#KHLNWKQENNPLYRSAITTTVNPQFQETGS 727 |
| Gallus gallus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 +|| |+| +|| ||+||||| |||||+# + | + |||+||| || +|||| Sbjct 717 IGLLLLLTWRLLTEIFDRREYRRFEKEK#SKAKWNEADNPLFKSATTTVMNPRF 769 |
| Danio rerio | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+| ++| + |+||||+ +||||+# | ||+||||+|| +|||++ Sbjct 730 VLIGLALLLIWKLLMVIHDRREFDKFEKEK#NNAKWDTGENPIYKSAVTTVVNPRYE 785 |
| N/A | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+| ++| + |+||||+++||||+# | ||+||||+|| +||+++ Sbjct 718 VLIGLALLLIWKLLMIIHDRREFAKFEKEK#MNAKWDTGENPIYKSAVTTVVNPKYE 773 |
| Sus scrofa | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+| ++| + |+||||+++||||+# | ||+||||+|| +||+++ Sbjct 741 VLIGLALLLIWKLLMIIHDRREFAKFEKEK#MNAKWDTGENPIYKSAVTTVVNPKYE 796 |
| Monodelphis domestica | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+| ++| + |+||||+++||||+# | ||+||||+|| +||+++ Sbjct 742 VLIGLALLLIWKLLMIIHDRREFAKFEKEK#MNAKWDTGENPIYKSAVTTVVNPKYE 797 |
| Tetraodon nigroviridis | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+| ++| + |+||||+++||||+# | ||+||||+|| +||+++ Sbjct 734 VLIGLALLLIWKLLMIIHDRREFAKFEKEK#MNAKWDTGENPIYKSAVTTVVNPKYE 789 |
| Xenopus laevis | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+| ++| + |+||||+++||||+# | ||+||||+|| +||+++ Sbjct 741 VLIGLALLLIWKLLMIIHDRREFAKFEKEK#MNAKWDTGENPIYKSAVTTVVNPKYE 796 |
| synthetic construct | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+| ++| + |+||||+++||||+# | ||+||||+|| +||+++ Sbjct 741 VLIGLALLLIWKLLMIIHDRREFAKFEKEK#MNAKWDTGENPIYKSAVTTVVNPKYE 796 |
| Oryctolagus cuniculus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+| ++| + |+||||+++||||+# | ||+||||+|| +||+++ Sbjct 6 VLIGLALLLIWKLLMIIHDRREFAKFEKEK#MNAKWDTGENPIYKSAVTTVVNPKYE 61 |
| Pongo pygmaeus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+| ++| + |+||||+++||||+# | ||+||||+|| +||+++ Sbjct 741 VLIGLALLLIWKLLMIIHDRREFAKFEKEK#MNAKWDTGENPIYKSAVTTVVNPKYE 796 |
| Xenopus tropicalis | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+| ++| + |+||||+++||||+# | ||+||||+|| +||+++ Sbjct 741 VLIGLALLLIWKLLMIIHDRREFAKFEKEK#MNAKWDTGENPIYKSAVTTVVNPKYE 796 |
| Ictalurus punctatus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+ ++| + |+||||+++||||+# | ||+||||+|| +||+++ Sbjct 750 VLIGLALLFIWKLLMIIHDRREFAKFEKEK#MNAKWDAGENPIYKSAVTTVVNPKYE 805 |
| Felis catus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQ 667 | +|| |+| ++| + |+| ||+++||||+# | ||+||||+|| +||+++ Sbjct 741 VLIGLALLLIWKLLMIIHDTREFAKFEKEK#MNAKWDTGENPIYKSAVTTVVNPKYE 796 |
| Aedes aegypti | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 | +|| ++| ++| |+||||++|||||+# | ||+|| | || || + Sbjct 778 VLIGLAVLLLWKLLTTIHDRREFARFEKER#MMAKWDTGENPIYKQATTTFKNPTY 832 |
| Pacifastacus leniusculus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEA 669 ||+|| +| ++| ++|||||++||||+#+ + |+ ||||||| || || | ++ Sbjct 743 VAIGLLTLLIWKLLTTLHDRREYAKFEKER#KLPSGKRAENPLYKSAKTTFQNPAFAQS 800 |
| Drosophila melanogaster | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 | ||| ++| ++| |+||||++|||||+# | ||+|| | +| || + Sbjct 789 VLVGLAILLLWKLLTTIHDRREFARFEKER#MNAKWDTGENPIYKQATSTFKNPMY 843 |
| Strongylocentrotus purpuratus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQE 668 + ||| |+| +|| | + | ||++ ||||+# +| |+ ||+|| + +| || +|+ Sbjct 727 IIVGLALLLVWRLLVYVQDSREFASFEKER#AGTHWGQNENPIYKPSTSTFKNPTYQK 783 |
| Anopheles gambiae | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 | +|+ ++| +++ |+||||++|||||+# | ||+|| | || || + Sbjct 780 VLIGMAVLLLWKVLTSIHDRREFARFEKER#MMAKWDTGENPIYKQATTTFKNPTY 834 |
| Anopheles gambiae str. PEST | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 | +|+ ++| +++ |+||||++|||||+# | ||+|| | || || + Sbjct 780 VLIGMAVLLLWKVLTSIHDRREFARFEKER#MMAKWDTGENPIYKQATTTFKNPTY 834 |
| Biomphalaria glabrata | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 ||||| |+| ++| |+| ||+++||||+#| | ||+|| | +| || + Sbjct 731 VAVGLFLLLLWKLLTFIHDTREFAKFEKER#QNAKWDTGENPIYKQATSTFKNPTY 785 |
| Ostrinia furnacalis | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 | ||| |++ +++ |+||||++|||||+# | ||+|| | +| || + Sbjct 774 VLVGLALLMLWKMVTTIHDRREFARFEKER#MMAKWDTGENPIYKQATSTFKNPTY 828 |
| Podocoryne carnea | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 | +|| |+| ++| | ||||+++|||++#| | ||+|| | +| || + Sbjct 724 VGIGLALLLIWKLLATIQDRREFAKFEKDR#QNPKWDSGENPIYKKATSTFQNPMY 778 |
| Bubalus bubalis | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEA 669 | ||+ |++ ++ + | ||| |||||+# + | | |||+||| || +||+| |+ Sbjct 713 VLVGILLLVIWKALTHLSDLREYHRFEKEK#LKSQWNND-NPLFKSATTTVMNPKFAES 769 |
| Capra hircus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEA 669 | ||+ |++ ++ + | ||| |||||+# + | | |||+||| || +||+| |+ Sbjct 714 VLVGILLLVIWKALTHLSDLREYHRFEKEK#LKSQWNND-NPLFKSATTTVMNPKFAES 770 |
| Pseudoplusia includens | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 | ||| |++ ++++ +||||++|||||+# | ||+|| | +| || + Sbjct 780 VLVGLALLMLWKMATTSHDRREFARFEKER#MMAKWDTGENPIYKQATSTFKNPTY 834 |
| Ovis aries | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEA 669 | ||+ |+ ++ + | ||| |||||+# + | | |||+||| || +||+| |+ Sbjct 714 VLVGILLLAIWKALTHLSDLREYHRFEKEK#LKSQWNND-NPLFKSATTTVMNPKFAES 770 |
| Ovis canadensis | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEA 669 | ||+ |+ ++ + | ||| |||||+# + | | |||+||| || +||+| |+ Sbjct 714 VLVGILLLAIWKALTHLSDLREYHRFEKEK#LKSQWNND-NPLFKSATTTVMNPKFAES 770 |
| Tribolium castaneum | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 | +|+ ++| ++| |+||||+++|||| # | ||+|| | +| || + Sbjct 772 VLLGMAILLLWKLFTTIHDRREFAKFEKEA#MMARWDTGENPIYKQATSTFKNPTY 826 |
| Sigmodon hispidus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQEA 669 | +|+ |++ ++ + | ||| ||||+# + | | |||+||| || +||+| |+ Sbjct 713 VLIGVLLLVIWKALTHLTDLREYRHFEKEK#LKSQWNND-NPLFKSATTTVMNPKFAES 769 |
| Lytechinus variegatus | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 + ||| |+| +|| |+||||+ ||||+# |+ ||+|| + + || + Sbjct 749 ILVGLALLLIWRLLTYIHDRREFQNFEKER#ANATWEGGENPIYKPSTSVFKNPTY 803 |
| Crassostrea gigas | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQE 668 | |+ |+| ++| | |+|| +||||| # | ||+|| | +| +|| +++ Sbjct 743 VFFGIILLLIWKLFTTISDKRELARFEKEA#MNARWDTGENPIYKQATSTFVNPTYRQ 799 |
| Schistosoma japonicum | Query 615 GLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRFQE 668 || |+| |+| + | |||| + |+++ #+ + |+ ||+++| | +|| |+| Sbjct 232 GLILLLIYKLVITIDDRRELANFKQQG#ENMRWEMAENPIFESPTTNVLNPTFEE 285 |
| rats, Peptide Partial, 723 aa | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTIN 663 +|| +| ++| + |+||+|+++||+|+# + | +||||| | +| | Sbjct 668 IGLATLLIWKLLITIHDRKEFAKFEEER#ARAKWDTANNPLYKEATSTFTN 717 |
| Canis lupus familiaris | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTIN 663 +|| +| ++| + |+||+|+++||+|+# + | +||||| | +| | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEER#ARAKWDTANNPLYKEATSTFTN 778 |
| Equus caballus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTIN 663 +|| +| ++| + |+||+|+++||+|+# + | +||||| | +| | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEER#ARAKWDTANNPLYKEATSTFTN 778 |
| mice, Peptide Partial, 680 aa | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTIN 663 +|| +| ++| + |+||+|+++||+|+# + | +||||| | +| | Sbjct 629 IGLATLLIWKLLITIHDRKEFAKFEEER#ARAKWDTANNPLYKEATSTFTN 678 |
| Caenorhabditis briggsae | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINP 664 | +|+ |+| ++| ++|| ||++| |+# | + ||+|| | || || Sbjct 751 VILGILLLLLWKLLTVLHDRAEYAKFNNER#LMAKWDTNENPIYKQATTTFKNP 803 |
| Caenorhabditis elegans | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINP 664 | +|+ |+| ++| ++|| ||+ | |+# | + ||+|| | || || Sbjct 750 VILGILLLLLWKLLTVLHDRSEYATFNNER#LMAKWDTNENPIYKQATTTFKNP 802 |
| Apis mellifera | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLN-WKQDSNPLYKSAITTTINPRF 666 | +| ++| +++ | | ||+ +||+|+# | | + ||||| | +| || | Sbjct 758 VIIGFAILLTWKILTMIRDNREFKKFERER#ILANKWNRRDNPLYKEATSTFKNPTF 813 |
| Oncorhynchus mykiss | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 +|| ++| + + + | +|+ ||| ||#++ | |||+ +| || || | Sbjct 722 IGLLILLIIKAILYMRDVKEFRRFENEQ#KKGKWSPGDNPLFMNATTTVANPTF 774 |
| Acropora millepora | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 +|| +++ + + || || +||+|+# | ++ ||||++| || || + Sbjct 735 LGLLILIVIKGLFTMVDRIEYQKFERER#MHSKWTREKNPLYQAAKTTFENPTY 787 |
| Manduca sexta | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINP 664 | +|| ++|+++ |+++|+|||++||+| #+ + ||||+ || Sbjct 707 VLIGLLTLIAWKILVDLHDKREYAKFEEES#RSRGFDVSLNPLYQEPEINFSNP 759 |
| Halocynthia roretzi | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 | +|+ ++ +++ + |+||| +|+||+# | + ||+| +| + || | Sbjct 777 VLIGIIALIIWKVIQTLRDKREYEKFKKEE#DLRKWTKGENPVYVNASSKFDNPMF 831 |
| Cyprinus carpio | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 +|| ++| + + | ||+ ||||++#+ +|||+++| || || | Sbjct 712 IGLLILLIIKAVLYASDLREWKRFEKDR#KHEK-TSGTNPLFQNATTTVQNPTF 763 |
| Spodoptera frugiperda | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINP 664 +|+ |+ +++ |+++|++|| +|+ | # + ||||+ || Sbjct 724 IGILTVIIWKILVDMHDKKEYKKFQDEA#LAAGYDVSLNPLYQDPSINFSNP 774 |
| Ophlitaspongia tenuis | Query 612 VAVGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINPRF 666 + +|| |+| + + ++| | +||+| #+ + ++ ||||+|| || + Sbjct 782 IVLGLLLLLLLKGLLLLWDVVEVRKFEREI#KNAKYTKNENPLYRSATKDYQNPLY 836 |
| Geodia cydonium | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINP 664 +|+ ++ ++ + + | | +|| | # ++ + ++ |||| || | || Sbjct 824 LGILALILLKILLMVLDYTELKKFENEL#AKVKYTKNENPLYHSATTEHKNP 874 |
| Suberites domuncula | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQ#QQLNWKQDSNPLYKSAITTTINP 664 +|+ +| +| + | | | +|||| # + + + ||||+|| || Sbjct 710 LGVIALLLLKLFLFIIDVIEVKKFEKEL#AKTKYPKHQNPLYRSAAKDYQNP 760 |
[Site 4] QQQLNWKQDS650-NPLYKSAITT
Ser650  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gln641 | Gln642 | Gln643 | Leu644 | Asn645 | Trp646 | Lys647 | Gln648 | Asp649 | Ser650 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn651 | Pro652 | Leu653 | Tyr654 | Lys655 | Ser656 | Ala657 | Ile658 | Thr659 | Thr660 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| AYRLSVEIYDRREYSRFEKEQQQLNWKQDSNPLYKSAITTTINPRFQEADSPIL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 112.00 | 11 | PREDICTED: integrin, beta 7 |
| 2 | Homo sapiens | 110.00 | 19 | integrin, beta 7 |
| 3 | Pan troglodytes | 110.00 | 13 | PREDICTED: integrin, beta 7 isoform 2 |
| 4 | Bos taurus | 101.00 | 13 | PREDICTED: similar to Integrin beta-7 |
| 5 | Mus musculus | 96.30 | 14 | integrin beta 7 |
| 6 | Mus sp. | 96.30 | 1 | beta 7 integrin |
| 7 | Rattus norvegicus | 94.40 | 10 | integrin beta-7 subunit |
| 8 | Canis familiaris | 92.00 | 10 | PREDICTED: similar to Integrin beta-7 precursor |
| 9 | N/A | 65.90 | 26 | C |
| 10 | Gallus gallus | 63.50 | 7 | integrin, beta 2 |
| 11 | Danio rerio | 58.50 | 14 | integrin beta1 subunit-like protein 3 |
| 12 | Xenopus laevis | 57.40 | 6 | integrin beta 1 |
| 13 | Monodelphis domestica | 57.40 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 14 | Tetraodon nigroviridis | 57.40 | 6 | unnamed protein product |
| 15 | Sus scrofa | 57.40 | 6 | integrin beta-1 subunit |
| 16 | synthetic construct | 57.40 | 5 | hypothetical protein |
| 17 | Ictalurus punctatus | 57.40 | 2 | beta-1 integrin |
| 18 | Oryctolagus cuniculus | 57.40 | 2 | integrin beta 1 |
| 19 | Pongo pygmaeus | 57.40 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 20 | Xenopus tropicalis | 57.40 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 21 | Felis catus | 55.10 | 1 | integrin beta 1 |
| 22 | Pacifastacus leniusculus | 50.80 | 1 | integrin |
| 23 | Aedes aegypti | 50.80 | 1 | integrin beta subunit |
| 24 | Ovis aries | 50.40 | 2 | antigen CD18 |
| 25 | Bubalus bubalis | 50.40 | 1 | integrin beta-2 |
| 26 | Ovis canadensis | 50.40 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 27 | Capra hircus | 50.40 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 28 | Anopheles gambiae str. PEST | 50.10 | 2 | ENSANGP00000021373 |
| 29 | Anopheles gambiae | 50.10 | 2 | integrin beta subunit |
| 30 | Tribolium castaneum | 49.30 | 3 | PREDICTED: similar to CG1560-PA |
| 31 | Drosophila melanogaster | 49.30 | 1 | myospheroid CG1560-PA |
| 32 | Sigmodon hispidus | 48.50 | 1 | AF445415_1 integrin beta-2 precursor |
| 33 | Ostrinia furnacalis | 48.50 | 1 | integrin beta 1 precursor |
| 34 | Strongylocentrotus purpuratus | 48.10 | 5 | integrin beta G subunit |
| 35 | Crassostrea gigas | 47.80 | 1 | integrin beta cgh |
| 36 | Podocoryne carnea | 47.40 | 1 | AF308652_1 integrin beta chain |
| 37 | Pseudoplusia includens | 47.40 | 1 | integrin beta 1 |
| 38 | Biomphalaria glabrata | 47.00 | 1 | beta integrin subunit |
| 39 | Schistosoma japonicum | 47.00 | 1 | SJCHGC06221 protein |
| 40 | rats, Peptide Partial, 723 aa | 44.70 | 1 | beta 3 integrin, GPIIIA |
| 41 | mice, Peptide Partial, 680 aa | 44.70 | 1 | beta 3 integrin, GPIIIA |
| 42 | Canis lupus familiaris | 44.70 | 1 | integrin beta chain, beta 3 |
| 43 | Equus caballus | 44.70 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 44 | Lytechinus variegatus | 44.30 | 1 | beta-C integrin subunit |
| 45 | Caenorhabditis briggsae | 43.10 | 1 | Hypothetical protein CBG03601 |
| 46 | Acropora millepora | 42.70 | 1 | integrin subunit betaCn1 |
| 47 | Caenorhabditis elegans | 42.00 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 48 | Oncorhynchus mykiss | 41.20 | 1 | CD18 protein |
| 49 | Apis mellifera | 40.40 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 50 | Halocynthia roretzi | 38.10 | 2 | integrin beta Hr2 precursor |
| 51 | Manduca sexta | 38.10 | 1 | plasmatocyte-specific integrin beta 1 |
| 52 | Cyprinus carpio | 33.50 | 2 | CD18 type 2 |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 621 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||| Query 621 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEADSPIL 674 |
| Homo sapiens | Query 621 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||||||||||||| | Sbjct 745 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEADSPTL 798 |
| Pan troglodytes | Query 621 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||||||||||||| | Sbjct 623 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEADSPTL 676 |
| Bos taurus | Query 621 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEADSPIL 674 |||||||||||||+ |||||+| |||||| #||||+||||||+||||||||||+| Sbjct 747 AYRLSVEIYDRREFHRFEKERQHLNWKQDH#NPLYQSAITTTVNPRFQEADSPVL 800 |
| Mus musculus | Query 621 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 |||||||||||||| ||||||||||||||+#|||||||||||+||||| Sbjct 743 AYRLSVEIYDRREYRRFEKEQQQLNWKQDN#NPLYKSAITTTVNPRFQ 789 |
| Mus sp. | Query 621 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 |||||||||||||| ||||||||||||||+#|||||||||||+||||| Sbjct 744 AYRLSVEIYDRREYRRFEKEQQQLNWKQDN#NPLYKSAITTTVNPRFQ 790 |
| Rattus norvegicus | Query 621 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 |||||||+||| ||||||||+|||||||||#|||||||+|||+||||| Sbjct 578 AYRLSVEVYDRLEYSRFEKERQQLNWKQDS#NPLYKSAVTTTVNPRFQ 624 |
| Canis familiaris | Query 621 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEADS 671 |||||||||||||+|||||||+ |||||++#||||+||||||+||+||| | Sbjct 677 AYRLSVEIYDRREFSRFEKEQKHLNWKQEN#NPLYRSAITTTVNPQFQETGS 727 |
| N/A | Query 644 LNWKQDS#NPLYKSAITTTINPRFQEADSPIL 674 |||||||#|||||||||||||||||||||| | Sbjct 1 LNWKQDS#NPLYKSAITTTINPRFQEADSPTL 31 |
| Gallus gallus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 +|| ||+||||| |||||+ + | + #|||+||| || +|||| Sbjct 725 WRLLTEIFDRREYRRFEKEKSKAKWNEAD#NPLFKSATTTVMNPRF 769 |
| Danio rerio | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 ++| + |+||||+ +||||+ | #||+||||+|| +|||++ Sbjct 740 WKLLMVIHDRREFDKFEKEKNNAKWDTGE#NPIYKSAVTTVVNPRYE 785 |
| Xenopus laevis | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 ++| + |+||||+++||||+ | #||+||||+|| +||+++ Sbjct 751 WKLLMIIHDRREFAKFEKEKMNAKWDTGE#NPIYKSAVTTVVNPKYE 796 |
| Monodelphis domestica | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 ++| + |+||||+++||||+ | #||+||||+|| +||+++ Sbjct 752 WKLLMIIHDRREFAKFEKEKMNAKWDTGE#NPIYKSAVTTVVNPKYE 797 |
| Tetraodon nigroviridis | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 ++| + |+||||+++||||+ | #||+||||+|| +||+++ Sbjct 744 WKLLMIIHDRREFAKFEKEKMNAKWDTGE#NPIYKSAVTTVVNPKYE 789 |
| Sus scrofa | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 ++| + |+||||+++||||+ | #||+||||+|| +||+++ Sbjct 751 WKLLMIIHDRREFAKFEKEKMNAKWDTGE#NPIYKSAVTTVVNPKYE 796 |
| synthetic construct | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 ++| + |+||||+++||||+ | #||+||||+|| +||+++ Sbjct 751 WKLLMIIHDRREFAKFEKEKMNAKWDTGE#NPIYKSAVTTVVNPKYE 796 |
| Ictalurus punctatus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 ++| + |+||||+++||||+ | #||+||||+|| +||+++ Sbjct 760 WKLLMIIHDRREFAKFEKEKMNAKWDAGE#NPIYKSAVTTVVNPKYE 805 |
| Oryctolagus cuniculus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 ++| + |+||||+++||||+ | #||+||||+|| +||+++ Sbjct 16 WKLLMIIHDRREFAKFEKEKMNAKWDTGE#NPIYKSAVTTVVNPKYE 61 |
| Pongo pygmaeus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 ++| + |+||||+++||||+ | #||+||||+|| +||+++ Sbjct 751 WKLLMIIHDRREFAKFEKEKMNAKWDTGE#NPIYKSAVTTVVNPKYE 796 |
| Xenopus tropicalis | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 ++| + |+||||+++||||+ | #||+||||+|| +||+++ Sbjct 751 WKLLMIIHDRREFAKFEKEKMNAKWDTGE#NPIYKSAVTTVVNPKYE 796 |
| Felis catus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 ++| + |+| ||+++||||+ | #||+||||+|| +||+++ Sbjct 751 WKLLMIIHDTREFAKFEKEKMNAKWDTGE#NPIYKSAVTTVVNPKYE 796 |
| Pacifastacus leniusculus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEA 669 ++| ++|||||++||||++ + |+ #||||||| || || | ++ Sbjct 753 WKLLTTLHDRREYAKFEKERKLPSGKRAE#NPLYKSAKTTFQNPAFAQS 800 |
| Aedes aegypti | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 ++| |+||||++|||||+ | #||+|| | || || + Sbjct 788 WKLLTTIHDRREFARFEKERMMAKWDTGE#NPIYKQATTTFKNPTY 832 |
| Ovis aries | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEA 669 ++ + | ||| |||||+ + | | #|||+||| || +||+| |+ Sbjct 724 WKALTHLSDLREYHRFEKEKLKSQWNND-#NPLFKSATTTVMNPKFAES 770 |
| Bubalus bubalis | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEA 669 ++ + | ||| |||||+ + | | #|||+||| || +||+| |+ Sbjct 723 WKALTHLSDLREYHRFEKEKLKSQWNND-#NPLFKSATTTVMNPKFAES 769 |
| Ovis canadensis | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEA 669 ++ + | ||| |||||+ + | | #|||+||| || +||+| |+ Sbjct 724 WKALTHLSDLREYHRFEKEKLKSQWNND-#NPLFKSATTTVMNPKFAES 770 |
| Capra hircus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEA 669 ++ + | ||| |||||+ + | | #|||+||| || +||+| |+ Sbjct 724 WKALTHLSDLREYHRFEKEKLKSQWNND-#NPLFKSATTTVMNPKFAES 770 |
| Anopheles gambiae str. PEST | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 +++ |+||||++|||||+ | #||+|| | || || + Sbjct 790 WKVLTSIHDRREFARFEKERMMAKWDTGE#NPIYKQATTTFKNPTY 834 |
| Anopheles gambiae | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 +++ |+||||++|||||+ | #||+|| | || || + Sbjct 790 WKVLTSIHDRREFARFEKERMMAKWDTGE#NPIYKQATTTFKNPTY 834 |
| Tribolium castaneum | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQ 667 +++ ++||+||++|| |+|++ | ++ #||||+ | +| || ++ Sbjct 707 WKVCTTVHDRQEYAKFENERQKMQWHRND#NPLYRQATSTFKNPVYR 752 |
| Drosophila melanogaster | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 ++| |+||||++|||||+ | #||+|| | +| || + Sbjct 799 WKLLTTIHDRREFARFEKERMNAKWDTGE#NPIYKQATSTFKNPMY 843 |
| Sigmodon hispidus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQEA 669 ++ + | ||| ||||+ + | | #|||+||| || +||+| |+ Sbjct 723 WKALTHLTDLREYRHFEKEKLKSQWNND-#NPLFKSATTTVMNPKFAES 769 |
| Ostrinia furnacalis | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 +++ |+||||++|||||+ | #||+|| | +| || + Sbjct 784 WKMVTTIHDRREFARFEKERMMAKWDTGE#NPIYKQATSTFKNPTY 828 |
| Strongylocentrotus purpuratus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQE 668 +|| | + | ||++ ||||+ +| |+ #||+|| + +| || +|+ Sbjct 737 WRLLVYVQDSREFASFEKERAGTHWGQNE#NPIYKPSTSTFKNPTYQK 783 |
| Crassostrea gigas | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQE 668 ++| | |+|| +||||| | #||+|| | +| +|| +++ Sbjct 753 WKLFTTISDKRELARFEKEAMNARWDTGE#NPIYKQATSTFVNPTYRQ 799 |
| Podocoryne carnea | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 ++| | ||||+++|||++| | #||+|| | +| || + Sbjct 734 WKLLATIQDRREFAKFEKDRQNPKWDSGE#NPIYKKATSTFQNPMY 778 |
| Pseudoplusia includens | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 ++++ +||||++|||||+ | #||+|| | +| || + Sbjct 790 WKMATTSHDRREFARFEKERMMAKWDTGE#NPIYKQATSTFKNPTY 834 |
| Biomphalaria glabrata | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 ++| |+| ||+++||||+| | #||+|| | +| || + Sbjct 741 WKLLTFIHDTREFAKFEKERQNAKWDTGE#NPIYKQATSTFKNPTY 785 |
| Schistosoma japonicum | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRFQE 668 |+| + | |||| + |+++ + + |+ #||+++| | +|| |+| Sbjct 239 YKLVITIDDRRELANFKQQGENMRWEMAE#NPIFESPTTNVLNPTFEE 285 |
| rats, Peptide Partial, 723 aa | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTIN 663 ++| + |+||+|+++||+|+ + | +#||||| | +| | Sbjct 676 WKLLITIHDRKEFAKFEEERARAKWDTAN#NPLYKEATSTFTN 717 |
| mice, Peptide Partial, 680 aa | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTIN 663 ++| + |+||+|+++||+|+ + | +#||||| | +| | Sbjct 637 WKLLITIHDRKEFAKFEEERARAKWDTAN#NPLYKEATSTFTN 678 |
| Canis lupus familiaris | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTIN 663 ++| + |+||+|+++||+|+ + | +#||||| | +| | Sbjct 737 WKLLITIHDRKEFAKFEEERARAKWDTAN#NPLYKEATSTFTN 778 |
| Equus caballus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTIN 663 ++| + |+||+|+++||+|+ + | +#||||| | +| | Sbjct 737 WKLLITIHDRKEFAKFEEERARAKWDTAN#NPLYKEATSTFTN 778 |
| Lytechinus variegatus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 +|| |+||||+ ||||+ |+ #||+|| + + || + Sbjct 759 WRLLTYIHDRREFQNFEKERANATWEGGE#NPIYKPSTSVFKNPTY 803 |
| Caenorhabditis briggsae | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINP 664 ++| ++|| ||++| |+ | + #||+|| | || || Sbjct 761 WKLLTVLHDRAEYAKFNNERLMAKWDTNE#NPIYKQATTTFKNP 803 |
| Acropora millepora | Query 630 DRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 || || +||+|+ | ++ #||||++| || || + Sbjct 751 DRIEYQKFERERMHSKWTREK#NPLYQAAKTTFENPTY 787 |
| Caenorhabditis elegans | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINP 664 ++| ++|| ||+ | |+ | + #||+|| | || || Sbjct 760 WKLLTVLHDRSEYATFNNERLMAKWDTNE#NPIYKQATTTFKNP 802 |
| Oncorhynchus mykiss | Query 630 DRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 | +|+ ||| ||++ | #|||+ +| || || | Sbjct 738 DVKEFRRFENEQKKGKWSPGD#NPLFMNATTTVANPTF 774 |
| Apis mellifera | Query 622 YRLSVEIYDRREYSRFEKEQQQLN-WKQDS#NPLYKSAITTTINPRF 666 +++ | | ||+ +||+|+ | | + #||||| | +| || | Sbjct 768 WKILTMIRDNREFKKFERERILANKWNRRD#NPLYKEATSTFKNPTF 813 |
| Halocynthia roretzi | Query 630 DRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 |+||| +|+||+ | + #||+| +| + || | Sbjct 795 DKREYEKFKKEEDLRKWTKGE#NPVYVNASSKFDNPMF 831 |
| Manduca sexta | Query 621 AYRLSVEIYDRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINP 664 |+++ |+++|+|||++||+| + + #||||+ || Sbjct 716 AWKILVDLHDKREYAKFEEESRSRGFDVSL#NPLYQEPEINFSNP 759 |
| Cyprinus carpio | Query 630 DRREYSRFEKEQQQLNWKQDS#NPLYKSAITTTINPRF 666 | ||+ ||||+++ +#|||+++| || || | Sbjct 728 DLREWKRFEKDRKHEK-TSGT#NPLFQNATTTVQNPTF 763 |
[Site 5] EIYDRREYSR636-FEKEQQQLNW
Arg636  Phe
Phe
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu627 | Ile628 | Tyr629 | Asp630 | Arg631 | Arg632 | Glu633 | Tyr634 | Ser635 | Arg636 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Phe637 | Glu638 | Lys639 | Glu640 | Gln641 | Gln642 | Gln643 | Leu644 | Asn645 | Trp646 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| CVGGIVAVGLGLVLAYRLSVEIYDRREYSRFEKEQQQLNWKQDSNPLYKSAITTTINPRF |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 96.30 | 19 | integrin, beta 7 |
| 2 | Pan troglodytes | 96.30 | 13 | PREDICTED: integrin, beta 7 isoform 2 |
| 3 | Macaca mulatta | 96.30 | 11 | PREDICTED: integrin, beta 7 |
| 4 | Mus musculus | 92.80 | 14 | integrin beta 7 |
| 5 | Mus sp. | 92.80 | 1 | beta 7 integrin |
| 6 | Rattus norvegicus | 90.90 | 10 | integrin beta-7 subunit |
| 7 | Bos taurus | 85.50 | 13 | PREDICTED: similar to Integrin beta-7 |
| 8 | Canis familiaris | 85.50 | 10 | PREDICTED: similar to Integrin beta-7 precursor |
| 9 | Gallus gallus | 63.50 | 7 | integrin, beta 2 |
| 10 | Danio rerio | 57.80 | 14 | integrin beta1 subunit-like protein 3 |
| 11 | N/A | 56.60 | 26 | - |
| 12 | Monodelphis domestica | 56.60 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 13 | Xenopus laevis | 56.60 | 6 | integrin beta 1 |
| 14 | Tetraodon nigroviridis | 56.60 | 6 | unnamed protein product |
| 15 | Sus scrofa | 56.60 | 6 | integrin beta-1 subunit |
| 16 | synthetic construct | 56.60 | 5 | hypothetical protein |
| 17 | Oryctolagus cuniculus | 56.60 | 2 | integrin beta 1 |
| 18 | Pongo pygmaeus | 56.60 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 19 | Ictalurus punctatus | 56.60 | 2 | beta-1 integrin |
| 20 | Xenopus tropicalis | 56.60 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 21 | Felis catus | 54.30 | 1 | integrin beta 1 |
| 22 | Aedes aegypti | 50.80 | 1 | integrin beta subunit |
| 23 | Anopheles gambiae | 50.10 | 2 | integrin beta subunit |
| 24 | Anopheles gambiae str. PEST | 50.10 | 2 | ENSANGP00000021373 |
| 25 | Pacifastacus leniusculus | 50.10 | 1 | integrin |
| 26 | Drosophila melanogaster | 49.30 | 1 | myospheroid CG1560-PA |
| 27 | Tribolium castaneum | 48.90 | 3 | PREDICTED: similar to CG1560-PA |
| 28 | Ovis aries | 48.50 | 2 | antigen CD18 |
| 29 | Capra hircus | 48.50 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 30 | Ostrinia furnacalis | 48.50 | 1 | integrin beta 1 precursor |
| 31 | Bubalus bubalis | 48.50 | 1 | integrin beta-2 |
| 32 | Ovis canadensis | 48.50 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 33 | Pseudoplusia includens | 47.40 | 1 | integrin beta 1 |
| 34 | Podocoryne carnea | 47.40 | 1 | AF308652_1 integrin beta chain |
| 35 | Biomphalaria glabrata | 47.00 | 1 | beta integrin subunit |
| 36 | Sigmodon hispidus | 46.60 | 1 | AF445415_1 integrin beta-2 precursor |
| 37 | Crassostrea gigas | 46.60 | 1 | integrin beta cgh |
| 38 | Strongylocentrotus purpuratus | 46.20 | 5 | integrin beta L subunit |
| 39 | Canis lupus familiaris | 44.70 | 1 | integrin beta chain, beta 3 |
| 40 | Equus caballus | 44.70 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 41 | rats, Peptide Partial, 723 aa | 44.70 | 1 | beta 3 integrin, GPIIIA |
| 42 | mice, Peptide Partial, 680 aa | 44.70 | 1 | beta 3 integrin, GPIIIA |
| 43 | Lytechinus variegatus | 44.30 | 1 | beta-C integrin subunit |
| 44 | Schistosoma japonicum | 44.30 | 1 | SJCHGC06221 protein |
| 45 | Caenorhabditis briggsae | 43.10 | 1 | Hypothetical protein CBG03601 |
| 46 | Acropora millepora | 42.70 | 1 | integrin subunit betaCn1 |
| 47 | Caenorhabditis elegans | 42.00 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 48 | Oncorhynchus mykiss | 41.20 | 1 | CD18 protein |
| 49 | Apis mellifera | 40.40 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 50 | Halocynthia roretzi | 38.10 | 2 | integrin beta Hr2 precursor |
| 51 | Manduca sexta | 36.60 | 1 | plasmatocyte-specific integrin beta 1 |
| 52 | Cyprinus carpio | 33.50 | 2 | CD18 type 2 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 |||||||||||||||#|||||||||||||||||||||||||||||| Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 |
| Pan troglodytes | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 |||||||||||||||#|||||||||||||||||||||||||||||| Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 |
| Macaca mulatta | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 |||||||||||||||#|||||||||||||||||||||||||||||| Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 |
| Mus musculus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ||||||||||||| |#|||||||||||||+|||||||||||+|||| Sbjct 744 YRLSVEIYDRREYRR#FEKEQQQLNWKQDNNPLYKSAITTTVNPRF 788 |
| Mus sp. | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ||||||||||||| |#|||||||||||||+|||||||||||+|||| Sbjct 745 YRLSVEIYDRREYRR#FEKEQQQLNWKQDNNPLYKSAITTTVNPRF 789 |
| Rattus norvegicus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ||||||+||| ||||#||||+||||||||||||||||+|||+|||| Sbjct 579 YRLSVEVYDRLEYSR#FEKERQQLNWKQDSNPLYKSAVTTTVNPRF 623 |
| Bos taurus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ||||||||||||+ |#||||+| |||||| ||||+||||||+|||| Sbjct 748 YRLSVEIYDRREFHR#FEKERQHLNWKQDHNPLYQSAITTTVNPRF 792 |
| Canis familiaris | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ||||||||||||+||#|||||+ |||||++||||+||||||+||+| Sbjct 678 YRLSVEIYDRREFSR#FEKEQKHLNWKQENNPLYRSAITTTVNPQF 722 |
| Gallus gallus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 +|| ||+||||| |#||||+ + | + |||+||| || +|||| Sbjct 725 WRLLTEIFDRREYRR#FEKEKSKAKWNEADNPLFKSATTTVMNPRF 769 |
| Danio rerio | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+||||+ +#||||+ | ||+||||+|| +|||+ Sbjct 740 WKLLMVIHDRREFDK#FEKEKNNAKWDTGENPIYKSAVTTVVNPRY 784 |
| N/A | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+||||+++#||||+ | ||+||||+|| +||++ Sbjct 728 WKLLMIIHDRREFAK#FEKEKMNAKWDTGENPIYKSAVTTVVNPKY 772 |
| Monodelphis domestica | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+||||+++#||||+ | ||+||||+|| +||++ Sbjct 752 WKLLMIIHDRREFAK#FEKEKMNAKWDTGENPIYKSAVTTVVNPKY 796 |
| Xenopus laevis | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+||||+++#||||+ | ||+||||+|| +||++ Sbjct 751 WKLLMIIHDRREFAK#FEKEKMNAKWDTGENPIYKSAVTTVVNPKY 795 |
| Tetraodon nigroviridis | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+||||+++#||||+ | ||+||||+|| +||++ Sbjct 744 WKLLMIIHDRREFAK#FEKEKMNAKWDTGENPIYKSAVTTVVNPKY 788 |
| Sus scrofa | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+||||+++#||||+ | ||+||||+|| +||++ Sbjct 751 WKLLMIIHDRREFAK#FEKEKMNAKWDTGENPIYKSAVTTVVNPKY 795 |
| synthetic construct | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+||||+++#||||+ | ||+||||+|| +||++ Sbjct 751 WKLLMIIHDRREFAK#FEKEKMNAKWDTGENPIYKSAVTTVVNPKY 795 |
| Oryctolagus cuniculus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+||||+++#||||+ | ||+||||+|| +||++ Sbjct 16 WKLLMIIHDRREFAK#FEKEKMNAKWDTGENPIYKSAVTTVVNPKY 60 |
| Pongo pygmaeus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+||||+++#||||+ | ||+||||+|| +||++ Sbjct 751 WKLLMIIHDRREFAK#FEKEKMNAKWDTGENPIYKSAVTTVVNPKY 795 |
| Ictalurus punctatus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+||||+++#||||+ | ||+||||+|| +||++ Sbjct 760 WKLLMIIHDRREFAK#FEKEKMNAKWDAGENPIYKSAVTTVVNPKY 804 |
| Xenopus tropicalis | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+||||+++#||||+ | ||+||||+|| +||++ Sbjct 751 WKLLMIIHDRREFAK#FEKEKMNAKWDTGENPIYKSAVTTVVNPKY 795 |
| Felis catus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| + |+| ||+++#||||+ | ||+||||+|| +||++ Sbjct 751 WKLLMIIHDTREFAK#FEKEKMNAKWDTGENPIYKSAVTTVVNPKY 795 |
| Aedes aegypti | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| |+||||++|#||||+ | ||+|| | || || + Sbjct 788 WKLLTTIHDRREFAR#FEKERMMAKWDTGENPIYKQATTTFKNPTY 832 |
| Anopheles gambiae | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 +++ |+||||++|#||||+ | ||+|| | || || + Sbjct 790 WKVLTSIHDRREFAR#FEKERMMAKWDTGENPIYKQATTTFKNPTY 834 |
| Anopheles gambiae str. PEST | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 +++ |+||||++|#||||+ | ||+|| | || || + Sbjct 790 WKVLTSIHDRREFAR#FEKERMMAKWDTGENPIYKQATTTFKNPTY 834 |
| Pacifastacus leniusculus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| ++|||||++#||||++ + |+ ||||||| || || | Sbjct 753 WKLLTTLHDRREYAK#FEKERKLPSGKRAENPLYKSAKTTFQNPAF 797 |
| Drosophila melanogaster | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| |+||||++|#||||+ | ||+|| | +| || + Sbjct 799 WKLLTTIHDRREFAR#FEKERMNAKWDTGENPIYKQATSTFKNPMY 843 |
| Tribolium castaneum | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINP 664 +++ ++||+||++#|| |+|++ | ++ ||||+ | +| || Sbjct 707 WKVCTTVHDRQEYAK#FENERQKMQWHRNDNPLYRQATSTFKNP 749 |
| Ovis aries | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++ + | ||| |#||||+ + | | |||+||| || +||+| Sbjct 724 WKALTHLSDLREYHR#FEKEKLKSQWNND-NPLFKSATTTVMNPKF 767 |
| Capra hircus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++ + | ||| |#||||+ + | | |||+||| || +||+| Sbjct 724 WKALTHLSDLREYHR#FEKEKLKSQWNND-NPLFKSATTTVMNPKF 767 |
| Ostrinia furnacalis | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 +++ |+||||++|#||||+ | ||+|| | +| || + Sbjct 784 WKMVTTIHDRREFAR#FEKERMMAKWDTGENPIYKQATSTFKNPTY 828 |
| Bubalus bubalis | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++ + | ||| |#||||+ + | | |||+||| || +||+| Sbjct 723 WKALTHLSDLREYHR#FEKEKLKSQWNND-NPLFKSATTTVMNPKF 766 |
| Ovis canadensis | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++ + | ||| |#||||+ + | | |||+||| || +||+| Sbjct 724 WKALTHLSDLREYHR#FEKEKLKSQWNND-NPLFKSATTTVMNPKF 767 |
| Pseudoplusia includens | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++++ +||||++|#||||+ | ||+|| | +| || + Sbjct 790 WKMATTSHDRREFAR#FEKERMMAKWDTGENPIYKQATSTFKNPTY 834 |
| Podocoryne carnea | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| | ||||+++#|||++| | ||+|| | +| || + Sbjct 734 WKLLATIQDRREFAK#FEKDRQNPKWDSGENPIYKKATSTFQNPMY 778 |
| Biomphalaria glabrata | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| |+| ||+++#||||+| | ||+|| | +| || + Sbjct 741 WKLLTFIHDTREFAK#FEKERQNAKWDTGENPIYKQATSTFKNPTY 785 |
| Sigmodon hispidus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++ + | ||| #||||+ + | | |||+||| || +||+| Sbjct 723 WKALTHLTDLREYRH#FEKEKLKSQWNND-NPLFKSATTTVMNPKF 766 |
| Crassostrea gigas | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 ++| | |+|| +|#|||| | ||+|| | +| +|| + Sbjct 753 WKLFTTISDKRELAR#FEKEAMNARWDTGENPIYKQATSTFVNPTY 797 |
| Strongylocentrotus purpuratus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 +|| + |+|||++#+| + ++ | | ||+|||+ || || + Sbjct 756 WRLYTYVQDKREYAQ#WENDCKKAQWDQSDNPIYKSSTTTFKNPTY 800 |
| Canis lupus familiaris | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTIN 663 ++| + |+||+|+++#||+|+ + | +||||| | +| | Sbjct 737 WKLLITIHDRKEFAK#FEEERARAKWDTANNPLYKEATSTFTN 778 |
| Equus caballus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTIN 663 ++| + |+||+|+++#||+|+ + | +||||| | +| | Sbjct 737 WKLLITIHDRKEFAK#FEEERARAKWDTANNPLYKEATSTFTN 778 |
| rats, Peptide Partial, 723 aa | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTIN 663 ++| + |+||+|+++#||+|+ + | +||||| | +| | Sbjct 676 WKLLITIHDRKEFAK#FEEERARAKWDTANNPLYKEATSTFTN 717 |
| mice, Peptide Partial, 680 aa | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTIN 663 ++| + |+||+|+++#||+|+ + | +||||| | +| | Sbjct 637 WKLLITIHDRKEFAK#FEEERARAKWDTANNPLYKEATSTFTN 678 |
| Lytechinus variegatus | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 +|| |+||||+ #||||+ |+ ||+|| + + || + Sbjct 759 WRLLTYIHDRREFQN#FEKERANATWEGGENPIYKPSTSVFKNPTY 803 |
| Schistosoma japonicum | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 |+| + | |||| + #|+++ + + |+ ||+++| | +|| | Sbjct 239 YKLVITIDDRRELAN#FKQQGENMRWEMAENPIFESPTTNVLNPTF 283 |
| Caenorhabditis briggsae | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINP 664 ++| ++|| ||++#| |+ | + ||+|| | || || Sbjct 761 WKLLTVLHDRAEYAK#FNNERLMAKWDTNENPIYKQATTTFKNP 803 |
| Acropora millepora | Query 630 DRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 || || +#||+|+ | ++ ||||++| || || + Sbjct 751 DRIEYQK#FERERMHSKWTREKNPLYQAAKTTFENPTY 787 |
| Caenorhabditis elegans | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINP 664 ++| ++|| ||+ #| |+ | + ||+|| | || || Sbjct 760 WKLLTVLHDRSEYAT#FNNERLMAKWDTNENPIYKQATTTFKNP 802 |
| Oncorhynchus mykiss | Query 630 DRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 | +|+ |#|| ||++ | |||+ +| || || | Sbjct 738 DVKEFRR#FENEQKKGKWSPGDNPLFMNATTTVANPTF 774 |
| Apis mellifera | Query 622 YRLSVEIYDRREYSR#FEKEQQQLN-WKQDSNPLYKSAITTTINPRF 666 +++ | | ||+ +#||+|+ | | + ||||| | +| || | Sbjct 768 WKILTMIRDNREFKK#FERERILANKWNRRDNPLYKEATSTFKNPTF 813 |
| Halocynthia roretzi | Query 630 DRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 |+||| +#|+||+ | + ||+| +| + || | Sbjct 795 DKREYEK#FKKEEDLRKWTKGENPVYVNASSKFDNPMF 831 |
| Manduca sexta | Query 622 YRLSVEIYDRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINP 664 +++ |+++|+|||++#||+| + + ||||+ || Sbjct 717 WKILVDLHDKREYAK#FEEESRSRGFDVSLNPLYQEPEINFSNP 759 |
| Cyprinus carpio | Query 630 DRREYSR#FEKEQQQLNWKQDSNPLYKSAITTTINPRF 666 | ||+ |#|||+++ +|||+++| || || | Sbjct 728 DLREWKR#FEKDRKHEK-TSGTNPLFQNATTTVQNPTF 763 |
[Site 6] RFEKEQQQLN645-WKQDSNPLYK
Asn645  Trp
Trp
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg636 | Phe637 | Glu638 | Lys639 | Glu640 | Gln641 | Gln642 | Gln643 | Leu644 | Asn645 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Trp646 | Lys647 | Gln648 | Asp649 | Ser650 | Asn651 | Pro652 | Leu653 | Tyr654 | Lys655 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LGLVLAYRLSVEIYDRREYSRFEKEQQQLNWKQDSNPLYKSAITTTINPRFQEADSPIL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 121.00 | 13 | PREDICTED: integrin, beta 7 |
| 2 | Homo sapiens | 119.00 | 25 | integrin, beta 7 |
| 3 | Pan troglodytes | 119.00 | 15 | PREDICTED: integrin, beta 7 isoform 2 |
| 4 | Bos taurus | 110.00 | 13 | PREDICTED: similar to Integrin beta-7 |
| 5 | Mus musculus | 104.00 | 14 | integrin beta 7 |
| 6 | Mus sp. | 104.00 | 1 | beta 7 integrin |
| 7 | Rattus norvegicus | 102.00 | 10 | integrin beta-7 subunit |
| 8 | Canis familiaris | 100.00 | 10 | PREDICTED: similar to Integrin beta-7 precursor |
| 9 | Gallus gallus | 67.00 | 7 | integrin, beta 2 |
| 10 | N/A | 65.90 | 30 | C |
| 11 | Danio rerio | 63.20 | 14 | integrin beta1 subunit-like protein 3 |
| 12 | Sus scrofa | 62.00 | 6 | integrin beta-1 subunit |
| 13 | Monodelphis domestica | 62.00 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 14 | Tetraodon nigroviridis | 62.00 | 6 | unnamed protein product |
| 15 | Xenopus laevis | 62.00 | 6 | integrin beta 1 |
| 16 | synthetic construct | 62.00 | 5 | hypothetical protein |
| 17 | Oryctolagus cuniculus | 62.00 | 2 | integrin beta 1 |
| 18 | Pongo pygmaeus | 62.00 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 19 | Xenopus tropicalis | 62.00 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 20 | Ictalurus punctatus | 60.50 | 2 | beta-1 integrin |
| 21 | Felis catus | 59.70 | 1 | integrin beta 1 |
| 22 | Aedes aegypti | 54.30 | 1 | integrin beta subunit |
| 23 | Strongylocentrotus purpuratus | 53.10 | 5 | integrin beta G subunit |
| 24 | Drosophila melanogaster | 53.10 | 1 | myospheroid CG1560-PA |
| 25 | Tribolium castaneum | 52.80 | 3 | PREDICTED: similar to CG1560-PA |
| 26 | Anopheles gambiae str. PEST | 52.80 | 2 | ENSANGP00000021373 |
| 27 | Anopheles gambiae | 52.80 | 2 | integrin beta subunit |
| 28 | Pacifastacus leniusculus | 52.40 | 1 | integrin |
| 29 | Capra hircus | 52.40 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 30 | Ostrinia furnacalis | 52.40 | 1 | integrin beta 1 precursor |
| 31 | Bubalus bubalis | 52.40 | 1 | integrin beta-2 |
| 32 | Podocoryne carnea | 52.00 | 1 | AF308652_1 integrin beta chain |
| 33 | Ovis aries | 51.60 | 2 | antigen CD18 |
| 34 | Ovis canadensis | 51.60 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 35 | Pseudoplusia includens | 51.20 | 1 | integrin beta 1 |
| 36 | Crassostrea gigas | 50.80 | 1 | integrin beta cgh |
| 37 | Biomphalaria glabrata | 50.40 | 1 | beta integrin subunit |
| 38 | Sigmodon hispidus | 50.40 | 1 | AF445415_1 integrin beta-2 precursor |
| 39 | Schistosoma japonicum | 50.10 | 1 | SJCHGC06221 protein |
| 40 | Lytechinus variegatus | 48.90 | 1 | beta-C integrin subunit |
| 41 | mice, Peptide Partial, 680 aa | 47.40 | 1 | beta 3 integrin, GPIIIA |
| 42 | rats, Peptide Partial, 723 aa | 47.40 | 1 | beta 3 integrin, GPIIIA |
| 43 | Canis lupus familiaris | 47.40 | 1 | integrin beta chain, beta 3 |
| 44 | Equus caballus | 47.40 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 45 | Caenorhabditis briggsae | 46.20 | 1 | Hypothetical protein CBG03601 |
| 46 | Caenorhabditis elegans | 45.10 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 47 | Apis mellifera | 43.10 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 48 | Oncorhynchus mykiss | 43.10 | 1 | CD18 protein |
| 49 | Acropora millepora | 42.70 | 1 | integrin subunit betaCn1 |
| 50 | Manduca sexta | 39.30 | 1 | plasmatocyte-specific integrin beta 1 |
| 51 | Halocynthia roretzi | 38.90 | 2 | integrin beta Hr2 precursor |
| 52 | Cyprinus carpio | 35.80 | 2 | CD18 type 2 |
| 53 | Spodoptera frugiperda | 33.90 | 1 | integrin beta 1 |
| 54 | Geodia cydonium | 33.10 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||| Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEADSPIL 674 |
| Homo sapiens | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||| | Sbjct 740 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEADSPTL 798 |
| Pan troglodytes | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||| | Sbjct 618 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEADSPTL 676 |
| Bos taurus | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||+ |||||+| ||#|||| ||||+||||||+||||||||||+| Sbjct 742 LGLVLAYRLSVEIYDRREFHRFEKERQHLN#WKQDHNPLYQSAITTTVNPRFQEADSPVL 800 |
| Mus musculus | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 ||||||||||||||||||| ||||||||||#||||+|||||||||||+||||| Sbjct 738 LGLVLAYRLSVEIYDRREYRRFEKEQQQLN#WKQDNNPLYKSAITTTVNPRFQ 789 |
| Mus sp. | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 ||||||||||||||||||| ||||||||||#||||+|||||||||||+||||| Sbjct 739 LGLVLAYRLSVEIYDRREYRRFEKEQQQLN#WKQDNNPLYKSAITTTVNPRFQ 790 |
| Rattus norvegicus | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 ||||||||||||+||| ||||||||+||||#||||||||||||+|||+||||| Sbjct 573 LGLVLAYRLSVEVYDRLEYSRFEKERQQLN#WKQDSNPLYKSAVTTTVNPRFQ 624 |
| Canis familiaris | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEADS 671 ||||||||||||||||||+|||||||+ ||#|||++||||+||||||+||+||| | Sbjct 672 LGLVLAYRLSVEIYDRREFSRFEKEQKHLN#WKQENNPLYRSAITTTVNPQFQETGS 727 |
| Gallus gallus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 |+| +|| ||+||||| |||||+ + #| + |||+||| || +|||| Sbjct 721 LLLTWRLLTEIFDRREYRRFEKEKSKAK#WNEADNPLFKSATTTVMNPRF 769 |
| N/A | Query 644 LN#WKQDSNPLYKSAITTTINPRFQEADSPIL 674 ||#||||||||||||||||||||||||||| | Sbjct 1 LN#WKQDSNPLYKSAITTTINPRFQEADSPTL 31 |
| Danio rerio | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 | |+| ++| + |+||||+ +||||+ #| ||+||||+|| +|||++ Sbjct 734 LALLLIWKLLMVIHDRREFDKFEKEKNNAK#WDTGENPIYKSAVTTVVNPRYE 785 |
| Sus scrofa | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 | |+| ++| + |+||||+++||||+ #| ||+||||+|| +||+++ Sbjct 745 LALLLIWKLLMIIHDRREFAKFEKEKMNAK#WDTGENPIYKSAVTTVVNPKYE 796 |
| Monodelphis domestica | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 | |+| ++| + |+||||+++||||+ #| ||+||||+|| +||+++ Sbjct 746 LALLLIWKLLMIIHDRREFAKFEKEKMNAK#WDTGENPIYKSAVTTVVNPKYE 797 |
| Tetraodon nigroviridis | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 | |+| ++| + |+||||+++||||+ #| ||+||||+|| +||+++ Sbjct 738 LALLLIWKLLMIIHDRREFAKFEKEKMNAK#WDTGENPIYKSAVTTVVNPKYE 789 |
| Xenopus laevis | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 | |+| ++| + |+||||+++||||+ #| ||+||||+|| +||+++ Sbjct 745 LALLLIWKLLMIIHDRREFAKFEKEKMNAK#WDTGENPIYKSAVTTVVNPKYE 796 |
| synthetic construct | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 | |+| ++| + |+||||+++||||+ #| ||+||||+|| +||+++ Sbjct 745 LALLLIWKLLMIIHDRREFAKFEKEKMNAK#WDTGENPIYKSAVTTVVNPKYE 796 |
| Oryctolagus cuniculus | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 | |+| ++| + |+||||+++||||+ #| ||+||||+|| +||+++ Sbjct 10 LALLLIWKLLMIIHDRREFAKFEKEKMNAK#WDTGENPIYKSAVTTVVNPKYE 61 |
| Pongo pygmaeus | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 | |+| ++| + |+||||+++||||+ #| ||+||||+|| +||+++ Sbjct 745 LALLLIWKLLMIIHDRREFAKFEKEKMNAK#WDTGENPIYKSAVTTVVNPKYE 796 |
| Xenopus tropicalis | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 | |+| ++| + |+||||+++||||+ #| ||+||||+|| +||+++ Sbjct 745 LALLLIWKLLMIIHDRREFAKFEKEKMNAK#WDTGENPIYKSAVTTVVNPKYE 796 |
| Ictalurus punctatus | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 | |+ ++| + |+||||+++||||+ #| ||+||||+|| +||+++ Sbjct 754 LALLFIWKLLMIIHDRREFAKFEKEKMNAK#WDAGENPIYKSAVTTVVNPKYE 805 |
| Felis catus | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 | |+| ++| + |+| ||+++||||+ #| ||+||||+|| +||+++ Sbjct 745 LALLLIWKLLMIIHDTREFAKFEKEKMNAK#WDTGENPIYKSAVTTVVNPKYE 796 |
| Aedes aegypti | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 | ++| ++| |+||||++|||||+ #| ||+|| | || || + Sbjct 782 LAVLLLWKLLTTIHDRREFARFEKERMMAK#WDTGENPIYKQATTTFKNPTY 832 |
| Strongylocentrotus purpuratus | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQE 668 | |+| +|| | + | ||++ ||||+ +#| |+ ||+|| + +| || +|+ Sbjct 731 LALLLVWRLLVYVQDSREFASFEKERAGTH#WGQNENPIYKPSTSTFKNPTYQK 783 |
| Drosophila melanogaster | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 | ++| ++| |+||||++|||||+ #| ||+|| | +| || + Sbjct 793 LAILLLWKLLTTIHDRREFARFEKERMNAK#WDTGENPIYKQATSTFKNPMY 843 |
| Tribolium castaneum | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQ 667 |+| +++ ++||+||++|| |+|++ #| ++ ||||+ | +| || ++ Sbjct 703 LLLTWKVCTTVHDRQEYAKFENERQKMQ#WHRNDNPLYRQATSTFKNPVYR 752 |
| Anopheles gambiae str. PEST | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 + ++| +++ |+||||++|||||+ #| ||+|| | || || + Sbjct 784 MAVLLLWKVLTSIHDRREFARFEKERMMAK#WDTGENPIYKQATTTFKNPTY 834 |
| Anopheles gambiae | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 + ++| +++ |+||||++|||||+ #| ||+|| | || || + Sbjct 784 MAVLLLWKVLTSIHDRREFARFEKERMMAK#WDTGENPIYKQATTTFKNPTY 834 |
| Pacifastacus leniusculus | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEA 669 +| ++| ++|||||++||||++ +# |+ ||||||| || || | ++ Sbjct 750 LLIWKLLTTLHDRREYAKFEKERKLPS#GKRAENPLYKSAKTTFQNPAFAQS 800 |
| Capra hircus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEA 669 |++ ++ + | ||| |||||+ + #| | |||+||| || +||+| |+ Sbjct 720 LLVIWKALTHLSDLREYHRFEKEKLKSQ#WNND-NPLFKSATTTVMNPKFAES 770 |
| Ostrinia furnacalis | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 | |++ +++ |+||||++|||||+ #| ||+|| | +| || + Sbjct 778 LALLMLWKMVTTIHDRREFARFEKERMMAK#WDTGENPIYKQATSTFKNPTY 828 |
| Bubalus bubalis | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEA 669 |++ ++ + | ||| |||||+ + #| | |||+||| || +||+| |+ Sbjct 719 LLVIWKALTHLSDLREYHRFEKEKLKSQ#WNND-NPLFKSATTTVMNPKFAES 769 |
| Podocoryne carnea | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 | |+| ++| | ||||+++|||++| #| ||+|| | +| || + Sbjct 728 LALLLIWKLLATIQDRREFAKFEKDRQNPK#WDSGENPIYKKATSTFQNPMY 778 |
| Ovis aries | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEA 669 |+ ++ + | ||| |||||+ + #| | |||+||| || +||+| |+ Sbjct 720 LLAIWKALTHLSDLREYHRFEKEKLKSQ#WNND-NPLFKSATTTVMNPKFAES 770 |
| Ovis canadensis | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEA 669 |+ ++ + | ||| |||||+ + #| | |||+||| || +||+| |+ Sbjct 720 LLAIWKALTHLSDLREYHRFEKEKLKSQ#WNND-NPLFKSATTTVMNPKFAES 770 |
| Pseudoplusia includens | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 | |++ ++++ +||||++|||||+ #| ||+|| | +| || + Sbjct 784 LALLMLWKMATTSHDRREFARFEKERMMAK#WDTGENPIYKQATSTFKNPTY 834 |
| Crassostrea gigas | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQE 668 |+| ++| | |+|| +||||| #| ||+|| | +| +|| +++ Sbjct 749 LLLIWKLFTTISDKRELARFEKEAMNAR#WDTGENPIYKQATSTFVNPTYRQ 799 |
| Biomphalaria glabrata | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 | |+| ++| |+| ||+++||||+| #| ||+|| | +| || + Sbjct 735 LFLLLLWKLLTFIHDTREFAKFEKERQNAK#WDTGENPIYKQATSTFKNPTY 785 |
| Sigmodon hispidus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQEA 669 |++ ++ + | ||| ||||+ + #| | |||+||| || +||+| |+ Sbjct 719 LLVIWKALTHLTDLREYRHFEKEKLKSQ#WNND-NPLFKSATTTVMNPKFAES 769 |
| Schistosoma japonicum | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRFQE 668 |+| |+| + | |||| + |+++ + + #|+ ||+++| | +|| |+| Sbjct 235 LLLIYKLVITIDDRRELANFKQQGENMR#WEMAENPIFESPTTNVLNPTFEE 285 |
| Lytechinus variegatus | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 | |+| +|| |+||||+ ||||+ #|+ ||+|| + + || + Sbjct 753 LALLLIWRLLTYIHDRREFQNFEKERANAT#WEGGENPIYKPSTSVFKNPTY 803 |
| mice, Peptide Partial, 680 aa | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTIN 663 | +| ++| + |+||+|+++||+|+ + #| +||||| | +| | Sbjct 631 LATLLIWKLLITIHDRKEFAKFEEERARAK#WDTANNPLYKEATSTFTN 678 |
| rats, Peptide Partial, 723 aa | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTIN 663 | +| ++| + |+||+|+++||+|+ + #| +||||| | +| | Sbjct 670 LATLLIWKLLITIHDRKEFAKFEEERARAK#WDTANNPLYKEATSTFTN 717 |
| Canis lupus familiaris | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTIN 663 | +| ++| + |+||+|+++||+|+ + #| +||||| | +| | Sbjct 731 LATLLIWKLLITIHDRKEFAKFEEERARAK#WDTANNPLYKEATSTFTN 778 |
| Equus caballus | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTIN 663 | +| ++| + |+||+|+++||+|+ + #| +||||| | +| | Sbjct 731 LATLLIWKLLITIHDRKEFAKFEEERARAK#WDTANNPLYKEATSTFTN 778 |
| Caenorhabditis briggsae | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINP 664 |+| ++| ++|| ||++| |+ #| + ||+|| | || || Sbjct 757 LLLLWKLLTVLHDRAEYAKFNNERLMAK#WDTNENPIYKQATTTFKNP 803 |
| Caenorhabditis elegans | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINP 664 |+| ++| ++|| ||+ | |+ #| + ||+|| | || || Sbjct 756 LLLLWKLLTVLHDRSEYATFNNERLMAK#WDTNENPIYKQATTTFKNP 802 |
| Apis mellifera | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN-#WKQDSNPLYKSAITTTINPRF 666 ++| +++ | | ||+ +||+|+ | #| + ||||| | +| || | Sbjct 764 ILLTWKILTMIRDNREFKKFERERILANK#WNRRDNPLYKEATSTFKNPTF 813 |
| Oncorhynchus mykiss | Query 616 LGLVLAYRLSVEIY--DRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 +||++ + +| | +|+ ||| ||++ #| |||+ +| || || | Sbjct 722 IGLLILLIIKAILYMRDVKEFRRFENEQKKGK#WSPGDNPLFMNATTTVANPTF 774 |
| Acropora millepora | Query 630 DRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 || || +||+|+ #| ++ ||||++| || || + Sbjct 751 DRIEYQKFERERMHSK#WTREKNPLYQAAKTTFENPTY 787 |
| Manduca sexta | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINP 664 ++|+++ |+++|+|||++||+| + #+ ||||+ || Sbjct 714 LIAWKILVDLHDKREYAKFEEESRSRG#FDVSLNPLYQEPEINFSNP 759 |
| Halocynthia roretzi | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 ++ +++ + |+||| +|+||+ #| + ||+| +| + || | Sbjct 784 LIIWKVIQTLRDKREYEKFKKEEDLRK#WTKGENPVYVNASSKFDNPMF 831 |
| Cyprinus carpio | Query 616 LGLVLAYRLSVEIY--DRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINPRF 666 +||++ + +| | ||+ ||||+++ # +|||+++| || || | Sbjct 712 IGLLILLIIKAVLYASDLREWKRFEKDRKHEK#-TSGTNPLFQNATTTVQNPTF 763 |
| Spodoptera frugiperda | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINP 664 |+ +++ |+++|++|| +|+ | #+ ||||+ || Sbjct 729 VIIWKILVDMHDKKEYKKFQDEALAAG#YDVSLNPLYQDPSINFSNP 774 |
| Geodia cydonium | Query 616 LGLVLAYRLSVEIYDRREYSRFEKEQQQLN#WKQDSNPLYKSAITTTINP 664 | |+| ++ + + | | +|| | ++ #+ ++ |||| || | || Sbjct 827 LALILL-KILLMVLDYTELKKFENELAKVK#YTKNENPLYHSATTEHKNP 874 |
[Site 7] FEKEQQQLNW646-KQDSNPLYKS
Trp646  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe637 | Glu638 | Lys639 | Glu640 | Gln641 | Gln642 | Gln643 | Leu644 | Asn645 | Trp646 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys647 | Gln648 | Asp649 | Ser650 | Asn651 | Pro652 | Leu653 | Tyr654 | Lys655 | Ser656 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GLVLAYRLSVEIYDRREYSRFEKEQQQLNWKQDSNPLYKSAITTTINPRFQEADSPIL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 119.00 | 12 | PREDICTED: integrin, beta 7 |
| 2 | Homo sapiens | 117.00 | 22 | integrin, beta 7 |
| 3 | Pan troglodytes | 117.00 | 13 | PREDICTED: integrin, beta 7 isoform 2 |
| 4 | Bos taurus | 108.00 | 13 | PREDICTED: similar to Integrin beta-7 |
| 5 | Mus musculus | 103.00 | 14 | integrin beta 7 |
| 6 | Mus sp. | 103.00 | 1 | beta 7 integrin |
| 7 | Rattus norvegicus | 101.00 | 10 | integrin beta-7 subunit |
| 8 | Canis familiaris | 99.00 | 10 | PREDICTED: similar to Integrin beta-7 precursor |
| 9 | Gallus gallus | 67.00 | 7 | integrin, beta 2 |
| 10 | N/A | 65.90 | 28 | C |
| 11 | Danio rerio | 61.60 | 14 | integrin beta1 subunit-like protein 3 |
| 12 | Tetraodon nigroviridis | 60.50 | 6 | unnamed protein product |
| 13 | Monodelphis domestica | 60.50 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 14 | Xenopus laevis | 60.50 | 6 | integrin beta-1 subunit |
| 15 | Sus scrofa | 60.50 | 6 | integrin beta-1 subunit |
| 16 | synthetic construct | 60.50 | 5 | hypothetical protein |
| 17 | Oryctolagus cuniculus | 60.50 | 2 | integrin beta 1 |
| 18 | Pongo pygmaeus | 60.50 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 19 | Xenopus tropicalis | 60.50 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 20 | Ictalurus punctatus | 58.90 | 2 | beta-1 integrin |
| 21 | Felis catus | 58.20 | 1 | integrin beta 1 |
| 22 | Tribolium castaneum | 52.80 | 3 | PREDICTED: similar to CG1560-PA |
| 23 | Aedes aegypti | 52.80 | 1 | integrin beta subunit |
| 24 | Pacifastacus leniusculus | 52.40 | 1 | integrin |
| 25 | Capra hircus | 52.40 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 26 | Bubalus bubalis | 52.40 | 1 | integrin beta-2 |
| 27 | Anopheles gambiae | 52.00 | 2 | integrin beta subunit |
| 28 | Anopheles gambiae str. PEST | 52.00 | 2 | ENSANGP00000021373 |
| 29 | Strongylocentrotus purpuratus | 51.60 | 5 | integrin beta G subunit |
| 30 | Ovis aries | 51.60 | 2 | antigen CD18 |
| 31 | Drosophila melanogaster | 51.60 | 1 | myospheroid CG1560-PA |
| 32 | Ovis canadensis | 51.60 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 33 | Crassostrea gigas | 50.80 | 1 | integrin beta cgh |
| 34 | Ostrinia furnacalis | 50.80 | 1 | integrin beta 1 precursor |
| 35 | Sigmodon hispidus | 50.40 | 1 | AF445415_1 integrin beta-2 precursor |
| 36 | Podocoryne carnea | 50.40 | 1 | AF308652_1 integrin beta chain |
| 37 | Schistosoma japonicum | 50.10 | 1 | SJCHGC06221 protein |
| 38 | Biomphalaria glabrata | 50.10 | 1 | beta integrin subunit |
| 39 | Pseudoplusia includens | 49.70 | 1 | integrin beta 1 |
| 40 | Lytechinus variegatus | 47.40 | 1 | beta-C integrin subunit |
| 41 | Canis lupus familiaris | 46.20 | 1 | integrin beta chain, beta 3 |
| 42 | Caenorhabditis briggsae | 46.20 | 1 | Hypothetical protein CBG03601 |
| 43 | rats, Peptide Partial, 723 aa | 46.20 | 1 | beta 3 integrin, GPIIIA |
| 44 | Equus caballus | 46.20 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 45 | mice, Peptide Partial, 680 aa | 46.20 | 1 | beta 3 integrin, GPIIIA |
| 46 | Caenorhabditis elegans | 45.10 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 47 | Apis mellifera | 43.10 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 48 | Acropora millepora | 42.70 | 1 | integrin subunit betaCn1 |
| 49 | Oncorhynchus mykiss | 42.40 | 1 | CD18 protein |
| 50 | Manduca sexta | 39.30 | 1 | plasmatocyte-specific integrin beta 1 |
| 51 | Halocynthia roretzi | 38.90 | 2 | integrin beta Hr2 precursor |
| 52 | Cyprinus carpio | 35.00 | 2 | CD18 type 2 |
| 53 | Spodoptera frugiperda | 33.90 | 1 | integrin beta 1 |
| 54 | Geodia cydonium | 32.70 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 617 GLVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||| Query 617 GLVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEADSPIL 674 |
| Homo sapiens | Query 617 GLVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||| | Sbjct 741 GLVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEADSPTL 798 |
| Pan troglodytes | Query 617 GLVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||| | Sbjct 619 GLVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEADSPTL 676 |
| Bos taurus | Query 617 GLVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEADSPIL 674 |||||||||||||||||+ |||||+| |||#||| ||||+||||||+||||||||||+| Sbjct 743 GLVLAYRLSVEIYDRREFHRFEKERQHLNW#KQDHNPLYQSAITTTVNPRFQEADSPVL 800 |
| Mus musculus | Query 617 GLVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |||||||||||||||||| |||||||||||#|||+|||||||||||+||||| Sbjct 739 GLVLAYRLSVEIYDRREYRRFEKEQQQLNW#KQDNNPLYKSAITTTVNPRFQ 789 |
| Mus sp. | Query 617 GLVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |||||||||||||||||| |||||||||||#|||+|||||||||||+||||| Sbjct 740 GLVLAYRLSVEIYDRREYRRFEKEQQQLNW#KQDNNPLYKSAITTTVNPRFQ 790 |
| Rattus norvegicus | Query 617 GLVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |||||||||||+||| ||||||||+|||||#|||||||||||+|||+||||| Sbjct 574 GLVLAYRLSVEVYDRLEYSRFEKERQQLNW#KQDSNPLYKSAVTTTVNPRFQ 624 |
| Canis familiaris | Query 617 GLVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEADS 671 |||||||||||||||||+|||||||+ |||#||++||||+||||||+||+||| | Sbjct 673 GLVLAYRLSVEIYDRREFSRFEKEQKHLNW#KQENNPLYRSAITTTVNPQFQETGS 727 |
| Gallus gallus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 |+| +|| ||+||||| |||||+ + |# + |||+||| || +|||| Sbjct 721 LLLTWRLLTEIFDRREYRRFEKEKSKAKW#NEADNPLFKSATTTVMNPRF 769 |
| N/A | Query 644 LNW#KQDSNPLYKSAITTTINPRFQEADSPIL 674 |||#|||||||||||||||||||||||||| | Sbjct 1 LNW#KQDSNPLYKSAITTTINPRFQEADSPTL 31 |
| Danio rerio | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+| ++| + |+||||+ +||||+ |# ||+||||+|| +|||++ Sbjct 736 LLLIWKLLMVIHDRREFDKFEKEKNNAKW#DTGENPIYKSAVTTVVNPRYE 785 |
| Tetraodon nigroviridis | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+| ++| + |+||||+++||||+ |# ||+||||+|| +||+++ Sbjct 740 LLLIWKLLMIIHDRREFAKFEKEKMNAKW#DTGENPIYKSAVTTVVNPKYE 789 |
| Monodelphis domestica | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+| ++| + |+||||+++||||+ |# ||+||||+|| +||+++ Sbjct 748 LLLIWKLLMIIHDRREFAKFEKEKMNAKW#DTGENPIYKSAVTTVVNPKYE 797 |
| Xenopus laevis | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+| ++| + |+||||+++||||+ |# ||+||||+|| +||+++ Sbjct 747 LLLIWKLLMIIHDRREFAKFEKEKMNAKW#DTGENPIYKSAVTTVVNPKYE 796 |
| Sus scrofa | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+| ++| + |+||||+++||||+ |# ||+||||+|| +||+++ Sbjct 747 LLLIWKLLMIIHDRREFAKFEKEKMNAKW#DTGENPIYKSAVTTVVNPKYE 796 |
| synthetic construct | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+| ++| + |+||||+++||||+ |# ||+||||+|| +||+++ Sbjct 747 LLLIWKLLMIIHDRREFAKFEKEKMNAKW#DTGENPIYKSAVTTVVNPKYE 796 |
| Oryctolagus cuniculus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+| ++| + |+||||+++||||+ |# ||+||||+|| +||+++ Sbjct 12 LLLIWKLLMIIHDRREFAKFEKEKMNAKW#DTGENPIYKSAVTTVVNPKYE 61 |
| Pongo pygmaeus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+| ++| + |+||||+++||||+ |# ||+||||+|| +||+++ Sbjct 747 LLLIWKLLMIIHDRREFAKFEKEKMNAKW#DTGENPIYKSAVTTVVNPKYE 796 |
| Xenopus tropicalis | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+| ++| + |+||||+++||||+ |# ||+||||+|| +||+++ Sbjct 747 LLLIWKLLMIIHDRREFAKFEKEKMNAKW#DTGENPIYKSAVTTVVNPKYE 796 |
| Ictalurus punctatus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+ ++| + |+||||+++||||+ |# ||+||||+|| +||+++ Sbjct 756 LLFIWKLLMIIHDRREFAKFEKEKMNAKW#DAGENPIYKSAVTTVVNPKYE 805 |
| Felis catus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+| ++| + |+| ||+++||||+ |# ||+||||+|| +||+++ Sbjct 747 LLLIWKLLMIIHDTREFAKFEKEKMNAKW#DTGENPIYKSAVTTVVNPKYE 796 |
| Tribolium castaneum | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQ 667 |+| +++ ++||+||++|| |+|++ |# ++ ||||+ | +| || ++ Sbjct 703 LLLTWKVCTTVHDRQEYAKFENERQKMQW#HRNDNPLYRQATSTFKNPVYR 752 |
| Aedes aegypti | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 ++| ++| |+||||++|||||+ |# ||+|| | || || + Sbjct 784 VLLLWKLLTTIHDRREFARFEKERMMAKW#DTGENPIYKQATTTFKNPTY 832 |
| Pacifastacus leniusculus | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEA 669 +| ++| ++|||||++||||++ + #|+ ||||||| || || | ++ Sbjct 750 LLIWKLLTTLHDRREYAKFEKERKLPSG#KRAENPLYKSAKTTFQNPAFAQS 800 |
| Capra hircus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEA 669 |++ ++ + | ||| |||||+ + |# | |||+||| || +||+| |+ Sbjct 720 LLVIWKALTHLSDLREYHRFEKEKLKSQW#NND-NPLFKSATTTVMNPKFAES 770 |
| Bubalus bubalis | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEA 669 |++ ++ + | ||| |||||+ + |# | |||+||| || +||+| |+ Sbjct 719 LLVIWKALTHLSDLREYHRFEKEKLKSQW#NND-NPLFKSATTTVMNPKFAES 769 |
| Anopheles gambiae | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 ++| +++ |+||||++|||||+ |# ||+|| | || || + Sbjct 786 VLLLWKVLTSIHDRREFARFEKERMMAKW#DTGENPIYKQATTTFKNPTY 834 |
| Anopheles gambiae str. PEST | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 ++| +++ |+||||++|||||+ |# ||+|| | || || + Sbjct 786 VLLLWKVLTSIHDRREFARFEKERMMAKW#DTGENPIYKQATTTFKNPTY 834 |
| Strongylocentrotus purpuratus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQE 668 |+| +|| | + | ||++ ||||+ +|# |+ ||+|| + +| || +|+ Sbjct 733 LLLVWRLLVYVQDSREFASFEKERAGTHW#GQNENPIYKPSTSTFKNPTYQK 783 |
| Ovis aries | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEA 669 |+ ++ + | ||| |||||+ + |# | |||+||| || +||+| |+ Sbjct 720 LLAIWKALTHLSDLREYHRFEKEKLKSQW#NND-NPLFKSATTTVMNPKFAES 770 |
| Drosophila melanogaster | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 ++| ++| |+||||++|||||+ |# ||+|| | +| || + Sbjct 795 ILLLWKLLTTIHDRREFARFEKERMNAKW#DTGENPIYKQATSTFKNPMY 843 |
| Ovis canadensis | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEA 669 |+ ++ + | ||| |||||+ + |# | |||+||| || +||+| |+ Sbjct 720 LLAIWKALTHLSDLREYHRFEKEKLKSQW#NND-NPLFKSATTTVMNPKFAES 770 |
| Crassostrea gigas | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQE 668 |+| ++| | |+|| +||||| |# ||+|| | +| +|| +++ Sbjct 749 LLLIWKLFTTISDKRELARFEKEAMNARW#DTGENPIYKQATSTFVNPTYRQ 799 |
| Ostrinia furnacalis | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 |++ +++ |+||||++|||||+ |# ||+|| | +| || + Sbjct 780 LLMLWKMVTTIHDRREFARFEKERMMAKW#DTGENPIYKQATSTFKNPTY 828 |
| Sigmodon hispidus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQEA 669 |++ ++ + | ||| ||||+ + |# | |||+||| || +||+| |+ Sbjct 719 LLVIWKALTHLTDLREYRHFEKEKLKSQW#NND-NPLFKSATTTVMNPKFAES 769 |
| Podocoryne carnea | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 |+| ++| | ||||+++|||++| |# ||+|| | +| || + Sbjct 730 LLLIWKLLATIQDRREFAKFEKDRQNPKW#DSGENPIYKKATSTFQNPMY 778 |
| Schistosoma japonicum | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRFQE 668 |+| |+| + | |||| + |+++ + + |#+ ||+++| | +|| |+| Sbjct 235 LLLIYKLVITIDDRRELANFKQQGENMRW#EMAENPIFESPTTNVLNPTFEE 285 |
| Biomphalaria glabrata | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 |+| ++| |+| ||+++||||+| |# ||+|| | +| || + Sbjct 737 LLLLWKLLTFIHDTREFAKFEKERQNAKW#DTGENPIYKQATSTFKNPTY 785 |
| Pseudoplusia includens | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 |++ ++++ +||||++|||||+ |# ||+|| | +| || + Sbjct 786 LLMLWKMATTSHDRREFARFEKERMMAKW#DTGENPIYKQATSTFKNPTY 834 |
| Lytechinus variegatus | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 |+| +|| |+||||+ ||||+ |#+ ||+|| + + || + Sbjct 755 LLLIWRLLTYIHDRREFQNFEKERANATW#EGGENPIYKPSTSVFKNPTY 803 |
| Canis lupus familiaris | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTIN 663 +| ++| + |+||+|+++||+|+ + |# +||||| | +| | Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKW#DTANNPLYKEATSTFTN 778 |
| Caenorhabditis briggsae | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINP 664 |+| ++| ++|| ||++| |+ |# + ||+|| | || || Sbjct 757 LLLLWKLLTVLHDRAEYAKFNNERLMAKW#DTNENPIYKQATTTFKNP 803 |
| rats, Peptide Partial, 723 aa | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTIN 663 +| ++| + |+||+|+++||+|+ + |# +||||| | +| | Sbjct 673 LLIWKLLITIHDRKEFAKFEEERARAKW#DTANNPLYKEATSTFTN 717 |
| Equus caballus | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTIN 663 +| ++| + |+||+|+++||+|+ + |# +||||| | +| | Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKW#DTANNPLYKEATSTFTN 778 |
| mice, Peptide Partial, 680 aa | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTIN 663 +| ++| + |+||+|+++||+|+ + |# +||||| | +| | Sbjct 634 LLIWKLLITIHDRKEFAKFEEERARAKW#DTANNPLYKEATSTFTN 678 |
| Caenorhabditis elegans | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINP 664 |+| ++| ++|| ||+ | |+ |# + ||+|| | || || Sbjct 756 LLLLWKLLTVLHDRSEYATFNNERLMAKW#DTNENPIYKQATTTFKNP 802 |
| Apis mellifera | Query 618 LVLAYRLSVEIYDRREYSRFEKEQQQLN-W#KQDSNPLYKSAITTTINPRF 666 ++| +++ | | ||+ +||+|+ | |# + ||||| | +| || | Sbjct 764 ILLTWKILTMIRDNREFKKFERERILANKW#NRRDNPLYKEATSTFKNPTF 813 |
| Acropora millepora | Query 630 DRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 || || +||+|+ |# ++ ||||++| || || + Sbjct 751 DRIEYQKFERERMHSKW#TREKNPLYQAAKTTFENPTY 787 |
| Oncorhynchus mykiss | Query 617 GLVLAYRLSVEIY--DRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 ||++ + +| | +|+ ||| ||++ |# |||+ +| || || | Sbjct 723 GLLILLIIKAILYMRDVKEFRRFENEQKKGKW#SPGDNPLFMNATTTVANPTF 774 |
| Manduca sexta | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINP 664 ++|+++ |+++|+|||++||+| + +# ||||+ || Sbjct 714 LIAWKILVDLHDKREYAKFEEESRSRGF#DVSLNPLYQEPEINFSNP 759 |
| Halocynthia roretzi | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 ++ +++ + |+||| +|+||+ |# + ||+| +| + || | Sbjct 784 LIIWKVIQTLRDKREYEKFKKEEDLRKW#TKGENPVYVNASSKFDNPMF 831 |
| Cyprinus carpio | Query 617 GLVLAYRLSVEIY--DRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINPRF 666 ||++ + +| | ||+ ||||+++ # +|||+++| || || | Sbjct 713 GLLILLIIKAVLYASDLREWKRFEKDRKHEK-#TSGTNPLFQNATTTVQNPTF 763 |
| Spodoptera frugiperda | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINP 664 |+ +++ |+++|++|| +|+ | +# ||||+ || Sbjct 729 VIIWKILVDMHDKKEYKKFQDEALAAGY#DVSLNPLYQDPSINFSNP 774 |
| Geodia cydonium | Query 619 VLAYRLSVEIYDRREYSRFEKEQQQLNW#KQDSNPLYKSAITTTINP 664 ++ ++ + + | | +|| | ++ +# ++ |||| || | || Sbjct 829 LILLKILLMVLDYTELKKFENELAKVKY#TKNENPLYHSATTEHKNP 874 |
[Site 8] NWKQDSNPLY654-KSAITTTINP
Tyr654  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn645 | Trp646 | Lys647 | Gln648 | Asp649 | Ser650 | Asn651 | Pro652 | Leu653 | Tyr654 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys655 | Ser656 | Ala657 | Ile658 | Thr659 | Thr660 | Thr661 | Ile662 | Asn663 | Pro664 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| SVEIYDRREYSRFEKEQQQLNWKQDSNPLYKSAITTTINPRFQEADSPIL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 105.00 | 11 | PREDICTED: integrin, beta 7 |
| 2 | Homo sapiens | 103.00 | 19 | integrin, beta 7 |
| 3 | Pan troglodytes | 103.00 | 13 | PREDICTED: integrin, beta 7 isoform 2 |
| 4 | Bos taurus | 94.00 | 13 | PREDICTED: similar to Integrin beta-7 |
| 5 | Mus musculus | 88.60 | 14 | integrin beta 7 |
| 6 | Mus sp. | 88.60 | 1 | beta 7 integrin |
| 7 | Rattus norvegicus | 86.70 | 10 | integrin beta-7 subunit |
| 8 | Canis familiaris | 84.30 | 10 | PREDICTED: similar to Integrin beta-7 precursor |
| 9 | N/A | 65.90 | 26 | C |
| 10 | Gallus gallus | 60.10 | 7 | integrin, beta 2 |
| 11 | Danio rerio | 56.60 | 14 | integrin beta1 subunit-like protein 3 |
| 12 | Sus scrofa | 55.80 | 6 | integrin beta-1 subunit |
| 13 | Monodelphis domestica | 55.80 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 14 | Xenopus laevis | 55.80 | 6 | integrin beta-1 subunit |
| 15 | Tetraodon nigroviridis | 55.80 | 5 | unnamed protein product |
| 16 | synthetic construct | 55.80 | 5 | hypothetical protein |
| 17 | Oryctolagus cuniculus | 55.80 | 2 | integrin beta 1 |
| 18 | Ictalurus punctatus | 55.80 | 2 | beta-1 integrin |
| 19 | Pongo pygmaeus | 55.80 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 20 | Xenopus tropicalis | 55.80 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 21 | Felis catus | 53.50 | 1 | integrin beta 1 |
| 22 | Ovis aries | 50.10 | 2 | antigen CD18 |
| 23 | Bubalus bubalis | 50.10 | 1 | integrin beta-2 |
| 24 | Capra hircus | 50.10 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 25 | Ovis canadensis | 50.10 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 26 | Anopheles gambiae | 48.90 | 2 | integrin beta subunit |
| 27 | Pacifastacus leniusculus | 48.90 | 1 | integrin |
| 28 | Anopheles gambiae str. PEST | 48.90 | 1 | ENSANGP00000021373 |
| 29 | Aedes aegypti | 48.90 | 1 | integrin beta subunit |
| 30 | Tribolium castaneum | 48.10 | 3 | PREDICTED: similar to CG1560-PA |
| 31 | Sigmodon hispidus | 48.10 | 1 | AF445415_1 integrin beta-2 precursor |
| 32 | Drosophila melanogaster | 47.40 | 1 | myospheroid CG1560-PA |
| 33 | Ostrinia furnacalis | 47.40 | 1 | integrin beta 1 precursor |
| 34 | Biomphalaria glabrata | 45.80 | 1 | beta integrin subunit |
| 35 | Crassostrea gigas | 45.80 | 1 | integrin beta cgh |
| 36 | Pseudoplusia includens | 45.80 | 1 | integrin beta 1 |
| 37 | Podocoryne carnea | 45.40 | 1 | AF308652_1 integrin beta chain |
| 38 | Strongylocentrotus purpuratus | 44.70 | 5 | integrin beta G subunit |
| 39 | Acropora millepora | 42.70 | 1 | integrin subunit betaCn1 |
| 40 | Schistosoma japonicum | 42.70 | 1 | SJCHGC06221 protein |
| 41 | Canis lupus familiaris | 42.40 | 1 | integrin beta chain, beta 3 |
| 42 | rats, Peptide Partial, 723 aa | 42.40 | 1 | beta 3 integrin, GPIIIA |
| 43 | Equus caballus | 42.40 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 44 | mice, Peptide Partial, 680 aa | 42.40 | 1 | beta 3 integrin, GPIIIA |
| 45 | Lytechinus variegatus | 41.60 | 1 | beta-C integrin subunit |
| 46 | Caenorhabditis briggsae | 41.60 | 1 | Hypothetical protein CBG03601 |
| 47 | Oncorhynchus mykiss | 41.20 | 1 | CD18 protein |
| 48 | Caenorhabditis elegans | 40.40 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 49 | Apis mellifera | 39.70 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 50 | Halocynthia roretzi | 38.10 | 2 | integrin beta Hr2 precursor |
| 51 | Manduca sexta | 35.00 | 1 | plasmatocyte-specific integrin beta 1 |
| 52 | Cyprinus carpio | 33.50 | 2 | CD18 type 2 |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 625 SVEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 625 SVEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEADSPIL 674 |
| Homo sapiens | Query 625 SVEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||||||||| | Sbjct 749 SVEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEADSPTL 798 |
| Pan troglodytes | Query 625 SVEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#|||||||||||||||||| | Sbjct 627 SVEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEADSPTL 676 |
| Bos taurus | Query 625 SVEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEADSPIL 674 |||||||||+ |||||+| |||||| ||||#+||||||+||||||||||+| Sbjct 751 SVEIYDRREFHRFEKERQHLNWKQDHNPLY#QSAITTTVNPRFQEADSPVL 800 |
| Mus musculus | Query 625 SVEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |||||||||| ||||||||||||||+||||#|||||||+||||| Sbjct 747 SVEIYDRREYRRFEKEQQQLNWKQDNNPLY#KSAITTTVNPRFQ 789 |
| Mus sp. | Query 625 SVEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |||||||||| ||||||||||||||+||||#|||||||+||||| Sbjct 748 SVEIYDRREYRRFEKEQQQLNWKQDNNPLY#KSAITTTVNPRFQ 790 |
| Rattus norvegicus | Query 625 SVEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |||+||| ||||||||+|||||||||||||#|||+|||+||||| Sbjct 582 SVEVYDRLEYSRFEKERQQLNWKQDSNPLY#KSAVTTTVNPRFQ 624 |
| Canis familiaris | Query 625 SVEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEADS 671 |||||||||+|||||||+ |||||++||||#+||||||+||+||| | Sbjct 681 SVEIYDRREFSRFEKEQKHLNWKQENNPLY#RSAITTTVNPQFQETGS 727 |
| N/A | Query 644 LNWKQDSNPLY#KSAITTTINPRFQEADSPIL 674 |||||||||||#|||||||||||||||||| | Sbjct 1 LNWKQDSNPLY#KSAITTTINPRFQEADSPTL 31 |
| Gallus gallus | Query 627 EIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 ||+||||| |||||+ + | + |||+#||| || +|||| Sbjct 730 EIFDRREYRRFEKEKSKAKWNEADNPLF#KSATTTVMNPRF 769 |
| Danio rerio | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |+||||+ +||||+ | ||+|#|||+|| +|||++ Sbjct 746 IHDRREFDKFEKEKNNAKWDTGENPIY#KSAVTTVVNPRYE 785 |
| Sus scrofa | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |+||||+++||||+ | ||+|#|||+|| +||+++ Sbjct 757 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYE 796 |
| Monodelphis domestica | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |+||||+++||||+ | ||+|#|||+|| +||+++ Sbjct 758 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYE 797 |
| Xenopus laevis | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |+||||+++||||+ | ||+|#|||+|| +||+++ Sbjct 757 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYE 796 |
| Tetraodon nigroviridis | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |+||||+++||||+ | ||+|#|||+|| +||+++ Sbjct 750 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYE 789 |
| synthetic construct | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |+||||+++||||+ | ||+|#|||+|| +||+++ Sbjct 757 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYE 796 |
| Oryctolagus cuniculus | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |+||||+++||||+ | ||+|#|||+|| +||+++ Sbjct 22 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYE 61 |
| Ictalurus punctatus | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |+||||+++||||+ | ||+|#|||+|| +||+++ Sbjct 766 IHDRREFAKFEKEKMNAKWDAGENPIY#KSAVTTVVNPKYE 805 |
| Pongo pygmaeus | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |+||||+++||||+ | ||+|#|||+|| +||+++ Sbjct 757 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYE 796 |
| Xenopus tropicalis | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |+||||+++||||+ | ||+|#|||+|| +||+++ Sbjct 757 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYE 796 |
| Felis catus | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 |+| ||+++||||+ | ||+|#|||+|| +||+++ Sbjct 757 IHDTREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYE 796 |
| Ovis aries | Query 630 DRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEA 669 | ||| |||||+ + | | |||+#||| || +||+| |+ Sbjct 732 DLREYHRFEKEKLKSQWNND-NPLF#KSATTTVMNPKFAES 770 |
| Bubalus bubalis | Query 630 DRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEA 669 | ||| |||||+ + | | |||+#||| || +||+| |+ Sbjct 731 DLREYHRFEKEKLKSQWNND-NPLF#KSATTTVMNPKFAES 769 |
| Capra hircus | Query 630 DRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEA 669 | ||| |||||+ + | | |||+#||| || +||+| |+ Sbjct 732 DLREYHRFEKEKLKSQWNND-NPLF#KSATTTVMNPKFAES 770 |
| Ovis canadensis | Query 630 DRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEA 669 | ||| |||||+ + | | |||+#||| || +||+| |+ Sbjct 732 DLREYHRFEKEKLKSQWNND-NPLF#KSATTTVMNPKFAES 770 |
| Anopheles gambiae | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 |+||||++|||||+ | ||+|#| | || || + Sbjct 796 IHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTY 834 |
| Pacifastacus leniusculus | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEA 669 ++|||||++||||++ + |+ ||||#||| || || | ++ Sbjct 759 LHDRREYAKFEKERKLPSGKRAENPLY#KSAKTTFQNPAFAQS 800 |
| Anopheles gambiae str. PEST | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 |+||||++|||||+ | ||+|#| | || || + Sbjct 796 IHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTY 834 |
| Aedes aegypti | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 |+||||++|||||+ | ||+|#| | || || + Sbjct 794 IHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTY 832 |
| Tribolium castaneum | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQ 667 ++||+||++|| |+|++ | ++ ||||#+ | +| || ++ Sbjct 713 VHDRQEYAKFENERQKMQWHRNDNPLY#RQATSTFKNPVYR 752 |
| Sigmodon hispidus | Query 630 DRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEA 669 | ||| ||||+ + | | |||+#||| || +||+| |+ Sbjct 731 DLREYRHFEKEKLKSQWNND-NPLF#KSATTTVMNPKFAES 769 |
| Drosophila melanogaster | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 |+||||++|||||+ | ||+|#| | +| || + Sbjct 805 IHDRREFARFEKERMNAKWDTGENPIY#KQATSTFKNPMY 843 |
| Ostrinia furnacalis | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 |+||||++|||||+ | ||+|#| | +| || + Sbjct 790 IHDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTY 828 |
| Biomphalaria glabrata | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 |+| ||+++||||+| | ||+|#| | +| || + Sbjct 747 IHDTREFAKFEKERQNAKWDTGENPIY#KQATSTFKNPTY 785 |
| Crassostrea gigas | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQE 668 | |+|| +||||| | ||+|#| | +| +|| +++ Sbjct 759 ISDKRELARFEKEAMNARWDTGENPIY#KQATSTFVNPTYRQ 799 |
| Pseudoplusia includens | Query 629 YDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 +||||++|||||+ | ||+|#| | +| || + Sbjct 797 HDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTY 834 |
| Podocoryne carnea | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 | ||||+++|||++| | ||+|#| | +| || + Sbjct 740 IQDRREFAKFEKDRQNPKWDSGENPIY#KKATSTFQNPMY 778 |
| Strongylocentrotus purpuratus | Query 626 VEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQE 668 | + | ||++ ||||+ +| |+ ||+|#| + +| || +|+ Sbjct 741 VYVQDSREFASFEKERAGTHWGQNENPIY#KPSTSTFKNPTYQK 783 |
| Acropora millepora | Query 630 DRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 || || +||+|+ | ++ ||||#++| || || + Sbjct 751 DRIEYQKFERERMHSKWTREKNPLY#QAAKTTFENPTY 787 |
| Schistosoma japonicum | Query 626 VEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQE 668 + | |||| + |+++ + + |+ ||++#+| | +|| |+| Sbjct 243 ITIDDRRELANFKQQGENMRWEMAENPIF#ESPTTNVLNPTFEE 285 |
| Canis lupus familiaris | Query 626 VEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTIN 663 + |+||+|+++||+|+ + | +||||#| | +| | Sbjct 741 ITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 778 |
| rats, Peptide Partial, 723 aa | Query 626 VEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTIN 663 + |+||+|+++||+|+ + | +||||#| | +| | Sbjct 680 ITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 717 |
| Equus caballus | Query 626 VEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTIN 663 + |+||+|+++||+|+ + | +||||#| | +| | Sbjct 741 ITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 778 |
| mice, Peptide Partial, 680 aa | Query 626 VEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTIN 663 + |+||+|+++||+|+ + | +||||#| | +| | Sbjct 641 ITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 678 |
| Lytechinus variegatus | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 |+||||+ ||||+ |+ ||+|#| + + || + Sbjct 765 IHDRREFQNFEKERANATWEGGENPIY#KPSTSVFKNPTY 803 |
| Caenorhabditis briggsae | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINP 664 ++|| ||++| |+ | + ||+|#| | || || Sbjct 767 LHDRAEYAKFNNERLMAKWDTNENPIY#KQATTTFKNP 803 |
| Oncorhynchus mykiss | Query 630 DRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 | +|+ ||| ||++ | |||+# +| || || | Sbjct 738 DVKEFRRFENEQKKGKWSPGDNPLF#MNATTTVANPTF 774 |
| Caenorhabditis elegans | Query 628 IYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINP 664 ++|| ||+ | |+ | + ||+|#| | || || Sbjct 766 LHDRSEYATFNNERLMAKWDTNENPIY#KQATTTFKNP 802 |
| Apis mellifera | Query 628 IYDRREYSRFEKEQQQLN-WKQDSNPLY#KSAITTTINPRF 666 | | ||+ +||+|+ | | + ||||#| | +| || | Sbjct 774 IRDNREFKKFERERILANKWNRRDNPLY#KEATSTFKNPTF 813 |
| Halocynthia roretzi | Query 630 DRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 |+||| +|+||+ | + ||+|# +| + || | Sbjct 795 DKREYEKFKKEEDLRKWTKGENPVY#VNASSKFDNPMF 831 |
| Manduca sexta | Query 626 VEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINP 664 |+++|+|||++||+| + + ||||#+ || Sbjct 721 VDLHDKREYAKFEEESRSRGFDVSLNPLY#QEPEINFSNP 759 |
| Cyprinus carpio | Query 630 DRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRF 666 | ||+ ||||+++ +|||+#++| || || | Sbjct 728 DLREWKRFEKDRKHEK-TSGTNPLF#QNATTTVQNPTF 763 |
[Site 9] EYSRFEKEQQ642-QLNWKQDSNP
Gln642  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu633 | Tyr634 | Ser635 | Arg636 | Phe637 | Glu638 | Lys639 | Glu640 | Gln641 | Gln642 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln643 | Leu644 | Asn645 | Trp646 | Lys647 | Gln648 | Asp649 | Ser650 | Asn651 | Pro652 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| AVGLGLVLAYRLSVEIYDRREYSRFEKEQQQLNWKQDSNPLYKSAITTTINPRFQEADSP |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 123.00 | 25 | integrin, beta 7 |
| 2 | Pan troglodytes | 123.00 | 15 | PREDICTED: integrin, beta 7 isoform 2 |
| 3 | Macaca mulatta | 123.00 | 13 | PREDICTED: integrin, beta 7 |
| 4 | Bos taurus | 112.00 | 13 | PREDICTED: similar to Integrin beta-7 |
| 5 | Mus musculus | 110.00 | 14 | integrin beta 7 |
| 6 | Mus sp. | 110.00 | 1 | beta 7 integrin |
| 7 | Rattus norvegicus | 108.00 | 10 | integrin beta-7 subunit |
| 8 | Canis familiaris | 105.00 | 10 | PREDICTED: similar to Integrin beta-7 precursor |
| 9 | Gallus gallus | 70.50 | 7 | integrin, beta 2 |
| 10 | Danio rerio | 66.60 | 14 | integrin beta1 subunit-like protein 3 |
| 11 | N/A | 65.50 | 30 | - |
| 12 | Sus scrofa | 65.50 | 6 | integrin beta-1 subunit |
| 13 | Monodelphis domestica | 65.50 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 14 | Tetraodon nigroviridis | 65.50 | 6 | unnamed protein product |
| 15 | Xenopus laevis | 65.50 | 6 | integrin beta 1 |
| 16 | synthetic construct | 65.50 | 5 | hypothetical protein |
| 17 | Oryctolagus cuniculus | 65.50 | 2 | integrin beta 1 |
| 18 | Pongo pygmaeus | 65.50 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 19 | Xenopus tropicalis | 65.50 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 20 | Ictalurus punctatus | 63.90 | 2 | beta-1 integrin |
| 21 | Felis catus | 63.20 | 1 | integrin beta 1 |
| 22 | Aedes aegypti | 57.80 | 1 | integrin beta subunit |
| 23 | Strongylocentrotus purpuratus | 57.00 | 6 | integrin beta G subunit |
| 24 | Drosophila melanogaster | 57.00 | 1 | myospheroid CG1560-PA |
| 25 | Pacifastacus leniusculus | 57.00 | 1 | integrin |
| 26 | Anopheles gambiae | 56.20 | 2 | integrin beta subunit |
| 27 | Anopheles gambiae str. PEST | 56.20 | 2 | ENSANGP00000021373 |
| 28 | Ostrinia furnacalis | 56.20 | 1 | integrin beta 1 precursor |
| 29 | Biomphalaria glabrata | 55.80 | 1 | beta integrin subunit |
| 30 | Podocoryne carnea | 55.50 | 1 | AF308652_1 integrin beta chain |
| 31 | Bubalus bubalis | 55.50 | 1 | integrin beta-2 |
| 32 | Capra hircus | 55.50 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 33 | Pseudoplusia includens | 55.10 | 1 | integrin beta 1 |
| 34 | Ovis aries | 54.70 | 2 | antigen CD18 |
| 35 | Ovis canadensis | 54.70 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 36 | Tribolium castaneum | 53.90 | 3 | PREDICTED: similar to CG1560-PA |
| 37 | Lytechinus variegatus | 52.80 | 1 | beta-C integrin subunit |
| 38 | Sigmodon hispidus | 52.80 | 1 | AF445415_1 integrin beta-2 precursor |
| 39 | Schistosoma japonicum | 52.40 | 1 | SJCHGC06221 protein |
| 40 | Crassostrea gigas | 52.40 | 1 | integrin beta cgh |
| 41 | rats, Peptide Partial, 723 aa | 50.80 | 1 | beta 3 integrin, GPIIIA |
| 42 | Canis lupus familiaris | 50.80 | 1 | integrin beta chain, beta 3 |
| 43 | Equus caballus | 50.80 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 44 | mice, Peptide Partial, 680 aa | 50.80 | 1 | beta 3 integrin, GPIIIA |
| 45 | Caenorhabditis briggsae | 48.10 | 1 | Hypothetical protein CBG03601 |
| 46 | Caenorhabditis elegans | 47.00 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 47 | Apis mellifera | 46.60 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 48 | Oncorhynchus mykiss | 45.40 | 1 | CD18 protein |
| 49 | Acropora millepora | 44.70 | 1 | integrin subunit betaCn1 |
| 50 | Manduca sexta | 42.40 | 1 | plasmatocyte-specific integrin beta 1 |
| 51 | Halocynthia roretzi | 41.20 | 2 | integrin beta Hr2 precursor |
| 52 | Cyprinus carpio | 37.00 | 2 | CD18 type 2 |
| 53 | Spodoptera frugiperda | 36.20 | 1 | integrin beta 1 |
| 54 | Geodia cydonium | 34.30 | 1 | integrin beta subunit |
| 55 | Suberites domuncula | 33.50 | 1 | integrin beta subunit |
| 56 | Ophlitaspongia tenuis | 33.50 | 1 | integrin subunit betaPo1 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEADSP 672 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEADSP 672 |
| Pan troglodytes | Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEADSP 672 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEADSP 672 |
| Macaca mulatta | Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEADSP 672 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEADSP 672 |
| Bos taurus | Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEADSP 672 |||||||||||||||||||||+ |||||+|# |||||| ||||+||||||+|||||||||| Sbjct 739 AVGLGLVLAYRLSVEIYDRREFHRFEKERQ#HLNWKQDHNPLYQSAITTTVNPRFQEADSP 798 |
| Mus musculus | Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 |||||||||||||||||||||| |||||||#|||||||+|||||||||||+||||| Sbjct 735 AVGLGLVLAYRLSVEIYDRREYRRFEKEQQ#QLNWKQDNNPLYKSAITTTVNPRFQ 789 |
| Mus sp. | Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 |||||||||||||||||||||| |||||||#|||||||+|||||||||||+||||| Sbjct 736 AVGLGLVLAYRLSVEIYDRREYRRFEKEQQ#QLNWKQDNNPLYKSAITTTVNPRFQ 790 |
| Rattus norvegicus | Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 |||||||||||||||+||| ||||||||+|#|||||||||||||||+|||+||||| Sbjct 570 AVGLGLVLAYRLSVEVYDRLEYSRFEKERQ#QLNWKQDSNPLYKSAVTTTVNPRFQ 624 |
| Canis familiaris | Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEADS 671 |||||||||||||||||||||+|||||||+# |||||++||||+||||||+||+||| | Sbjct 669 AVGLGLVLAYRLSVEIYDRREFSRFEKEQK#HLNWKQENNPLYRSAITTTVNPQFQETGS 727 |
| Gallus gallus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 +|| |+| +|| ||+||||| |||||+ #+ | + |||+||| || +|||| Sbjct 717 IGLLLLLTWRLLTEIFDRREYRRFEKEKS#KAKWNEADNPLFKSATTTVMNPRF 769 |
| Danio rerio | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+| ++| + |+||||+ +||||+ # | ||+||||+|| +|||++ Sbjct 732 IGLALLLIWKLLMVIHDRREFDKFEKEKN#NAKWDTGENPIYKSAVTTVVNPRYE 785 |
| N/A | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+| ++| + |+||||+++||||+ # | ||+||||+|| +||+++ Sbjct 720 IGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYE 773 |
| Sus scrofa | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+| ++| + |+||||+++||||+ # | ||+||||+|| +||+++ Sbjct 743 IGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYE 796 |
| Monodelphis domestica | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+| ++| + |+||||+++||||+ # | ||+||||+|| +||+++ Sbjct 744 IGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYE 797 |
| Tetraodon nigroviridis | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+| ++| + |+||||+++||||+ # | ||+||||+|| +||+++ Sbjct 736 IGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYE 789 |
| Xenopus laevis | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+| ++| + |+||||+++||||+ # | ||+||||+|| +||+++ Sbjct 743 IGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYE 796 |
| synthetic construct | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+| ++| + |+||||+++||||+ # | ||+||||+|| +||+++ Sbjct 743 IGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYE 796 |
| Oryctolagus cuniculus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+| ++| + |+||||+++||||+ # | ||+||||+|| +||+++ Sbjct 8 IGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYE 61 |
| Pongo pygmaeus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+| ++| + |+||||+++||||+ # | ||+||||+|| +||+++ Sbjct 743 IGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYE 796 |
| Xenopus tropicalis | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+| ++| + |+||||+++||||+ # | ||+||||+|| +||+++ Sbjct 743 IGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYE 796 |
| Ictalurus punctatus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+ ++| + |+||||+++||||+ # | ||+||||+|| +||+++ Sbjct 752 IGLALLFIWKLLMIIHDRREFAKFEKEKM#NAKWDAGENPIYKSAVTTVVNPKYE 805 |
| Felis catus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 +|| |+| ++| + |+| ||+++||||+ # | ||+||||+|| +||+++ Sbjct 743 IGLALLLIWKLLMIIHDTREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYE 796 |
| Aedes aegypti | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 +|| ++| ++| |+||||++|||||+ # | ||+|| | || || + Sbjct 780 IGLAVLLLWKLLTTIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTY 832 |
| Strongylocentrotus purpuratus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQE 668 ||| |+| +|| | + | ||++ ||||+ # +| |+ ||+|| + +| || +|+ Sbjct 729 VGLALLLVWRLLVYVQDSREFASFEKERA#GTHWGQNENPIYKPSTSTFKNPTYQK 783 |
| Drosophila melanogaster | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 ||| ++| ++| |+||||++|||||+ # | ||+|| | +| || + Sbjct 791 VGLAILLLWKLLTTIHDRREFARFEKERM#NAKWDTGENPIYKQATSTFKNPMY 843 |
| Pacifastacus leniusculus | Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEA 669 |+|| +| ++| ++|||||++||||++# + |+ ||||||| || || | ++ Sbjct 744 AIGLLTLLIWKLLTTLHDRREYAKFEKERK#LPSGKRAENPLYKSAKTTFQNPAFAQS 800 |
| Anopheles gambiae | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 +|+ ++| +++ |+||||++|||||+ # | ||+|| | || || + Sbjct 782 IGMAVLLLWKVLTSIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTY 834 |
| Anopheles gambiae str. PEST | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 +|+ ++| +++ |+||||++|||||+ # | ||+|| | || || + Sbjct 782 IGMAVLLLWKVLTSIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTY 834 |
| Ostrinia furnacalis | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 ||| |++ +++ |+||||++|||||+ # | ||+|| | +| || + Sbjct 776 VGLALLMLWKMVTTIHDRREFARFEKERM#MAKWDTGENPIYKQATSTFKNPTY 828 |
| Biomphalaria glabrata | Query 613 AVGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 |||| |+| ++| |+| ||+++||||+|# | ||+|| | +| || + Sbjct 732 AVGLFLLLLWKLLTFIHDTREFAKFEKERQ#NAKWDTGENPIYKQATSTFKNPTY 785 |
| Podocoryne carnea | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 +|| |+| ++| | ||||+++|||++|# | ||+|| | +| || + Sbjct 726 IGLALLLIWKLLATIQDRREFAKFEKDRQ#NPKWDSGENPIYKKATSTFQNPMY 778 |
| Bubalus bubalis | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEA 669 ||+ |++ ++ + | ||| |||||+ #+ | | |||+||| || +||+| |+ Sbjct 715 VGILLLVIWKALTHLSDLREYHRFEKEKL#KSQWNND-NPLFKSATTTVMNPKFAES 769 |
| Capra hircus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEA 669 ||+ |++ ++ + | ||| |||||+ #+ | | |||+||| || +||+| |+ Sbjct 716 VGILLLVIWKALTHLSDLREYHRFEKEKL#KSQWNND-NPLFKSATTTVMNPKFAES 770 |
| Pseudoplusia includens | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 ||| |++ ++++ +||||++|||||+ # | ||+|| | +| || + Sbjct 782 VGLALLMLWKMATTSHDRREFARFEKERM#MAKWDTGENPIYKQATSTFKNPTY 834 |
| Ovis aries | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEA 669 ||+ |+ ++ + | ||| |||||+ #+ | | |||+||| || +||+| |+ Sbjct 716 VGILLLAIWKALTHLSDLREYHRFEKEKL#KSQWNND-NPLFKSATTTVMNPKFAES 770 |
| Ovis canadensis | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEA 669 ||+ |+ ++ + | ||| |||||+ #+ | | |||+||| || +||+| |+ Sbjct 716 VGILLLAIWKALTHLSDLREYHRFEKEKL#KSQWNND-NPLFKSATTTVMNPKFAES 770 |
| Tribolium castaneum | Query 615 GLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQ 667 |+ |+| +++ ++||+||++|| |+|#++ | ++ ||||+ | +| || ++ Sbjct 700 GVILLLTWKVCTTVHDRQEYAKFENERQ#KMQWHRNDNPLYRQATSTFKNPVYR 752 |
| Lytechinus variegatus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 ||| |+| +|| |+||||+ ||||+ # |+ ||+|| + + || + Sbjct 751 VGLALLLIWRLLTYIHDRREFQNFEKERA#NATWEGGENPIYKPSTSVFKNPTY 803 |
| Sigmodon hispidus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQEA 669 +|+ |++ ++ + | ||| ||||+ #+ | | |||+||| || +||+| |+ Sbjct 715 IGVLLLVIWKALTHLTDLREYRHFEKEKL#KSQWNND-NPLFKSATTTVMNPKFAES 769 |
| Schistosoma japonicum | Query 615 GLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQE 668 || |+| |+| + | |||| + |+++ +# + |+ ||+++| | +|| |+| Sbjct 232 GLILLLIYKLVITIDDRRELANFKQQGE#NMRWEMAENPIFESPTTNVLNPTFEE 285 |
| Crassostrea gigas | Query 615 GLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRFQE 668 |+ |+| ++| | |+|| +||||| # | ||+|| | +| +|| +++ Sbjct 746 GIILLLIWKLFTTISDKRELARFEKEAM#NARWDTGENPIYKQATSTFVNPTYRQ 799 |
| rats, Peptide Partial, 723 aa | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTIN 663 +|| +| ++| + |+||+|+++||+|+ #+ | +||||| | +| | Sbjct 668 IGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTN 717 |
| Canis lupus familiaris | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTIN 663 +|| +| ++| + |+||+|+++||+|+ #+ | +||||| | +| | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTN 778 |
| Equus caballus | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTIN 663 +|| +| ++| + |+||+|+++||+|+ #+ | +||||| | +| | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTN 778 |
| mice, Peptide Partial, 680 aa | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTIN 663 +|| +| ++| + |+||+|+++||+|+ #+ | +||||| | +| | Sbjct 629 IGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTN 678 |
| Caenorhabditis briggsae | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINP 664 +|+ |+| ++| ++|| ||++| |+ # | + ||+|| | || || Sbjct 753 LGILLLLLWKLLTVLHDRAEYAKFNNERL#MAKWDTNENPIYKQATTTFKNP 803 |
| Caenorhabditis elegans | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINP 664 +|+ |+| ++| ++|| ||+ | |+ # | + ||+|| | || || Sbjct 752 LGILLLLLWKLLTVLHDRSEYATFNNERL#MAKWDTNENPIYKQATTTFKNP 802 |
| Apis mellifera | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLN-WKQDSNPLYKSAITTTINPRF 666 +| ++| +++ | | ||+ +||+|+ # | | + ||||| | +| || | Sbjct 760 IGFAILLTWKILTMIRDNREFKKFERERI#LANKWNRRDNPLYKEATSTFKNPTF 813 |
| Oncorhynchus mykiss | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 +|| ++| + + + | +|+ ||| ||+#+ | |||+ +| || || | Sbjct 722 IGLLILLIIKAILYMRDVKEFRRFENEQK#KGKWSPGDNPLFMNATTTVANPTF 774 |
| Acropora millepora | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 +|| +++ + + || || +||+|+ # | ++ ||||++| || || + Sbjct 735 LGLLILIVIKGLFTMVDRIEYQKFERERM#HSKWTREKNPLYQAAKTTFENPTY 787 |
| Manduca sexta | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINP 664 +|| ++|+++ |+++|+|||++||+| +# + ||||+ || Sbjct 709 IGLLTLIAWKILVDLHDKREYAKFEEESR#SRGFDVSLNPLYQEPEINFSNP 759 |
| Halocynthia roretzi | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 +|+ ++ +++ + |+||| +|+||+ # | + ||+| +| + || | Sbjct 779 IGIIALIIWKVIQTLRDKREYEKFKKEED#LRKWTKGENPVYVNASSKFDNPMF 831 |
| Cyprinus carpio | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 +|| ++| + + | ||+ ||||+++# +|||+++| || || | Sbjct 712 IGLLILLIIKAVLYASDLREWKRFEKDRK#HEK-TSGTNPLFQNATTTVQNPTF 763 |
| Spodoptera frugiperda | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINP 664 +|+ |+ +++ |+++|++|| +|+ | # + ||||+ || Sbjct 724 IGILTVIIWKILVDMHDKKEYKKFQDEAL#AAGYDVSLNPLYQDPSINFSNP 774 |
| Geodia cydonium | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINP 664 +|+ ++ ++ + + | | +|| | #++ + ++ |||| || | || Sbjct 824 LGILALILLKILLMVLDYTELKKFENELA#KVKYTKNENPLYHSATTEHKNP 874 |
| Suberites domuncula | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINP 664 +|+ +| +| + | | | +|||| #+ + + ||||+|| || Sbjct 710 LGVIALLLLKLFLFIIDVIEVKKFEKELA#KTKYPKHQNPLYRSAAKDYQNP 760 |
| Ophlitaspongia tenuis | Query 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQ#QLNWKQDSNPLYKSAITTTINPRF 666 +|| |+| + + ++| | +||+| +# + ++ ||||+|| || + Sbjct 784 LGLLLLLLLKGLLLLWDVVEVRKFEREIK#NAKYTKNENPLYRSATKDYQNPLY 836 |
[Site 10] EQQQLNWKQD649-SNPLYKSAIT
Asp649  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu640 | Gln641 | Gln642 | Gln643 | Leu644 | Asn645 | Trp646 | Lys647 | Gln648 | Asp649 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser650 | Asn651 | Pro652 | Leu653 | Tyr654 | Lys655 | Ser656 | Ala657 | Ile658 | Thr659 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LAYRLSVEIYDRREYSRFEKEQQQLNWKQDSNPLYKSAITTTINPRFQEADSPIL |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 114.00 | 12 | PREDICTED: integrin, beta 7 |
| 2 | Homo sapiens | 112.00 | 22 | integrin, beta 7 |
| 3 | Pan troglodytes | 112.00 | 13 | PREDICTED: integrin, beta 7 isoform 2 |
| 4 | Bos taurus | 103.00 | 13 | PREDICTED: similar to Integrin beta-7 |
| 5 | Mus musculus | 97.80 | 14 | integrin beta 7 |
| 6 | Mus sp. | 97.80 | 1 | beta 7 integrin |
| 7 | Rattus norvegicus | 95.90 | 10 | integrin beta-7 subunit |
| 8 | Canis familiaris | 93.60 | 10 | PREDICTED: similar to Integrin beta-7 precursor |
| 9 | N/A | 65.90 | 27 | C |
| 10 | Gallus gallus | 65.10 | 7 | integrin, beta 2 |
| 11 | Danio rerio | 59.70 | 14 | integrin beta1 subunit-like protein 3 |
| 12 | Tetraodon nigroviridis | 58.50 | 6 | unnamed protein product |
| 13 | Xenopus laevis | 58.50 | 6 | integrin beta 1 |
| 14 | Monodelphis domestica | 58.50 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 15 | Sus scrofa | 58.50 | 6 | integrin beta-1 subunit |
| 16 | synthetic construct | 58.50 | 5 | hypothetical protein |
| 17 | Pongo pygmaeus | 58.50 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 18 | Oryctolagus cuniculus | 58.50 | 2 | integrin beta 1 |
| 19 | Xenopus tropicalis | 58.50 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 20 | Ictalurus punctatus | 57.40 | 2 | beta-1 integrin |
| 21 | Felis catus | 56.20 | 1 | integrin beta 1 |
| 22 | Pacifastacus leniusculus | 52.00 | 1 | integrin |
| 23 | Aedes aegypti | 52.00 | 1 | integrin beta subunit |
| 24 | Anopheles gambiae | 51.20 | 2 | integrin beta subunit |
| 25 | Anopheles gambiae str. PEST | 51.20 | 2 | ENSANGP00000021373 |
| 26 | Tribolium castaneum | 50.80 | 3 | PREDICTED: similar to CG1560-PA |
| 27 | Ovis aries | 50.40 | 2 | antigen CD18 |
| 28 | Ovis canadensis | 50.40 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 29 | Bubalus bubalis | 50.40 | 1 | integrin beta-2 |
| 30 | Capra hircus | 50.40 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 31 | Drosophila melanogaster | 50.40 | 1 | myospheroid CG1560-PA |
| 32 | Strongylocentrotus purpuratus | 49.70 | 5 | integrin beta G subunit |
| 33 | Crassostrea gigas | 48.90 | 1 | integrin beta cgh |
| 34 | Ostrinia furnacalis | 48.90 | 1 | integrin beta 1 precursor |
| 35 | Podocoryne carnea | 48.50 | 1 | AF308652_1 integrin beta chain |
| 36 | Sigmodon hispidus | 48.50 | 1 | AF445415_1 integrin beta-2 precursor |
| 37 | Biomphalaria glabrata | 48.10 | 1 | beta integrin subunit |
| 38 | Schistosoma japonicum | 48.10 | 1 | SJCHGC06221 protein |
| 39 | Pseudoplusia includens | 47.80 | 1 | integrin beta 1 |
| 40 | Canis lupus familiaris | 45.80 | 1 | integrin beta chain, beta 3 |
| 41 | rats, Peptide Partial, 723 aa | 45.80 | 1 | beta 3 integrin, GPIIIA |
| 42 | Equus caballus | 45.80 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 43 | mice, Peptide Partial, 680 aa | 45.80 | 1 | beta 3 integrin, GPIIIA |
| 44 | Lytechinus variegatus | 45.40 | 1 | beta-C integrin subunit |
| 45 | Caenorhabditis briggsae | 44.30 | 1 | Hypothetical protein CBG03601 |
| 46 | Caenorhabditis elegans | 43.10 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 47 | Acropora millepora | 42.70 | 1 | integrin subunit betaCn1 |
| 48 | Apis mellifera | 42.00 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 49 | Oncorhynchus mykiss | 41.20 | 1 | CD18 protein |
| 50 | Manduca sexta | 38.90 | 1 | plasmatocyte-specific integrin beta 1 |
| 51 | Halocynthia roretzi | 38.50 | 2 | integrin beta Hr2 precursor |
| 52 | Cyprinus carpio | 33.50 | 2 | CD18 type 2 |
| 53 | Spodoptera frugiperda | 32.30 | 1 | integrin beta 1 |
| 54 | Geodia cydonium | 32.30 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEADSPIL 674 |
| Homo sapiens | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#||||||||||||||||||||||| | Sbjct 744 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEADSPTL 798 |
| Pan troglodytes | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||||||||||||||||||#||||||||||||||||||||||| | Sbjct 622 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEADSPTL 676 |
| Bos taurus | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEADSPIL 674 ||||||||||||||+ |||||+| ||||||# ||||+||||||+||||||||||+| Sbjct 746 LAYRLSVEIYDRREFHRFEKERQHLNWKQD#HNPLYQSAITTTVNPRFQEADSPVL 800 |
| Mus musculus | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 ||||||||||||||| ||||||||||||||#+|||||||||||+||||| Sbjct 742 LAYRLSVEIYDRREYRRFEKEQQQLNWKQD#NNPLYKSAITTTVNPRFQ 789 |
| Mus sp. | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 ||||||||||||||| ||||||||||||||#+|||||||||||+||||| Sbjct 743 LAYRLSVEIYDRREYRRFEKEQQQLNWKQD#NNPLYKSAITTTVNPRFQ 790 |
| Rattus norvegicus | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 ||||||||+||| ||||||||+||||||||#||||||||+|||+||||| Sbjct 577 LAYRLSVEVYDRLEYSRFEKERQQLNWKQD#SNPLYKSAVTTTVNPRFQ 624 |
| Canis familiaris | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEADS 671 ||||||||||||||+|||||||+ |||||+#+||||+||||||+||+||| | Sbjct 676 LAYRLSVEIYDRREFSRFEKEQKHLNWKQE#NNPLYRSAITTTVNPQFQETGS 727 |
| N/A | Query 644 LNWKQD#SNPLYKSAITTTINPRFQEADSPIL 674 ||||||#||||||||||||||||||||||| | Sbjct 1 LNWKQD#SNPLYKSAITTTINPRFQEADSPTL 31 |
| Gallus gallus | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 | +|| ||+||||| |||||+ + | + # |||+||| || +|||| Sbjct 723 LTWRLLTEIFDRREYRRFEKEKSKAKWNEA#DNPLFKSATTTVMNPRF 769 |
| Danio rerio | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 | ++| + |+||||+ +||||+ | # ||+||||+|| +|||++ Sbjct 738 LIWKLLMVIHDRREFDKFEKEKNNAKWDTG#ENPIYKSAVTTVVNPRYE 785 |
| Tetraodon nigroviridis | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 | ++| + |+||||+++||||+ | # ||+||||+|| +||+++ Sbjct 742 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYE 789 |
| Xenopus laevis | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 | ++| + |+||||+++||||+ | # ||+||||+|| +||+++ Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYE 796 |
| Monodelphis domestica | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 | ++| + |+||||+++||||+ | # ||+||||+|| +||+++ Sbjct 750 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYE 797 |
| Sus scrofa | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 | ++| + |+||||+++||||+ | # ||+||||+|| +||+++ Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYE 796 |
| synthetic construct | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 | ++| + |+||||+++||||+ | # ||+||||+|| +||+++ Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYE 796 |
| Pongo pygmaeus | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 | ++| + |+||||+++||||+ | # ||+||||+|| +||+++ Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYE 796 |
| Oryctolagus cuniculus | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 | ++| + |+||||+++||||+ | # ||+||||+|| +||+++ Sbjct 14 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYE 61 |
| Xenopus tropicalis | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 | ++| + |+||||+++||||+ | # ||+||||+|| +||+++ Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYE 796 |
| Ictalurus punctatus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 ++| + |+||||+++||||+ | # ||+||||+|| +||+++ Sbjct 760 WKLLMIIHDRREFAKFEKEKMNAKWDAG#ENPIYKSAVTTVVNPKYE 805 |
| Felis catus | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 | ++| + |+| ||+++||||+ | # ||+||||+|| +||+++ Sbjct 749 LIWKLLMIIHDTREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYE 796 |
| Pacifastacus leniusculus | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEA 669 | ++| ++|||||++||||++ + |+ # ||||||| || || | ++ Sbjct 751 LIWKLLTTLHDRREYAKFEKERKLPSGKRA#ENPLYKSAKTTFQNPAFAQS 800 |
| Aedes aegypti | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 | ++| |+||||++|||||+ | # ||+|| | || || + Sbjct 786 LLWKLLTTIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTY 832 |
| Anopheles gambiae | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 | +++ |+||||++|||||+ | # ||+|| | || || + Sbjct 788 LLWKVLTSIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTY 834 |
| Anopheles gambiae str. PEST | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 | +++ |+||||++|||||+ | # ||+|| | || || + Sbjct 788 LLWKVLTSIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTY 834 |
| Tribolium castaneum | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQ 667 | +++ ++||+||++|| |+|++ | ++# ||||+ | +| || ++ Sbjct 705 LTWKVCTTVHDRQEYAKFENERQKMQWHRN#DNPLYRQATSTFKNPVYR 752 |
| Ovis aries | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEA 669 ++ + | ||| |||||+ + | |# |||+||| || +||+| |+ Sbjct 724 WKALTHLSDLREYHRFEKEKLKSQWNND#-NPLFKSATTTVMNPKFAES 770 |
| Ovis canadensis | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEA 669 ++ + | ||| |||||+ + | |# |||+||| || +||+| |+ Sbjct 724 WKALTHLSDLREYHRFEKEKLKSQWNND#-NPLFKSATTTVMNPKFAES 770 |
| Bubalus bubalis | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEA 669 ++ + | ||| |||||+ + | |# |||+||| || +||+| |+ Sbjct 723 WKALTHLSDLREYHRFEKEKLKSQWNND#-NPLFKSATTTVMNPKFAES 769 |
| Capra hircus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEA 669 ++ + | ||| |||||+ + | |# |||+||| || +||+| |+ Sbjct 724 WKALTHLSDLREYHRFEKEKLKSQWNND#-NPLFKSATTTVMNPKFAES 770 |
| Drosophila melanogaster | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 | ++| |+||||++|||||+ | # ||+|| | +| || + Sbjct 797 LLWKLLTTIHDRREFARFEKERMNAKWDTG#ENPIYKQATSTFKNPMY 843 |
| Strongylocentrotus purpuratus | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQE 668 | +|| | + | ||++ ||||+ +| |+# ||+|| + +| || +|+ Sbjct 735 LVWRLLVYVQDSREFASFEKERAGTHWGQN#ENPIYKPSTSTFKNPTYQK 783 |
| Crassostrea gigas | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQE 668 | ++| | |+|| +||||| | # ||+|| | +| +|| +++ Sbjct 751 LIWKLFTTISDKRELARFEKEAMNARWDTG#ENPIYKQATSTFVNPTYRQ 799 |
| Ostrinia furnacalis | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 + +++ |+||||++|||||+ | # ||+|| | +| || + Sbjct 782 MLWKMVTTIHDRREFARFEKERMMAKWDTG#ENPIYKQATSTFKNPTY 828 |
| Podocoryne carnea | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 | ++| | ||||+++|||++| | # ||+|| | +| || + Sbjct 732 LIWKLLATIQDRREFAKFEKDRQNPKWDSG#ENPIYKKATSTFQNPMY 778 |
| Sigmodon hispidus | Query 622 YRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQEA 669 ++ + | ||| ||||+ + | |# |||+||| || +||+| |+ Sbjct 723 WKALTHLTDLREYRHFEKEKLKSQWNND#-NPLFKSATTTVMNPKFAES 769 |
| Biomphalaria glabrata | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 | ++| |+| ||+++||||+| | # ||+|| | +| || + Sbjct 739 LLWKLLTFIHDTREFAKFEKERQNAKWDTG#ENPIYKQATSTFKNPTY 785 |
| Schistosoma japonicum | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRFQE 668 | |+| + | |||| + |+++ + + |+ # ||+++| | +|| |+| Sbjct 237 LIYKLVITIDDRRELANFKQQGENMRWEMA#ENPIFESPTTNVLNPTFEE 285 |
| Pseudoplusia includens | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 + ++++ +||||++|||||+ | # ||+|| | +| || + Sbjct 788 MLWKMATTSHDRREFARFEKERMMAKWDTG#ENPIYKQATSTFKNPTY 834 |
| Canis lupus familiaris | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTIN 663 | ++| + |+||+|+++||+|+ + | #+||||| | +| | Sbjct 735 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTN 778 |
| rats, Peptide Partial, 723 aa | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTIN 663 | ++| + |+||+|+++||+|+ + | #+||||| | +| | Sbjct 674 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTN 717 |
| Equus caballus | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTIN 663 | ++| + |+||+|+++||+|+ + | #+||||| | +| | Sbjct 735 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTN 778 |
| mice, Peptide Partial, 680 aa | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTIN 663 | ++| + |+||+|+++||+|+ + | #+||||| | +| | Sbjct 635 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTN 678 |
| Lytechinus variegatus | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 | +|| |+||||+ ||||+ |+ # ||+|| + + || + Sbjct 757 LIWRLLTYIHDRREFQNFEKERANATWEGG#ENPIYKPSTSVFKNPTY 803 |
| Caenorhabditis briggsae | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINP 664 | ++| ++|| ||++| |+ | +# ||+|| | || || Sbjct 759 LLWKLLTVLHDRAEYAKFNNERLMAKWDTN#ENPIYKQATTTFKNP 803 |
| Caenorhabditis elegans | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINP 664 | ++| ++|| ||+ | |+ | +# ||+|| | || || Sbjct 758 LLWKLLTVLHDRSEYATFNNERLMAKWDTN#ENPIYKQATTTFKNP 802 |
| Acropora millepora | Query 630 DRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 || || +||+|+ | ++# ||||++| || || + Sbjct 751 DRIEYQKFERERMHSKWTRE#KNPLYQAAKTTFENPTY 787 |
| Apis mellifera | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLN-WKQD#SNPLYKSAITTTINPRF 666 | +++ | | ||+ +||+|+ | | + # ||||| | +| || | Sbjct 766 LTWKILTMIRDNREFKKFERERILANKWNRR#DNPLYKEATSTFKNPTF 813 |
| Oncorhynchus mykiss | Query 630 DRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 | +|+ ||| ||++ | # |||+ +| || || | Sbjct 738 DVKEFRRFENEQKKGKWSPG#DNPLFMNATTTVANPTF 774 |
| Manduca sexta | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINP 664 +|+++ |+++|+|||++||+| + + # ||||+ || Sbjct 715 IAWKILVDLHDKREYAKFEEESRSRGFDVS#LNPLYQEPEINFSNP 759 |
| Halocynthia roretzi | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 + +++ + |+||| +|+||+ | + # ||+| +| + || | Sbjct 785 IIWKVIQTLRDKREYEKFKKEEDLRKWTKG#ENPVYVNASSKFDNPMF 831 |
| Cyprinus carpio | Query 630 DRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINPRF 666 | ||+ ||||+++ #+|||+++| || || | Sbjct 728 DLREWKRFEKDRKHEK-TSG#TNPLFQNATTTVQNPTF 763 |
| Spodoptera frugiperda | Query 620 LAYRLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINP 664 + +++ |+++|++|| +|+ | + # ||||+ || Sbjct 730 IIWKILVDMHDKKEYKKFQDEALAAGYDVS#LNPLYQDPSINFSNP 774 |
| Geodia cydonium | Query 623 RLSVEIYDRREYSRFEKEQQQLNWKQD#SNPLYKSAITTTINP 664 ++ + + | | +|| | ++ + ++# |||| || | || Sbjct 833 KILLMVLDYTELKKFENELAKVKYTKN#ENPLYHSATTEHKNP 874 |
* References
None.