XSB0035 : PREDICTED: similar to cortactin isoform a [Macaca mulatta]
[ CaMP Format ]
This entry is computationally expanded from SB0022
* Basic Information
| Organism | Macaca mulatta (rhesus monkey) |
| Protein Names | similar to cortactin isoform a |
| Gene Names | LOC708291; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 1 mRNA, 79 ESTs, 10 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001100193 | XM_001100193 | 708291 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
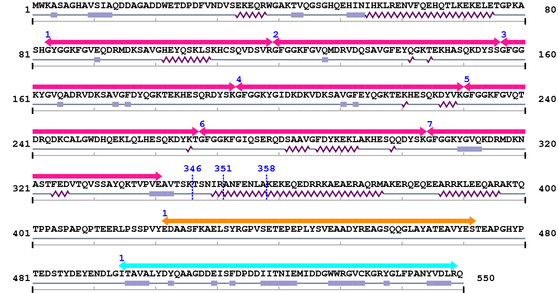
Length: 550 aa
Average Mass: 61.642 kDa
Monoisotopic Mass: 61.605 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| HS1_rep 1. | 83 | 119 | 68.2 | 3.1e-17 |
| HS1_rep 2. | 120 | 156 | 72.7 | 1.3e-18 |
| HS1_rep 3. | 157 | 193 | 74.4 | 4.1e-19 |
| HS1_rep 4. | 194 | 230 | 69.9 | 9.2e-18 |
| HS1_rep 5. | 231 | 267 | 68.5 | 2.5e-17 |
| HS1_rep 6. | 268 | 304 | 69.5 | 1.2e-17 |
| HS1_rep 7. | 305 | 341 | 29.7 | 1.2e-05 |
| --- cleavage 346 --- | ||||
| --- cleavage 358 --- | ||||
| --- cleavage 351 --- | ||||
| PSD5 1. | 422 | 472 | -24.2 | 3.5 |
| SH3_1 1. | 495 | 549 | 88.0 | 3.3e-23 |
3. Sequence Information
Fasta Sequence: XSB0035.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
3 [sites]
Cleavage sites (±10aa)
[Site 1] TVPVEAVTSK346-TSNIRANFEN
Lys346  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Thr337 | Val338 | Pro339 | Val340 | Glu341 | Ala342 | Val343 | Thr344 | Ser345 | Lys346 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr347 | Ser348 | Asn349 | Ile350 | Arg351 | Ala352 | Asn353 | Phe354 | Glu355 | Asn356 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| MDKNASTFEDVTQVSSAYQKTVPVEAVTSKTSNIRANFENLAKEKEQEDRRKAEAERAQR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 117.00 | 10 | cortactin isoform a |
| 2 | synthetic construct | 117.00 | 3 | cortactin |
| 3 | Macaca mulatta | 117.00 | 2 | PREDICTED: similar to cortactin isoform a |
| 4 | Macaca fascicularis | 117.00 | 2 | unnamed protein product |
| 5 | Pongo pygmaeus | 115.00 | 1 | hypothetical protein |
| 6 | Mus musculus | 110.00 | 6 | cortactin |
| 7 | Rattus norvegicus | 110.00 | 6 | cortactin isoform B |
| 8 | N/A | 110.00 | 2 | S |
| 9 | Bos taurus | 110.00 | 2 | cortactin |
| 10 | mice, BALB/c 3T3 cells, Peptide, 546 aa | 110.00 | 1 | cortactin, p80/p85 |
| 11 | Canis familiaris | 108.00 | 6 | PREDICTED: similar to cortactin isoform a isoform |
| 12 | Gallus gallus | 95.50 | 2 | cortactin |
| 13 | Xenopus laevis | 91.30 | 4 | MGC80961 protein |
| 14 | Danio rerio | 74.70 | 6 | cortactin |
| 15 | Tetraodon nigroviridis | 73.20 | 4 | unnamed protein product |
| 16 | Xenopus tropicalis | 64.30 | 1 | cortactin |
| 17 | Strongylocentrotus purpuratus | 52.00 | 1 | SRC8 protein |
| 18 | Aedes aegypti | 50.10 | 2 | cortactin |
| 19 | Pan troglodytes | 48.50 | 2 | PREDICTED: hypothetical protein isoform 1 |
| 20 | Drosophila melanogaster | 39.70 | 2 | Cortactin |
| 21 | Drosophila pseudoobscura | 38.50 | 1 | GA17577-PA |
| 22 | Monodelphis domestica | 33.90 | 1 | PREDICTED: similar to KIAA1561 protein |
| 23 | Arabidopsis thaliana | 33.50 | 2 | unnamed protein product |
| 24 | Trichomonas vaginalis G3 | 33.50 | 1 | hypothetical protein TVAG_334050 |
| 25 | Anopheles gambiae str. PEST | 33.50 | 1 | ENSANGP00000029554 |
| 26 | Tetrahymena thermophila SB210 | 32.30 | 1 | hypothetical protein TTHERM_00218900 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |
| synthetic construct | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |
| Macaca mulatta | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |
| Macaca fascicularis | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |
| Pongo pygmaeus | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||||||||||||||||||||||||||| ||#|||||||||||||||||||||||||||||| Sbjct 280 MDKNASTFEDVTQVSSAYQKTVPVEAVASK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 339 |
| Mus musculus | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||||||||+| || ||||||||+||||||#||||||||||||||+||||||||||||||| Sbjct 317 MDKNASTFEEVVQVPSAYQKTVPIEAVTSK#TSNIRANFENLAKEREQEDRRKAEAERAQR 376 |
| Rattus norvegicus | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||||||||+| || ||||||||+||||||#||||||||||||||+||||||||||||||| Sbjct 280 MDKNASTFEEVVQVPSAYQKTVPIEAVTSK#TSNIRANFENLAKEREQEDRRKAEAERAQR 339 |
| N/A | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||||||||+| || ||||||||+||||||#||||||||||||||+||||||||||||||| Sbjct 317 MDKNASTFEEVVQVPSAYQKTVPIEAVTSK#TSNIRANFENLAKEREQEDRRKAEAERAQR 376 |
| Bos taurus | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||||||||||| |||+|||||||||| ||#|||||||||||||||||||||||||||||| Sbjct 317 MDKNASTFEDVAAVSSSYQKTVPVEAVNSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |
| mice, BALB/c 3T3 cells, Peptide, 546 aa | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||||||||+| || ||||||||+||||||#||||||||||||||+||||||||||||||| Sbjct 317 MDKNASTFEEVVQVPSAYQKTVPIEAVTSK#TSNIRANFENLAKEREQEDRRKAEAERAQR 376 |
| Canis familiaris | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||||||||||| ||+ ||||||||||| |+#||||||||||||||||||||||||||+||| Sbjct 317 MDKNASTFEDVAQVAPAYQKTVPVEAVNSR#TSNIRANFENLAKEKEQEDRRKAEAEKAQR 376 |
| Gallus gallus | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||||+||||+ + +| |||| ||| | +|#||+|||| |||||||||||||||||||||| Sbjct 326 MDKNAATFEDIEKPTSTYQKTKPVERVANK#TSSIRANLENLAKEKEQEDRRKAEAERAQR 385 |
| Xenopus laevis | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||| |++|||| +|||+|||| ||| ||# |+||||||||||+||||||+||||||||| Sbjct 315 MDKAAASFEDVEKVSSSYQKTRPVEVEGSK#ASSIRANFENLAKDKEQEDRKKAEAERAQR 374 |
| Danio rerio | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||+| |||+| + |+||||| |||| +| # |+|+| |||+||+||+|++++ | ||++| Sbjct 316 MDKSAGTFEEVQKPSAAYQKTRPVEAASSS#ASSIKARFENIAKQKEEEEQQRLEEERSRR 375 |
| Tetraodon nigroviridis | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ||| | |||+| + + +|||| |||| | # +|+| |||+||+|| ||||+|| || +| Sbjct 325 MDKAAGTFEEVEKPTPSYQKTKPVEAAGSN#AGSIKARFENMAKQKEDEDRRRAEEERLRR 384 |
| Xenopus tropicalis | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 +|| || | ++ ||||+|| |+||+|| # |+|+ |||+|+ |+|+++||| || +| Sbjct 278 VDKCASGFGEIETPSSAYEKTQPLEAMTSG#AHNLRSRFENMARTAEEENKQKAEEERVRR 337 |
| Strongylocentrotus purpuratus | Query 318 DKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 | | | |+ |||+|+|| | | # |+| || +|+ |+| ||||| || +| Sbjct 281 DATAGGFGDMQGVSSSYKKTRPQPPAKSG#AGNMRNKFEQMAQAGEEESRRKAEEERGRR 339 |
| Aedes aegypti | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 |||+| |++ ++ + | || | + ++|# ||||| ||| | |+| |+||| ++ | Sbjct 241 MDKSALGFQEQDKIGTNYTKTKP-DIGSAK#PSNIRARFENFAMAAEEETRKKAEEQKRLR 299 |
| Pan troglodytes | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 +||+| | ++ ++||+|| |+|| +| # ++| ||++|+|| |++ | |+||+ Sbjct 246 VDKSAVGFNEMEAPTTAYKKTTPIEAASSG#ARGLKAKFESMAEEK----RKREEEEKAQQ 301 |
| Drosophila melanogaster | Query 345 SK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 +|# ||+|| |||||| |+| |++|| ++ | Sbjct 238 AK#PSNLRAKFENLAKNSEEESRKRAEEQKRLR 269 |
| Drosophila pseudoobscura | Query 345 SK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 +|# ||+|| |||||| |+| |+++| ++ | Sbjct 236 AK#PSNLRAKFENLAKNSEEESRKRSEEQKRLR 267 |
| Monodelphis domestica | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRKAEAERA 374 +++|| +| + || ||+ | + # +++ + | | +| + + || ||+|| Sbjct 1146 LEENAKQNSEVLAAQALLQKHVPLGQVEAL#KNSLSSTIEALREELKDKQRRHEEAQRA 1203 |
| Arabidopsis thaliana | Query 342 AVTSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 | |# | +|+ + + ||||+|||++ || + +| Sbjct 736 ARADK#LRNFKADLQKMKKEKEEEDRKRIEALKIER 770 |
| Trichomonas vaginalis G3 | Query 317 MDKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKE 362 + | | ||+|| + |+| | ++ + + # |++|| + || Sbjct 42 LKKYPSAFENVTMICSSYDKEQAIDYLMTS#IKNVQANLNQIFASKE 87 |
| Anopheles gambiae str. PEST | Query 344 TSK#TSNIRANFENLAKEKEQEDRRKAEAERAQR 376 ++|# ||+|| ||| | |+| |++|+ ++ | Sbjct 237 SAK#PSNLRAKFENFAATAEEEARKRADEQKRLR 269 |
| Tetrahymena thermophila SB210 | Query 318 DKNASTFEDVTQVSSAYQKTVPVEAVTSK#TSNIRANFENLAKEKEQEDRRK 368 |+|+| ++ |+||| +| ||+ | # +| + +| + ||+ ++| Sbjct 1543 DQNSSNIKNDTEVSSHEEKNNQNEAIQSI#KQQVRFDVQNENESDEQDQKQK 1593 |
[Site 2] NIRANFENLA358-KEKEQEDRRK
Ala358  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn349 | Ile350 | Arg351 | Ala352 | Asn353 | Phe354 | Glu355 | Asn356 | Leu357 | Ala358 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys359 | Glu360 | Lys361 | Glu362 | Gln363 | Glu364 | Asp365 | Arg366 | Arg367 | Lys368 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QVSSAYQKTVPVEAVTSKTSNIRANFENLAKEKEQEDRRKAEAERAQRMAKERQEQEEAR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 116.00 | 36 | SRC8_HUMAN Src substrate cortactin (Amplaxin) (Onc |
| 2 | Macaca mulatta | 116.00 | 6 | PREDICTED: similar to cortactin isoform a |
| 3 | synthetic construct | 116.00 | 3 | cortactin |
| 4 | Macaca fascicularis | 116.00 | 3 | unnamed protein product |
| 5 | Pongo pygmaeus | 114.00 | 1 | hypothetical protein |
| 6 | Mus musculus | 112.00 | 30 | cortactin |
| 7 | N/A | 112.00 | 2 | S |
| 8 | mice, BALB/c 3T3 cells, Peptide, 546 aa | 112.00 | 1 | cortactin, p80/p85 |
| 9 | Rattus norvegicus | 111.00 | 11 | cortactin isoform B |
| 10 | Canis familiaris | 108.00 | 15 | PREDICTED: similar to cortactin isoform a isoform |
| 11 | Bos taurus | 107.00 | 4 | cortactin |
| 12 | Gallus gallus | 96.30 | 3 | cortactin |
| 13 | Xenopus laevis | 91.30 | 4 | cortactin |
| 14 | Danio rerio | 78.20 | 14 | cortactin |
| 15 | Tetraodon nigroviridis | 77.40 | 7 | unnamed protein product |
| 16 | Xenopus tropicalis | 67.00 | 2 | cortactin |
| 17 | Strongylocentrotus purpuratus | 55.50 | 6 | SRC8 protein |
| 18 | Aedes aegypti | 51.60 | 2 | cortactin |
| 19 | Drosophila melanogaster | 50.80 | 5 | Cortactin |
| 20 | Drosophila pseudoobscura | 49.70 | 1 | GA17577-PA |
| 21 | Pan troglodytes | 49.30 | 11 | PREDICTED: hypothetical protein isoform 1 |
| 22 | Anopheles gambiae str. PEST | 42.40 | 2 | ENSANGP00000029554 |
| 23 | Oryza sativa (japonica cultivar-group) | 38.10 | 1 | Os02g0602300 |
| 24 | Babesia bovis | 37.40 | 1 | anonymous antigen-2 |
| 25 | Magnaporthe grisea 70-15 | 37.00 | 4 | hypothetical protein MGCH7_ch7g791 |
| 26 | Parvibaculum lavamentivorans DS-1 | 37.00 | 1 | translation initiation factor IF-2 |
| 27 | Felis catus | 37.00 | 1 | drebrin |
| 28 | Oryctolagus cuniculus | 37.00 | 1 | drebrin 1 isoform a |
| 29 | Lodderomyces elongisporus NRRL YB-4239 | 37.00 | 1 | hypothetical protein LELG_02889 |
| 30 | Entamoeba histolytica HM-1:IMSS | 36.20 | 2 | leucine rich repeat protein |
| 31 | Candida glabrata | 36.20 | 1 | unnamed protein product |
| 32 | Aspergillus nidulans FGSC A4 | 35.80 | 4 | hypothetical protein AN6060.2 |
| 33 | Monodelphis domestica | 35.80 | 3 | PREDICTED: similar to NAD(+) ADP-ribosyltransferas |
| 34 | Tetrahymena thermophila SB210 | 35.80 | 2 | hypothetical protein TTHERM_00043920 |
| 35 | Dichelobacter nodosus VCS1703A | 35.80 | 1 | TolA protein |
| 36 | Aspergillus clavatus NRRL 1 | 35.80 | 1 | ubiquitin-protein ligase (Tom1), putative |
| 37 | Reinekea sp. MED297 | 35.80 | 1 | hypothetical protein MED297_01825 |
| 38 | Giardia lamblia ATCC 50803 | 35.80 | 1 | hypothetical protein GLP_384_18902_20299 |
| 39 | Arabidopsis thaliana | 35.40 | 4 | heavy-metal-associated domain-containing protein |
| 40 | Borrelia burgdorferi | 35.40 | 3 | ErpB |
| 41 | Candida albicans SC5314 | 35.40 | 1 | putative Sir2 family histone deacetylase |
| 42 | Borrelia burgdorferi B31 | 35.40 | 1 | ErpB |
| 43 | Pichia guilliermondii ATCC 6260 | 35.40 | 1 | hypothetical protein PGUG_04149 |
| 44 | Trypanosoma cruzi strain CL Brener | 35.00 | 1 | cytoskeleton-associated protein CAP5.5 |
| 45 | Oryza sativa (indica cultivar-group) | 35.00 | 1 | hypothetical protein OsI_010140 |
| 46 | Tribolium castaneum | 35.00 | 1 | PREDICTED: similar to CG11905-PB, isoform B |
| 47 | Trypanosoma brucei TREU927 | 35.00 | 1 | hypothetical protein Tb11.01.7450 |
| 48 | Caenorhabditis elegans | 34.70 | 5 | T05F1.11 |
| 49 | Dictyostelium discoideum AX4 | 34.70 | 4 | YEATS family protein |
| 50 | Cryptosporidium hominis TU502 | 34.70 | 1 | hypothetical protein Chro.10068 |
| 51 | Dictyostelium discoideum | 34.70 | 1 | AC116032_6 hypothetical protein |
| 52 | Cryptosporidium parvum Iowa II | 34.70 | 1 | predicted secreted protein, signal peptide |
| 53 | Trichomonas vaginalis G3 | 34.30 | 7 | hypothetical protein TVAG_466120 |
| 54 | Paramecium tetraurelia | 34.30 | 2 | hypothetical protein |
| 55 | Plasmodium reichenowi | 34.30 | 1 | reticulocyte binding-like protein 2b |
| 56 | Myxococcus xanthus DK 1622 | 34.30 | 1 | hypothetical protein MXAN_5743 |
| 57 | Bacillus cereus ATCC 14579 | 34.30 | 1 | hypothetical protein BC0683 |
| 58 | Myxococcus xanthus | 34.30 | 1 | unknown |
| 59 | Aspergillus fumigatus Af293 | 34.30 | 1 | histone deacetylase complex subunit (Hos4), putati |
| 60 | Aspergillus niger | 34.30 | 1 | hypothetical protein An02g07370 |
| 61 | Coprinopsis cinerea okayama7#130 | 33.90 | 3 | predicted protein |
| 62 | Coccidioides immitis RS | 33.90 | 1 | hypothetical protein CIMG_06977 |
| 63 | Lyngbya sp. PCC 8106 | 33.90 | 1 | Multi-sensor Hybrid Histidine Kinase |
| 64 | Vibrio alginolyticus 12G01 | 33.90 | 1 | TolA protein |
| 65 | Neosartorya fischeri NRRL 181 | 33.90 | 1 | mitochondrial translation initiation factor IF-2, |
| 66 | Ostreococcus lucimarinus CCE9901 | 33.90 | 1 | predicted protein |
| 67 | Vibrio vulnificus YJ016 | 33.50 | 1 | outer membrane integrity protein TolA |
| 68 | Vibrio vulnificus CMCP6 | 33.50 | 1 | TolA protein |
| 69 | Alkalilimnicola ehrlichei MLHE-1 | 33.50 | 1 | TonB family protein |
| 70 | Shewanella woodyi ATCC 51908 | 33.10 | 1 | TolA |
| 71 | Nitrosococcus oceani ATCC 19707 | 32.30 | 1 | TonB-like |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| Macaca mulatta | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| synthetic construct | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| Macaca fascicularis | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| Pongo pygmaeus | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||| ||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 292 QVSSAYQKTVPVEAVASKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 351 |
| Mus musculus | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||||||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| N/A | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||||||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| mice, BALB/c 3T3 cells, Peptide, 546 aa | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||||||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| Rattus norvegicus | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||||||||||||||||||#||+|||||||||||||||||+||||||||| Sbjct 292 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAQERQEQEEAR 351 |
| Canis familiaris | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 ||+ ||||||||||| |+||||||||||||#||||||||||||||+||||||||||||||| Sbjct 329 QVAPAYQKTVPVEAVNSRTSNIRANFENLA#KEKEQEDRRKAEAEKAQRMAKERQEQEEAR 388 |
| Bos taurus | Query 330 VSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |||+|||||||||| ||||||||||||||#||||||||||||||||||||+|||||| || Sbjct 330 VSSSYQKTVPVEAVNSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAQERQEQEAAR 388 |
| Gallus gallus | Query 331 SSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 +| |||| ||| | +|||+|||| ||||#||||||||||||||||||||+|+||||||| Sbjct 340 TSTYQKTKPVERVANKTSSIRANLENLA#KEKEQEDRRKAEAERAQRMAREKQEQEEAR 397 |
| Xenopus laevis | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 +|||+|||| ||| || |+|||||||||#|+||||||+|||||||||+ +||+|||+|| Sbjct 327 KVSSSYQKTRPVEVEGSKASSIRANFENLA#KDKEQEDRKKAEAERAQRLERERREQEQAR 386 |
| Danio rerio | Query 331 SSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |+||||| |||| +| |+|+| |||+|#|+||+|++++ | ||++| |||+||||||| Sbjct 330 SAAYQKTRPVEAASSSASSIKARFENIA#KQKEEEEQQRLEEERSRRQAKEKQEQEEAR 387 |
| Tetraodon nigroviridis | Query 333 AYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 +|||| |||| | +|+| |||+|#|+|| ||||+|| || +| ||||||||||+ Sbjct 341 SYQKTKPVEAAGSNAGSIKARFENMA#KQKEDEDRRRAEEERLRRQAKERQEQEEAQ 396 |
| Xenopus tropicalis | Query 331 SSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQE 385 ||||+|| |+||+|| |+|+ |||+|#+ |+|+++||| || +| ||+||+| Sbjct 292 SSAYEKTQPLEAMTSGAHNLRSRFENMA#RTAEEENKQKAEEERVRRQKKEKQEKE 346 |
| Strongylocentrotus purpuratus | Query 330 VSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQE 385 |||+|+|| | | |+| || +|#+ |+| ||||| || +| |+|++|+| Sbjct 293 VSSSYKKTRPQPPAKSGAGNMRNKFEQMA#QAGEEESRRKAEEERGRRQAREQKEKE 348 |
| Aedes aegypti | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEA 387 ++ + | || | + ++| ||||| ||| |# |+| |+||| ++ |+ |+++++||| Sbjct 253 KIGTNYTKTKP-DIGSAKPSNIRARFENFA#MAAEEETRKKAEEQKRLRLEKDKKDREEA 310 |
| Drosophila melanogaster | Query 345 SKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEA 387 +| ||+|| |||||#| |+| |++|| ++ | ||+++++||| Sbjct 238 AKPSNLRAKFENLA#KNSEEESRKRAEEQKRLREAKDKRDREEA 280 |
| Drosophila pseudoobscura | Query 345 SKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEA 387 +| ||+|| |||||#| |+| |+++| ++ | ||+++++||| Sbjct 236 AKPSNLRAKFENLA#KNSEEESRKRSEEQKRLREAKDKRDREEA 278 |
| Pan troglodytes | Query 331 SSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQE 385 ++||+|| |+|| +| ++| ||++|#+|| |++ | |+||++|+ +||++ Sbjct 260 TTAYKKTTPIEAASSGARGLKAKFESMA#EEK----RKREEEEKAQQVARRQQERK 310 |
| Anopheles gambiae str. PEST | Query 344 TSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEA 387 ++| ||+|| ||| |# |+| |++|+ ++ | |+| ++||| Sbjct 237 SAKPSNLRAKFENFA#ATAEEEARKRADEQKRLREEKDRCDREEA 280 |
| Oryza sativa (japonica cultivar-group) | Query 336 KTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEE 386 || +|||+| + | + |+ +# |+|+|+ + | | + +| ||+|| Sbjct 75 KTALMEAVSSGKLSAREHLESCS#NEEEEEEEEEEEEEEEEEEEEEEQEEEE 125 |
| Babesia bovis | Query 333 AYQKTVPVEAVTSKTSNIRANFEN-LA#KEKEQEDRRKAEAERAQRMAKERQEQEE 386 | || + || + +| | | #|||| | +|+ | |+| |+| ||++||| Sbjct 455 ALQKRLAEEAAQKRLEQMRRQAEEKLR#KEKEAEMKRQEEEEQA-RIAAERKQQEE 508 |
| Magnaporthe grisea 70-15 | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQE 385 | + + | | | | | || + #+ +|+| ||| || | + |+||+| | Sbjct 356 QAPGSSRPTAPPAPVGSPASGIRGPKMIMQ#ERREREQRRKDEATRREAEAREREELE 412 |
| Parvibaculum lavamentivorans DS-1 | Query 338 VPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKE--RQEQEEA 387 | + +| + | || + |#+|+| ||||+|| | |+| |+| |+| | | Sbjct 129 VVLRTLTEEEKNARAQALDSA#REREGEDRRRAE-EEARRFAEEDARREAERA 179 |
| Felis catus | Query 330 VSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKE---QEDRRKAEAERA----QRMAKERQ 382 | + |||| || | | +| #||+| +|+|+|| || +|| +||| Sbjct 60 VGTTYQKTDA--AVEMKRINREQFWEQAK#KEEELRKEEERKKALDERLRFEQERMEQERQ 117 Query 383 EQEE 386 |||| Query 383 EQEE 386 |
| Oryctolagus cuniculus | Query 330 VSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKE---QEDRRKAEAERA----QRMAKERQ 382 | + |||| || | | +| #||+| +|+|+|| || +|| +||| Sbjct 159 VGTTYQKTDA--AVEMKRINREQFWEQAK#KEEELRKEEERKKALDERLRFEQERMEQERQ 216 Query 383 EQEE 386 |||| Query 383 EQEE 386 |
| Lodderomyces elongisporus NRRL YB-4239 | Query 341 EAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 || | + || | #+|||+++ | | | + + ||++|+|| + Sbjct 879 EAFTIEISNPTKQQRELE#EEKEEKEEEKEEKEEKEELEKEKEEEEEEK 926 |
| Entamoeba histolytica HM-1:IMSS | Query 335 QKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 | || |+ | | #|+||+| |+| | ++ |+ |++||++|| | Sbjct 954 QNEVPKESEEEKRHR-----EEER#KQKEEEKRQKEEEDKKQKEAEKRQKEEEKR 1002 |
| Candida glabrata | Query 355 ENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 | #|+||+|++|| | | +| ||++|+|+ | Sbjct 121 EKKK#KQKEEEEKRKKELEEQERKKKEQEEEEKRR 154 |
| Aspergillus nidulans FGSC A4 | Query 337 TVPVEAVTSKTSNIR-ANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 +| + | ++ | + +|#+++| | ||||| | | + || +|+| |+ Sbjct 519 SVSAKTAAEKKQELKDAVLQKMA#QDQEAEARRKAEEEEAAKRKKEEEEEEAAK 571 |
| Monodelphis domestica | Query 334 YQKTVPVEAVTSKTSNIRAN-FENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 +|+ +| | + ++ + +| | #||||++ |++ ||++ || |||+ | Sbjct 147 HQEKIPREKILTERAEMRLREVERRR#KEKEEKKRQQQEAQKRLEKLKEELEQEQQR 202 |
| Tetrahymena thermophila SB210 | Query 339 PVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |+++ +++ + + + | #||||+ |+ | | | ++ ||++|| +|+ Sbjct 760 PIKSFSAEQAGGQTAMQKLK#KEKEENDKVKKEKEDQDKLKKEKEEQLKAQ 809 |
| Dichelobacter nodosus VCS1703A | Query 350 IRANFENLA#KEK-EQEDRRKAEAERAQRMAKERQEQEE 386 + | | ||#||| ||| | + ||| |+|||| |||| Sbjct 97 LAAERERLA#KEKAEQERRARLEAE---RLAKERAEQEE 131 |
| Aspergillus clavatus NRRL 1 | Query 351 RANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 | | +|#||+|+++|++ | | | +|||+|| | Sbjct 2879 RERQERIA#KEEEEKERKRKEEEENARRQQERQQQEAER 2916 |
| Reinekea sp. MED297 | Query 355 ENLA#KEKEQEDRRKAEAERAQRMAKERQEQEE 386 | #+|+|+|+||+||||| ||+ +||| | | Sbjct 219 ERRR#REREEEERRQAEAER-QRLEQERQRQLE 249 |
| Giardia lamblia ATCC 50803 | Query 339 PVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 | | | | + | ||#+|| +++|++ | |+ +| |||+ ||+ | Sbjct 132 PRETVASPPQE---DLEALA#REKAEKERQRREQEQQERERKERERQEKER 178 |
| Arabidopsis thaliana | Query 341 EAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 | ++||| + | #|+||+||++| | |+ + |+++|+ + + Sbjct 176 EIISSKTEEEKKKEEEDK#KKKEEEDKKKKEDEKKKEEEKKKEEENKKK 223 |
| Borrelia burgdorferi | Query 329 QVSSAYQKTVPVEAVTSKTSNIRA-NFENLA#KEKEQEDRRKAEAERAQR-MAKERQEQEE 386 +| + || | | | + ||#|++|++ +|||| |+ +| +|||++|| Sbjct 121 KVKKVEESEAKVEGKEEKQENTEERNKQELA#KQEEEQQKRKAEQEKQKREEEQERQKREE 180 |
| Candida albicans SC5314 | Query 330 VSSAYQKTVPVEAVTSKTS--NIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEE 386 |+ + ||| ||+| +| | + | | #+|+|+|+ + | | + +| +|+|| Sbjct 54 VNKSNFSTVPFNAVSSSSSSDNDQDNDEEEE#EEEEEEEEEEEEEEEEEEEEEEEEEEEE 112 |
| Borrelia burgdorferi B31 | Query 329 QVSSAYQKTVPVEAVTSKTSNIRA-NFENLA#KEKEQEDRRKAEAERAQR-MAKERQEQEE 386 +| + || | | | + ||#|++|++ +|||| |+ +| +|||++|| Sbjct 121 KVKKVEESEAKVEGKEEKQENTEERNKQELA#KQEEEQQKRKAEQEKQKREEEQERQKREE 180 |
| Pichia guilliermondii ATCC 6260 | Query 333 AYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQE 385 | || || | | | | # +|| | +++ | || +| ||++|+| Sbjct 658 AEQKRRKEEARQKKEEEKRKRIEELR#LKKEAEKKKQEEKERKERELKEKKERE 710 |
| Trypanosoma cruzi strain CL Brener | Query 329 QVSSAYQKTVPVEA-VTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEA 387 +| || +| || ||| | + #+|+|+|++ + | || |+ +| + ||| Sbjct 694 EVIRPLQKKMPGEAAVTSAGSKLSTAMIPEK#QEEEEEEKEEEEEEREQQEQQEEELQEEE 753 Query 388 R 388 | Query 388 R 388 |
| Oryza sativa (indica cultivar-group) | Query 351 RANFENLA#KEKEQEDRRKAEAERAQR--MAKERQEQEEAR 388 | | #+|||+|+||+ | || +| |||+|+|| | Sbjct 483 REEEERRR#REKEEEERRRQEEERKRREEEEKERREREEER 522 |
| Tribolium castaneum | Query 344 TSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEE 386 ++| + |+ #|+||+|++|||| | +| +|++|+|| Sbjct 175 STKKEKDKKKKEDED#KKKEEEEKRKAEEEDRKRKEEEKKEEEE 217 |
| Trypanosoma brucei TREU927 | Query 341 EAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 | + + + + | |#|| |+|||++ + | +| |+|+|+|| | Sbjct 123 EVILKQKEEVEEDRELRA#KE-EEEDRKRKQREEEERKRKQREEEEEDR 169 |
| Caenorhabditis elegans | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 +| |++ +| + || | #|+||+|| ++ +|| ++|+ | ++|++ | Sbjct 416 EVQMEYERRWRIEE-EERLEEIRRKKEEER#KKKEEEDEKRRKAEESERVRKLKEEEKRKR 474 |
| Dictyostelium discoideum AX4 | Query 335 QKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEE 386 || | + |+ +|| + +| #||||+| ++ | |+ + ||++++++ Sbjct 576 QKIVNNDKVSENSSNNEDSQKNKE#KEKEKEKEKEKEKEKEKEKEKEKEKEKD 627 |
| Cryptosporidium hominis TU502 | Query 340 VEAVTSKTSNIRANFENLA#KEKEQEDRR-----KAEAERAQRMAKERQEQEEA 387 + |+ + +| | ||#|||| |+++ | |+ | ++|+| ||||+ Sbjct 166 LNAIRKQQEEMRLQKERLA#KEKELEEKKAKEKEKTESNLANKIAQELLEQEES 218 |
| Dictyostelium discoideum | Query 335 QKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEE 386 || | + |+ +|| + +| #||||+| ++ | |+ + ||++++++ Sbjct 576 QKIVNNDKVSENSSNNEDSQKNKE#KEKEKEKEKEKEKEKEKEKEKEKEKEKD 627 |
| Cryptosporidium parvum Iowa II | Query 340 VEAVTSKTSNIRANFENLA#KEKEQEDRR-----KAEAERAQRMAKERQEQEEA 387 + |+ + +| | ||#|||| |+++ | |+ | ++|+| ||||+ Sbjct 166 LNAIRKQQEEMRLQKERLA#KEKELEEKKAKEKEKTESNLANKIAQELLEQEES 218 |
| Trichomonas vaginalis G3 | Query 351 RANFENLA#KEKEQEDRR-KAEAERAQRMAKERQEQEEAR 388 | | | #+|||+ +|+ | | |+ +| |||||+|+ + Sbjct 319 REQEEKLK#REKEERERKEKEEREKKEREEKERQEREQEK 357 |
| Paramecium tetraurelia | Query 346 KTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEE 386 |+ | + + | # +||+|+|++ | | || | |+ ++|| Sbjct 296 KSENTKTEDDILK#AQKEEEERQRKEEEEKQRQALEKLQREE 336 |
| Plasmodium reichenowi | Query 351 RANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 | | #+|||+ +| | | || +| ||++||| | Sbjct 2685 RERIEKEK#QEKERLEREKQEEERKERERLEREKQEEER 2722 |
| Myxococcus xanthus DK 1622 | Query 332 SAYQKTVPVEAVTSKTSNIRANFENLA#KE-KEQEDRRKAEAERAQRMAKERQEQEEAR 388 | | + + +| | + #++ || |+||| | ||| +||+ ||| | Sbjct 532 STSNKRAQEDEARQREEELRKRREEVE#RQRKELEERRKQEEAAAQREQEERRRQEERR 589 |
| Bacillus cereus ATCC 14579 | Query 341 EAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 || + + ++ | | #|++ | +|||| || ||+|+|+++ |||| Sbjct 95 EAEKQRAAEVQRNAE-AE#KQRNAEAQRKAEEER-QRVAEEQRKAEEAR 140 |
| Myxococcus xanthus | Query 332 SAYQKTVPVEAVTSKTSNIRANFENLA#KE-KEQEDRRKAEAERAQRMAKERQEQEEAR 388 | | + + +| | + #++ || |+||| | ||| +||+ ||| | Sbjct 532 STSNKRAQEDEARQREEELRKRREEVE#RQRKELEERRKQEEAAAQREQEERRRQEERR 589 |
| Aspergillus fumigatus Af293 | Query 352 ANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 | | ||# | |+ | | ||| |+|+|| |+|| + Sbjct 965 AEEERLA#AEAEKARLAKEEEERAARVARERAEEEERK 1001 |
| Aspergillus niger | Query 355 ENLA#KEKEQEDRR-KAEAERAQRMAKERQEQEEAR 388 | #||||||++| | | +| || +|+| +|+ | Sbjct 462 EERE#KEKEQEEKRQKEEQDREQRQKEEKQREEKER 496 |
| Coprinopsis cinerea okayama7#130 | Query 359 KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 +|||+|+| | || + +||+|++||| Sbjct 983 REKEREEREKEREEREKEREEEREERDEAR 1012 |
| Coccidioides immitis RS | Query 341 EAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEA 387 || + ++|| | #+++|+| ||| + || ++ |||+|+||| Sbjct 76 EAEETLHQHLRAAEEERR#RKEEEERRRKEKIER-EKAEKERREREEA 121 |
| Lyngbya sp. PCC 8106 | Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEA 387 +| | | ||+ | + | ++ # +|++||||| | +|+|| +||| Sbjct 1407 EVRYIYGKAVPLRNELGKVRGVIGAFLDVT#------ERKQAEAEREQLLARERSAREEA 1459 |
| Vibrio alginolyticus 12G01 | Query 330 VSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAE-AERA--QRMAKERQEQEE 386 | | |+ + || +| | | |#|| |++ ||+ | ||+| +|+|||+ +| Sbjct 156 VRKAEQERLAKEAAIAKAEQERLAREKAA#KEAEEKARREREAAEKAEQERIAKEKAAKEA 215 Query 387 A 387 | Query 387 A 387 Query 359 KEKEQEDRRKAEAERAQRMAKERQEQEEA 387 + |++|+| + | ||| ++ |||+|+||| Sbjct 127 ERKQEEERVRKEKERAAKLEKERKEKEEA 155 |
| Neosartorya fischeri NRRL 181 | Query 350 IRANFENLA#KEKEQEDRR-----KAEAERAQRMAKERQEQEEAR 388 |+ | | #|++|+|+|| || || +| |+| +++|||| Sbjct 205 IQKQQEALR#KQREEEERRLAEEKAAEEERLRREAEEEKKREEAR 248 |
| Ostreococcus lucimarinus CCE9901 | Query 351 RANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQE 385 | +++ #|+|| |+||| + |||+ || + |+| Sbjct 98 RPVYDDRK#KKKEMEERRKQQQERAEAEAKAKAEEE 132 |
| Vibrio vulnificus YJ016 | Query 336 KTVPVEAVTSKTSNIRANFENLA#KEK---EQEDRRKAEAERAQRMAKERQEQEEA 387 | +| | + +| | +|#||| | ++ + | || +|+ +||+||| | Sbjct 185 KEAEEKARREKEAAAKAEQERIA#KEKAAKEAAEKARKEKERLERLERERKEQEAA 239 Query 330 VSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEA---ERAQRMAKERQEQEE 386 + | |+ + || +| | | |#|| |++ ||+ || +|+|||+ +| Sbjct 156 IKKAEQERLAKEAAIAKAEQERIAKEKAA#KEAEEKARREKEAAAKAEQERIAKEKAAKEA 215 Query 387 A 387 | Query 387 A 387 |
| Vibrio vulnificus CMCP6 | Query 336 KTVPVEAVTSKTSNIRANFENLA#KEK---EQEDRRKAEAERAQRMAKERQEQEEA 387 | +| | + +| | +|#||| | ++ + | || +|+ +||+||| | Sbjct 149 KEAEEKARREKEAAAKAEQERIA#KEKAAKEAAEKARKEKERLERLERERKEQEAA 203 Query 330 VSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEA---ERAQRMAKERQEQEE 386 + | |+ + || +| | | |#|| |++ ||+ || +|+|||+ +| Sbjct 120 IKKAEQERLAKEAAIAKAEQERIAKEKAA#KEAEEKARREKEAAAKAEQERIAKEKAAKEA 179 Query 387 A 387 | Query 387 A 387 |
| Alkalilimnicola ehrlichei MLHE-1 | Query 351 RANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 | | #+ +|+|+||+ | || ++ +||| ||| | Sbjct 142 RLRREEEQ#RRQEEEERRRQEEERRRQEEEERQRQEEER 179 Query 351 RANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEE 386 | | | #+++++|+||+ | || +||+ ||| Sbjct 179 RRRQEELE#RQRQEEERRRQEEAERQRQEEERRRQEE 214 |
| Shewanella woodyi ATCC 51908 | Query 341 EAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEE 386 | | + | | |#|+ |+| +||||||| ++ +| + |+| Sbjct 150 EEAARKAAEKRKAEEAAA#KKAEEERKRKAEAERKRKAEEEARRQQE 195 Query 340 VEAVTSKTSNIRANFENLA#KEKEQEDR----RKAEAERAQRMAKERQEQEEAR 388 |+|| | + + | | # ||++ +| |+ || | | |++ +|+|+|| Sbjct 44 VQAVVVDQSKVAQHVERLK#AEKKEAERKEKARQDEANRQVREARKEREREQAR 96 |
| Nitrosococcus oceani ATCC 19707 | Query 341 EAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEE 386 || | | | |#| | +|+|+|||| | |+ ++| | || Sbjct 170 EAAKRKAEEERKKIE-AA#KRKAEEERKKAEAARRQQELQDRIEAEE 214 Query 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KE---KEQEDRRKAEAERAQRMAKERQEQE 385 + | ++| +|+ | +| + | # | ||+ ||+||||+ + |+|| + Sbjct 98 RAEQARRETQEQQALEQKQQEEQARLKRLE#AERQAKEEAARRQAEAEKKRAEEKKRQAEA 157 Query 386 EAR 388 | | Sbjct 158 EKR 160 |
[Site 3] AVTSKTSNIR351-ANFENLAKEK
Arg351  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala342 | Val343 | Thr344 | Ser345 | Lys346 | Thr347 | Ser348 | Asn349 | Ile350 | Arg351 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala352 | Asn353 | Phe354 | Glu355 | Asn356 | Leu357 | Ala358 | Lys359 | Glu360 | Lys361 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| STFEDVTQVSSAYQKTVPVEAVTSKTSNIRANFENLAKEKEQEDRRKAEAERAQRMAKER |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 116.00 | 22 | cortactin isoform a |
| 2 | synthetic construct | 116.00 | 3 | cortactin |
| 3 | Macaca fascicularis | 116.00 | 2 | unnamed protein product |
| 4 | Macaca mulatta | 116.00 | 2 | PREDICTED: similar to cortactin isoform a |
| 5 | Pongo pygmaeus | 114.00 | 1 | hypothetical protein |
| 6 | Mus musculus | 109.00 | 7 | cortactin |
| 7 | N/A | 109.00 | 2 | S |
| 8 | mice, BALB/c 3T3 cells, Peptide, 546 aa | 109.00 | 1 | cortactin, p80/p85 |
| 9 | Rattus norvegicus | 108.00 | 6 | cortactin isoform B |
| 10 | Canis familiaris | 107.00 | 6 | PREDICTED: similar to cortactin isoform a isoform |
| 11 | Bos taurus | 107.00 | 3 | cortactin |
| 12 | Gallus gallus | 92.40 | 2 | cortactin |
| 13 | Xenopus laevis | 89.40 | 4 | cortactin |
| 14 | Tetraodon nigroviridis | 73.60 | 5 | unnamed protein product |
| 15 | Danio rerio | 72.80 | 6 | cortactin |
| 16 | Xenopus tropicalis | 63.50 | 1 | cortactin |
| 17 | Strongylocentrotus purpuratus | 53.90 | 1 | SRC8 protein |
| 18 | Aedes aegypti | 47.40 | 1 | cortactin |
| 19 | Pan troglodytes | 46.60 | 9 | PREDICTED: hypothetical protein isoform 1 |
| 20 | Drosophila melanogaster | 43.90 | 2 | Cortactin |
| 21 | Drosophila pseudoobscura | 42.70 | 1 | GA17577-PA |
| 22 | Anopheles gambiae str. PEST | 37.00 | 1 | ENSANGP00000029554 |
| 23 | Parvibaculum lavamentivorans DS-1 | 35.00 | 1 | translation initiation factor IF-2 |
| 24 | Arabidopsis thaliana | 34.30 | 2 | unnamed protein product |
| 25 | Dictyostelium discoideum AX4 | 33.50 | 2 | hypothetical protein DDBDRAFT_0205004 |
| 26 | Magnaporthe grisea 70-15 | 33.10 | 3 | hypothetical protein MGCH7_ch7g791 |
| 27 | Trichomonas vaginalis G3 | 33.10 | 3 | hypothetical protein TVAG_298320 |
| 28 | Monodelphis domestica | 33.10 | 1 | PREDICTED: similar to nexilin (F actin binding pro |
| 29 | Caenorhabditis briggsae | 33.10 | 1 | Hypothetical protein CBG17979 |
| 30 | Paramecium tetraurelia | 32.30 | 1 | hypothetical protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |
| synthetic construct | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |
| Macaca fascicularis | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |
| Macaca mulatta | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |
| Pongo pygmaeus | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |||||||||||||||||||||| |||||||#|||||||||||||||||||||||||||||| Sbjct 285 STFEDVTQVSSAYQKTVPVEAVASKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 344 |
| Mus musculus | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 |
| N/A | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 |
| mice, BALB/c 3T3 cells, Peptide, 546 aa | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 |
| Rattus norvegicus | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#|||||||||+|||||||||||||||||+|| Sbjct 285 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAQER 344 |
| Canis familiaris | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |||||| ||+ ||||||||||| |+|||||#|||||||||||||||||||||+|||||||| Sbjct 322 STFEDVAQVAPAYQKTVPVEAVNSRTSNIR#ANFENLAKEKEQEDRRKAEAEKAQRMAKER 381 |
| Bos taurus | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |||||| |||+|||||||||| |||||||#|||||||||||||||||||||||||||+|| Sbjct 322 STFEDVAAVSSSYQKTVPVEAVNSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAQER 381 |
| Gallus gallus | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 +||||+ + +| |||| ||| | +|||+||#|| ||||||||||||||||||||||||+|+ Sbjct 331 ATFEDIEKPTSTYQKTKPVERVANKTSSIR#ANLENLAKEKEQEDRRKAEAERAQRMAREK 390 |
| Xenopus laevis | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ++|||| +|||+|||| ||| || |+||#||||||||+||||||+|||||||||+ +|| Sbjct 320 ASFEDVEKVSSSYQKTRPVEVEGSKASSIR#ANFENLAKDKEQEDRKKAEAERAQRLERER 379 |
| Tetraodon nigroviridis | Query 323 TFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |||+| + + +|||| |||| | +|+#| |||+||+|| ||||+|| || +| |||| Sbjct 331 TFEEVEKPTPSYQKTKPVEAAGSNAGSIK#ARFENMAKQKEDEDRRRAEEERLRRQAKER 389 |
| Danio rerio | Query 323 TFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |||+| + |+||||| |||| +| |+|+#| |||+||+||+|++++ | ||++| |||+ Sbjct 322 TFEEVQKPSAAYQKTRPVEAASSSASSIK#ARFENIAKQKEEEEQQRLEEERSRRQAKEK 380 |
| Xenopus tropicalis | Query 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 | | ++ ||||+|| |+||+|| |+|#+ |||+|+ |+|+++||| || +| ||+ Sbjct 283 SGFGEIETPSSAYEKTQPLEAMTSGAHNLR#SRFENMARTAEEENKQKAEEERVRRQKKEK 342 |
| Strongylocentrotus purpuratus | Query 324 FEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 | |+ |||+|+|| | | |+|# || +|+ |+| ||||| || +| |+|+ Sbjct 287 FGDMQGVSSSYKKTRPQPPAKSGAGNMR#NKFEQMAQAGEEESRRKAEEERGRRQAREQ 344 |
| Aedes aegypti | Query 324 FEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |++ ++ + | || | + ++| ||||#| ||| | |+| |+||| ++ |+ |++ Sbjct 248 FQEQDKIGTNYTKTKP-DIGSAKPSNIR#ARFENFAMAAEEETRKKAEEQKRLRLEKDK 304 |
| Pan troglodytes | Query 324 FEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 | ++ ++||+|| |+|| +| ++#| ||++|+|| |++ | |+||++|+ + Sbjct 253 FNEMEAPTTAYKKTTPIEAASSGARGLK#AKFESMAEEK----RKREEEEKAQQVARRQ 306 |
| Drosophila melanogaster | Query 345 SKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 +| ||+|#| |||||| |+| |++|| ++ | ||++ Sbjct 238 AKPSNLR#AKFENLAKNSEEESRKRAEEQKRLREAKDK 274 |
| Drosophila pseudoobscura | Query 345 SKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 +| ||+|#| |||||| |+| |+++| ++ | ||++ Sbjct 236 AKPSNLR#AKFENLAKNSEEESRKRSEEQKRLREAKDK 272 |
| Anopheles gambiae str. PEST | Query 344 TSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ++| ||+|#| ||| | |+| |++|+ ++ | |+| Sbjct 237 SAKPSNLR#AKFENFAATAEEEARKRADEQKRLREEKDR 274 |
| Parvibaculum lavamentivorans DS-1 | Query 327 VTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKE 380 | + | + | + +| + | |#| + |+|+| ||||+|| | |+| |+| Sbjct 118 VEKKPSRSRSGVVLRTLTEEEKNAR#AQALDSAREREGEDRRRAE-EEARRFAEE 170 |
| Arabidopsis thaliana | Query 342 AVTSKTSNIR#ANFENLAKEKEQEDRRKAEA---ERAQRMAKE 380 | | | +#|+ + + ||||+|||++ || || +|+| + Sbjct 736 ARADKLRNFK#ADLQKMKKEKEEEDRKRIEALKIERQKRIASK 777 |
| Dictyostelium discoideum AX4 | Query 331 SSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 ||+ + | +++ ||+ + # | + |+||+| ++ | || + +|| Sbjct 883 SSSSSSSTPSQSMMSKSERMN#RENEKVIKQKEKEAEKQKEIEREKEKIRER 933 |
| Magnaporthe grisea 70-15 | Query 329 QVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 | + + | | | | | ||# + + +|+| ||| || | + |+|| Sbjct 356 QAPGSSRPTAPPAPVGSPASGIR#GPKMIMQERREREQRRKDEATRREAEARER 408 |
| Trichomonas vaginalis G3 | Query 323 TFEDVTQVSSAYQKTVPV---EAVTSKTSNIR#ANFENLAKEKEQEDRRK--------AEA 371 || + +|| |+++ || ++ +# | +||++|+|++|+ || Sbjct 369 TFSSFSPFNSAVQESLESQRREAEEAEKKEEQ#KRLEKIAKQREEEEKRRKEEEEKAAAER 428 Query 372 ERAQRMAKER 381 |+ +|+||+| Sbjct 429 EKQERIAKQR 438 |
| Monodelphis domestica | Query 341 EAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 || + |++#| || +|| +|+|++|+ | ++ || |+ Sbjct 429 EAPFTHKVNMK#ARFEQMAKAREEEEQRRIEQQKLLRMQFEQ 469 Query 350 IR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 + # ||| | |+| +|++|+ | || |+ |+ Sbjct 299 LE#MNFEELLKQKMEEEKRRTEEERKHRLEMEK 330 |
| Caenorhabditis briggsae | Query 326 DVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKE 380 || + | + |+| ++ |+# ||+|+ | +|| | | ||+ | Sbjct 90 DVISAQATYFAPLAQSAMTMSSALIK#LQEENIAERKRKEDEHKKERMRAEEKHSE 144 |
| Paramecium tetraurelia | Query 325 EDVTQVSSAYQKT--VPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRM 377 | + +++ |||| +|| + + #| |++||| ++| ||+++|++ Sbjct 213 EQILEMAPAYQKDEDLPVSKKVKRQKKLM#AKKFETNKKQEQESQQKIEAQQSQQL 267 |
* References
None.