XSB0041 : PREDICTED: protein kinase C, beta isoform 1 [Macaca mulatta]
[ CaMP Format ]
This entry is computationally expanded from SB0039
* Basic Information
| Organism | Macaca mulatta (rhesus monkey) |
| Protein Names | protein kinase C; beta isoform 1 |
| Gene Names | PRKCB1; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 15 mRNAs, 242 ESTs, 29 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001095880 | XM_001095880 | 701195 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
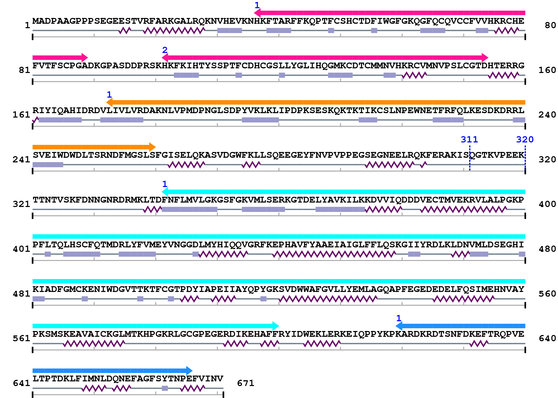
Length: 671 aa
Average Mass: 76.855 kDa
Monoisotopic Mass: 76.805 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| C1_1 1. | 37 | 89 | 80.1 | 8.2e-21 |
| C1_1 2. | 102 | 154 | 97.3 | 5.4e-26 |
| C2 1. | 173 | 260 | 126.7 | 7.5e-35 |
| --- cleavage 311 --- | ||||
| --- cleavage 320 --- | ||||
| Pkinase 1. | 342 | 600 | 262.2 | 1.2e-75 |
| Pkinase_C 1. | 620 | 666 | 71.9 | 2.4e-18 |
3. Sequence Information
Fasta Sequence: XSB0041.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites]
Cleavage sites (±10aa)
[Site 1] RQKFERAKIS311-QGTKVPEEKT
Ser311  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg302 | Gln303 | Lys304 | Phe305 | Glu306 | Arg307 | Ala308 | Lys309 | Ile310 | Ser311 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln312 | Gly313 | Thr314 | Lys315 | Val316 | Pro317 | Glu318 | Glu319 | Lys320 | Thr321 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EGEYFNVPVPPEGSEGNEELRQKFERAKISQGTKVPEEKTTNTVSKFDNNGNRDRMKLTD |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 126.00 | 6 | PREDICTED: protein kinase C, beta isoform 1 |
| 2 | Homo sapiens | 124.00 | 9 | protein kinase C, beta isoform 1 |
| 3 | Oryctolagus cuniculus | 122.00 | 3 | KPCB_RABIT Protein kinase C beta type (PKC-beta) ( |
| 4 | Canis familiaris | 122.00 | 2 | PREDICTED: similar to Protein kinase C, beta type |
| 5 | N/A | 120.00 | 10 | K |
| 6 | Mus musculus | 120.00 | 4 | protein kinase C, beta 1 |
| 7 | Rattus norvegicus | 120.00 | 4 | unnamed protein product |
| 8 | Bos taurus | 118.00 | 3 | Unknown (protein for MGC:148660) |
| 9 | Monodelphis domestica | 113.00 | 3 | PREDICTED: similar to Protein kinase C beta type ( |
| 10 | Gallus gallus | 107.00 | 3 | PREDICTED: similar to protein kinase C beta-1 |
| 11 | Latimeria chalumnae | 98.20 | 2 | protein kinase C beta 1 |
| 12 | Protopterus dolloi | 95.10 | 2 | protein kinase C beta 1 |
| 13 | Danio rerio | 92.00 | 7 | protein kinase C, beta 1 |
| 14 | Tetraodon nigroviridis | 91.30 | 3 | unnamed protein product |
| 15 | Scyliorhinus canicula | 85.50 | 2 | protein kinase C |
| 16 | Petromyzon marinus | 57.80 | 1 | protein kinase C |
| 17 | synthetic construct | 54.70 | 1 | protein kinase C alpha |
| 18 | Takifugu rubripes | 52.80 | 1 | protein kinase C, alpha type |
| 19 | Xenopus tropicalis | 52.40 | 1 | hypothetical protein LOC780209 |
| 20 | Xenopus laevis | 48.50 | 1 | protein kinase C, alpha |
| 21 | Myxine glutinosa | 47.00 | 1 | protein kinase C |
| 22 | Caenorhabditis briggsae | 40.00 | 1 | Hypothetical protein CBG16150 |
| 23 | Aplysia californica | 37.70 | 1 | KPC1_APLCA Calcium-dependent protein kinase C (APL |
| 24 | Aedes aegypti | 35.00 | 1 | protein kinase c |
| 25 | Apis mellifera | 33.50 | 1 | PREDICTED: similar to Protein C kinase 53E CG6622- |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 |
| Homo sapiens | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||| ||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 |
| Oryctolagus cuniculus | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||||||||||||||||| #|||| ||||||||+|||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKTPEEKTTNTISKFDNNGNRDRMKLTD 341 |
| Canis familiaris | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||||||||||||||||||#|||| ||||||||+|| ||||||||||||| Sbjct 274 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKAPEEKTTNTISKLDNNGNRDRMKLTD 333 |
| N/A | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||||||||||||||||| #|||| ||||| ||+|||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKAPEEKTANTISKFDNNGNRDRMKLTD 341 |
| Mus musculus | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||||||||||||||||| #|||| ||||| ||+|||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKAPEEKTANTISKFDNNGNRDRMKLTD 341 |
| Rattus norvegicus | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||||||||||||||||| #|||| ||||| ||+|||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKAPEEKTANTISKFDNNGNRDRMKLTD 341 |
| Bos taurus | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||||||||||||||||| # | | ||||||||+|||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#PGPKTPEEKTTNTISKFDNNGNRDRMKLTD 341 |
| Monodelphis domestica | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| ||||||||||||||| #||||| |||| | +|| ||||||||||||| Sbjct 279 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 |
| Gallus gallus | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| ||||||||||||||| # |+| +||||| +|||||||+||||||+| Sbjct 281 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#TGSKAADEKTTNAISKFDNNGSRDRMKLSD 340 |
| Latimeria chalumnae | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| |||||||+||||||| # |+| |+| |+ ||| |||||||+||+| Sbjct 204 EGEYFNVPVPPEGEEGNEELRKKFERAKIG#PGSKATEDKVKNSSSKFGNNGNRDRIKLSD 263 |
| Protopterus dolloi | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| ||||||||||||||| # | + |+| | +||||||||||+||+| Sbjct 213 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#PGAEAAEDKIAN-AAKFDNNGNRDRVKLSD 271 |
| Danio rerio | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| ||||||||||||||| # | + ++| +||||+|||||||||+| Sbjct 281 EGEYFNVPVPPEGEEGNEELRQKFERAKI-#-GPSKTDGSSSNAISKFDSNGNRDRMKLSD 338 |
| Tetraodon nigroviridis | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| ||||||||||||||| # | | + |+ | |+||+|||+||||| | Sbjct 218 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#PG-KNTDGKSANAASRFDSNGNQDRMKLAD 276 |
| Scyliorhinus canicula | Query 282 EGEYFNVPVPPE-GSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLT 340 ||||+||||||| ||||||||||||||+| # |+| ||| +++ || | ||||||||| Sbjct 202 EGEYYNVPVPPEEDSEGNEELRQKFERARIG#PGSKAAEEKRSSSWSKID-NGNRDRMKLA 260 Query 341 D 341 | Query 341 D 341 |
| Petromyzon marinus | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTK--VPEEKTTNTVSKFDNNGNRDRMKL 339 ||||+|||+ |+ +|+ |+|+|||+|++ # | +|+ |++| || | ||+| Sbjct 212 EGEYYNVPIAPDIEDGDTEMRRKFEKARLG#PSVKPRASDERRANSLSAILNNANVDRVKA 271 Query 340 TD 341 | Sbjct 272 DD 273 |
| synthetic construct | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS-#QGTKVPEEKTTNTVSKFDNNGNRDRMKLT 340 ||||+|||+| ||| |||||||+||+ # | || + + + + | ||+||| Sbjct 282 EGEYYNVPIPEGDEEGNMELRQKFEKAKLGP#AGNKV----ISPSEDRKQPSNNLDRVKLT 337 Query 341 D 341 | Query 341 D 341 |
| Takifugu rubripes | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||+|||+| | + | |||||||+||+ #|| || | +|| ||++| | Sbjct 280 EGEYYNVPIP-EVDDVNLELRQKFEKAKLG#QGKKVITPSDHRRFSL--PSGNMDRVRLND 336 |
| Xenopus tropicalis | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS-#QGTKVPEEKTTNTVSKFDNNGNRDRMKLT 340 ||||+|||+| | +|| |||||||+||+ # | || |+ + + | | ++|| Sbjct 286 EGEYYNVPIPEEEDDGNIELRQKFEKAKLGP#AGNKV--ISPTDERRPYVPSNNIDSVRLT 343 Query 341 D 341 | Query 341 D 341 |
| Xenopus laevis | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS-#QGTKVPEEKTTNTVSKFDNNGNRDRMKLT 340 ||||+|||+| | +|| |||||||+||+ # | || | + + | | ++|| Sbjct 286 EGEYYNVPIP-EADDGNLELRQKFEKAKLGP#AGNKV--INPTGERRPYIPSNNIDSIRLT 342 Query 341 D 341 | Query 341 D 341 |
| Myxine glutinosa | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||+|||| ||| || ||||+ +|++|+#+ | + + | + ||+ | | Sbjct 203 EGEYYNVPVAPEGEEG-LELRQRLQRSQIA#RSDKNKQSPAKDQPSDSASLSGLDRVNLED 261 |
| Caenorhabditis briggsae | Query 282 EGEYFNVPVPPEGSEGNEELRQKF-ERAKIS#QGTKV--PEEKTTNTVSKFDNNGNRDRMK 338 |||++|+ + || | |++|+| + |||#+ + | | | | | ||| +| Sbjct 352 EGEFYNINITPEYDEDMEKVRKKMNDNFKIS#RDSSAGKPRESTPRATSTSLTNTNRDVIK 411 Query 339 LTD 341 +| Sbjct 412 ASD 414 |
| Aplysia californica | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFD 329 |||++ ||| + +| +|++ | |+ ||# + || +|| | Sbjct 266 EGEFYGVPVTDDITESIQEIKSKMHRSSIS#SEKRYPEPDKVQNMSKQD 313 |
| Aedes aegypti | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||+||||| ||++ +|+ + + ||#+ | +| | | +| ++ || Sbjct 230 EGEYYNVPVPEEGTD-LVQLKSQMRKTSIS#KKAPVLCDKDVP-----HNMGKKDVIRATD 283 |
| Apis mellifera | Query 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKF--DNNGNRDRMKL 339 ||||+||||| || + ||+ | | |# +||| | | | | | ++ Sbjct 299 EGEYYNVPVPEEGVD-LAELKMKPSPQKTS#-----VTKKTTTTQDKEVPHNMGKSDLIRA 352 Query 340 TD 341 +| Sbjct 353 SD 354 |
[Site 2] SQGTKVPEEK320-TTNTVSKFDN
Lys320  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser311 | Gln312 | Gly313 | Thr314 | Lys315 | Val316 | Pro317 | Glu318 | Glu319 | Lys320 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr321 | Thr322 | Asn323 | Thr324 | Val325 | Ser326 | Lys327 | Phe328 | Asp329 | Asn330 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PPEGSEGNEELRQKFERAKISQGTKVPEEKTTNTVSKFDNNGNRDRMKLTDFNFLMVLGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Macaca mulatta | 124.00 | 7 | PREDICTED: protein kinase C, beta isoform 1 |
| 2 | Homo sapiens | 122.00 | 8 | protein kinase C, beta isoform 1 |
| 3 | Oryctolagus cuniculus | 121.00 | 3 | KPCB_RABIT Protein kinase C beta type (PKC-beta) ( |
| 4 | Canis familiaris | 120.00 | 3 | PREDICTED: similar to Protein kinase C, beta type |
| 5 | N/A | 119.00 | 10 | K |
| 6 | Mus musculus | 119.00 | 4 | protein kinase C, beta 1 |
| 7 | Rattus norvegicus | 119.00 | 4 | unnamed protein product |
| 8 | Bos taurus | 116.00 | 3 | Unknown (protein for MGC:148660) |
| 9 | Monodelphis domestica | 112.00 | 3 | PREDICTED: similar to Protein kinase C beta type ( |
| 10 | Gallus gallus | 106.00 | 3 | PREDICTED: similar to protein kinase C beta-1 |
| 11 | Latimeria chalumnae | 96.70 | 2 | protein kinase C beta 1 |
| 12 | Protopterus dolloi | 93.60 | 2 | protein kinase C beta 1 |
| 13 | Danio rerio | 90.50 | 6 | protein kinase C, beta 1 |
| 14 | Tetraodon nigroviridis | 89.70 | 2 | unnamed protein product |
| 15 | Scyliorhinus canicula | 86.70 | 2 | protein kinase C |
| 16 | Petromyzon marinus | 58.20 | 1 | protein kinase C |
| 17 | synthetic construct | 55.80 | 1 | protein kinase C alpha |
| 18 | Takifugu rubripes | 53.90 | 1 | protein kinase C, alpha type |
| 19 | Xenopus tropicalis | 50.40 | 1 | hypothetical protein LOC780209 |
| 20 | Xenopus laevis | 49.70 | 1 | protein kinase C, alpha |
| 21 | Caenorhabditis briggsae | 43.50 | 1 | Hypothetical protein CBG16150 |
| 22 | Myxine glutinosa | 41.20 | 1 | protein kinase C |
| 23 | Aplysia californica | 37.70 | 1 | KPC1_APLCA Calcium-dependent protein kinase C (APL |
| 24 | Branchiostoma lanceolatum | 35.80 | 1 | protein kinase C |
| 25 | Aedes aegypti | 35.40 | 1 | protein kinase c |
| 26 | Apis mellifera | 33.90 | 1 | PREDICTED: similar to Protein C kinase 53E CG6622- |
| 27 | Anopheles gambiae str. PEST | 32.70 | 1 | ENSANGP00000009078 |
Top-ranked sequences
| organism | matching |
|---|---|
| Macaca mulatta | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Homo sapiens | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||||| |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 291 PPEGSEANEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Oryctolagus cuniculus | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||||||||||||||||||| |||| ||||#||||+||||||||||||||||||||||||| Sbjct 291 PPEGSEGNEELRQKFERAKIGQGTKTPEEK#TTNTISKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Canis familiaris | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 ||||||||||||||||||||||||| ||||#||||+|| |||||||||||||||||||||| Sbjct 283 PPEGSEGNEELRQKFERAKISQGTKAPEEK#TTNTISKLDNNGNRDRMKLTDFNFLMVLGK 342 |
| N/A | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||||||||||||||||||| |||| ||||#| ||+||||||||||||||||||||||||| Sbjct 291 PPEGSEGNEELRQKFERAKIGQGTKAPEEK#TANTISKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Mus musculus | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||||||||||||||||||| |||| ||||#| ||+||||||||||||||||||||||||| Sbjct 291 PPEGSEGNEELRQKFERAKIGQGTKAPEEK#TANTISKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Rattus norvegicus | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||||||||||||||||||| |||| ||||#| ||+||||||||||||||||||||||||| Sbjct 291 PPEGSEGNEELRQKFERAKIGQGTKAPEEK#TANTISKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Bos taurus | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||||||||||||||||||| | | ||||#||||+||||||||||||||||||||||||| Sbjct 291 PPEGSEGNEELRQKFERAKIGPGPKTPEEK#TTNTISKFDNNGNRDRMKLTDFNFLMVLGK 350 |
| Monodelphis domestica | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||| ||||||||||||||| ||||| |||#| | +|| |||||||||||||||||||||| Sbjct 288 PPEGEEGNEELRQKFERAKIGQGTKVTEEK#TANAISKIDNNGNRDRMKLTDFNFLMVLGK 347 |
| Gallus gallus | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||| ||||||||||||||| |+| +||#||| +|||||||+||||||+|||||||||| Sbjct 290 PPEGEEGNEELRQKFERAKIGTGSKAADEK#TTNAISKFDNNGSRDRMKLSDFNFLMVLGK 349 |
| Latimeria chalumnae | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||| |||||||+||||||| |+| |+|# |+ ||| |||||||+||+|||||||||| Sbjct 213 PPEGEEGNEELRKKFERAKIGPGSKATEDK#VKNSSSKFGNNGNRDRIKLSDFNFLMVLGK 272 |
| Protopterus dolloi | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||| ||||||||||||||| | + |+|# | +||||||||||+||+|||||||||| Sbjct 222 PPEGEEGNEELRQKFERAKIGPGAEAAEDK#IAN-AAKFDNNGNRDRVKLSDFNFLMVLGK 280 |
| Danio rerio | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||| ||||||||||||||| | + #++| +||||+|||||||||+|||||||||| Sbjct 290 PPEGEEGNEELRQKFERAKI--GPSKTDGS#SSNAISKFDSNGNRDRMKLSDFNFLMVLGK 347 |
| Tetraodon nigroviridis | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 |||| ||||||||||||||| | | + |#+ | |+||+|||+||||| |||||||||| Sbjct 227 PPEGEEGNEELRQKFERAKIGPG-KNTDGK#SANAASRFDSNGNQDRMKLADFNFLMVLGK 285 |
| Scyliorhinus canicula | Query 291 PPEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 | | ||||||||||||||+| |+| |||# +++ || | ||||||||| |||||||||| Sbjct 212 PEEDSEGNEELRQKFERARIGPGSKAAEEK#RSSSWSKID-NGNRDRMKLADFNFLMVLGK 270 |
| Petromyzon marinus | Query 292 PEGSEGNEELRQKFERAKISQGTK--VPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLG 349 |+ +|+ |+|+|||+|++ | +|+# |++| || | ||+| ||||||||| Sbjct 222 PDIEDGDTEMRRKFEKARLGPSVKPRASDER#RANSLSAILNNANVDRVKADDFNFLMVLG 281 Query 350 K 350 | Query 350 K 350 |
| synthetic construct | Query 292 PEGSE-GNEELRQKFERAKISQ-GTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLG 349 ||| | || |||||||+||+ | || # + + + + | ||+|||||||||||| Sbjct 291 PEGDEEGNMELRQKFEKAKLGPAGNKV----#ISPSEDRKQPSNNLDRVKLTDFNFLMVLG 346 Query 350 K 350 | Query 350 K 350 |
| Takifugu rubripes | Query 292 PEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 || + | |||||||+||+ || || # | +|| ||++| ||||| +||| Sbjct 289 PEVDDVNLELRQKFEKAKLGQGKKVITPS#DHRRFSL--PSGNMDRVRLNDFNFLALLGK 345 |
| Xenopus tropicalis | Query 291 PPEGSEGNEELRQKFERAKIS-QGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLG 349 | | +|| |||||||+||+ | || # |+ + + | | ++||||+|||||| Sbjct 295 PEEEDDGNIELRQKFEKAKLGPAGNKV--IS#PTDERRPYVPSNNIDSVRLTDFSFLMVLG 352 Query 350 K 350 | Query 350 K 350 |
| Xenopus laevis | Query 292 PEGSEGNEELRQKFERAKIS-QGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 || +|| |||||||+||+ | || # | + + | | ++|||| ||||||| Sbjct 295 PEADDGNLELRQKFEKAKLGPAGNKV--IN#PTGERRPYIPSNNIDSIRLTDFCFLMVLGK 352 |
| Caenorhabditis briggsae | Query 292 PEGSEGNEELRQKF-ERAKISQGTKV--PEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVL 348 || | |++|+| + |||+ + | | #| | | ||| +| +||||| || Sbjct 362 PEYDEDMEKVRKKMNDNFKISRDSSAGKPRES#TPRATSTSLTNTNRDVIKASDFNFLTVL 421 Query 349 GK 350 || Query 349 GK 350 |
| Myxine glutinosa | Query 292 PEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 ||| || | |||+ +|++|++ | + # + | + ||+ | || || |||| Sbjct 213 PEGEEGLE-LRQRLQRSQIARSDKNKQSP#AKDQPSDSASLSGLDRVNLEDFVFLTVLGK 270 |
| Aplysia californica | Query 295 SEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 +| +|++ | |+ || + || # +|| +| ++ +||||| |||| Sbjct 279 TESIQEIKSKMHRSSISSEKRYPEPD#KVQNMSK------QDIVRASDFNFLTVLGK 328 |
| Branchiostoma lanceolatum | Query 296 EGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 | +|++ ++ |+ + | ++#+ | |+ | || |||||||||| Sbjct 226 ENVAKLKEHLQKQKLDEQRKQKSKR#SEEC-----NIGSLDHMKAADFNFLMVLGK 275 |
| Aedes aegypti | Query 292 PEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 | || + ++ | + | | | | # + | | +| ++ ||||||||||| Sbjct 237 PVPEEGTDLVQLKSQMRKTSISKKAPVLC#DKDVPH---NMGKKDVIRATDFNFLMVLGK 292 |
| Apis mellifera | Query 312 QGTKVPEEK#TTNTVSKFDNN-GNRDRMKLTDFNFLMVLGK 350 | | | ++ #|| + +| | | ++ +|||||||||| Sbjct 324 QKTSVTKKT#TTTQDKEVPHNMGKSDLIRASDFNFLMVLGK 363 |
| Anopheles gambiae str. PEST | Query 292 PEGSEGNEELRQKFERAKISQGTKVPEEK#TTNTVSKFDNNGNRDRMKLTDFNFLMVLGK 350 | || + ++ | + | | |+| # + | +| ++ ||||||||||| Sbjct 229 PVPEEGADLVQLKSQMRKTSITKKIPMMG#DKDVPH---NMTKKDVIRATDFNFLMVLGK 284 |
* References
None.