XSB0161 : integrin beta-2 precursor [Sigmodon hispidus]
[ CaMP Format ]
This entry is computationally expanded from SB0002
* Basic Information
| Organism | Sigmodon hispidus (hispid cotton rat) |
| Protein Names | Integrin beta; integrin beta-2 precursor |
| Gene Names | Not available |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
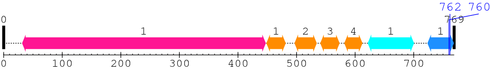
Length: 769 aa
Average Mass: 84.686 kDa
Monoisotopic Mass: 84.630 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Integrin_beta 1. | 32 | 447 | 1041.5 | 0 |
| EGF_2 1. | 449 | 481 | 17.2 | 0.03 |
| EGF_2 2. | 497 | 534 | 15.2 | 0.052 |
| EGF_2 3. | 541 | 573 | 21.0 | 0.0048 |
| EGF_2 4. | 582 | 612 | 29.4 | 1.5e-05 |
| Integrin_B_tail 1. | 622 | 700 | 131.9 | 2.1e-36 |
| Integrin_b_cyt 1. | 724 | 768 | 81.5 | 3e-21 |
| --- cleavage 762 (inside Integrin_b_cyt 724..768) --- | ||||
| --- cleavage 760 (inside Integrin_b_cyt 724..768) --- | ||||
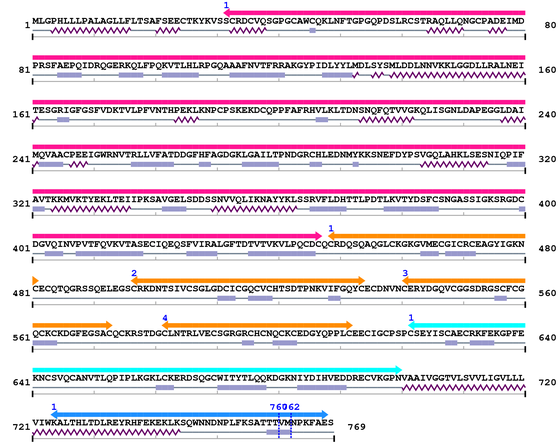
3. Sequence Information
Fasta Sequence: XSB0161.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites]
Cleavage sites (±10aa)
[Site 1] LFKSATTTVM762-NPKFAES
Met762  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu753 | Phe754 | Lys755 | Ser756 | Ala757 | Thr758 | Thr759 | Thr760 | Val761 | Met762 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn763 | Pro764 | Lys765 | Phe766 | Ala767 | Glu768 | Ser769 | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| REYRHFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Sigmodon hispidus | 81.60 | 1 | AF445415_1 integrin beta-2 precursor |
| 2 | N/A | 78.60 | 20 | - |
| 3 | Homo sapiens | 78.60 | 14 | integrin beta chain, beta 2 precursor variant |
| 4 | Mus musculus | 78.60 | 10 | integrin beta 2 |
| 5 | Pan troglodytes | 78.60 | 6 | integrin, beta 2 |
| 6 | synthetic construct | 78.60 | 3 | integrin, beta 2 (antigen CD18 (p95), lymphocyte f |
| 7 | Rattus norvegicus | 76.60 | 8 | PREDICTED: similar to Integrin beta-2 precursor (C |
| 8 | Bos taurus | 76.60 | 6 | integrin, beta 2 |
| 9 | Ovis aries | 76.60 | 1 | antigen CD18 |
| 10 | Capra hircus | 76.60 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 11 | Ovis canadensis | 76.60 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 12 | Bubalus bubalis | 76.60 | 1 | integrin beta-2 |
| 13 | Sus scrofa | 75.90 | 4 | CD18 leukocyte adhesion molecule |
| 14 | Canis familiaris | 73.60 | 4 | AF181965_1 B-2 integrin |
| 15 | Gallus gallus | 60.50 | 4 | integrin, beta 2 |
| 16 | Monodelphis domestica | 59.30 | 4 | PREDICTED: similar to Stromal interaction molecule |
| 17 | Danio rerio | 47.00 | 12 | integrin, beta 1b |
| 18 | Tetraodon nigroviridis | 47.00 | 5 | unnamed protein product |
| 19 | Xenopus laevis | 47.00 | 5 | hypothetical protein LOC379708 |
| 20 | Ictalurus punctatus | 47.00 | 2 | beta-1 integrin |
| 21 | Oryctolagus cuniculus | 47.00 | 2 | integrin beta 1 |
| 22 | Xenopus tropicalis | 47.00 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 23 | Pongo pygmaeus | 47.00 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 24 | Felis catus | 47.00 | 1 | integrin beta 1 |
| 25 | Mus sp. | 47.00 | 1 | beta 7 integrin |
| 26 | Macaca mulatta | 46.60 | 2 | PREDICTED: integrin, beta 7 |
| 27 | Oncorhynchus mykiss | 45.10 | 1 | CD18 protein |
| 28 | Pacifastacus leniusculus | 43.10 | 1 | integrin |
| 29 | Anopheles gambiae | 42.70 | 1 | integrin beta subunit |
| 30 | Anopheles gambiae str. PEST | 42.70 | 1 | ENSANGP00000021373 |
| 31 | Aedes aegypti | 42.70 | 1 | integrin beta subunit |
| 32 | Tribolium castaneum | 42.00 | 2 | PREDICTED: similar to CG1560-PA |
| 33 | Cyprinus carpio | 42.00 | 2 | CD18 type 2 |
| 34 | Drosophila melanogaster | 41.60 | 1 | myospheroid CG1560-PA |
| 35 | Pseudoplusia includens | 41.20 | 1 | integrin beta 1 |
| 36 | Ostrinia furnacalis | 41.20 | 1 | integrin beta 1 precursor |
| 37 | Strongylocentrotus purpuratus | 40.80 | 5 | integrin beta L subunit |
| 38 | Caenorhabditis briggsae | 40.80 | 1 | Hypothetical protein CBG03601 |
| 39 | Apis mellifera | 40.80 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 40 | Caenorhabditis elegans | 40.40 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 41 | Acropora millepora | 39.30 | 1 | integrin subunit betaCn1 |
| 42 | Crassostrea gigas | 38.50 | 1 | integrin beta cgh |
| 43 | Biomphalaria glabrata | 38.10 | 1 | beta integrin subunit |
| 44 | Podocoryne carnea | 37.00 | 1 | AF308652_1 integrin beta chain |
| 45 | Lytechinus variegatus | 35.40 | 1 | beta-C integrin subunit |
| 46 | Schistosoma japonicum | 34.30 | 1 | SJCHGC06221 protein |
| 47 | rats, Peptide Partial, 723 aa | 33.50 | 1 | beta 3 integrin, GPIIIA |
| 48 | Canis lupus familiaris | 33.50 | 1 | integrin beta chain, beta 3 |
| 49 | mice, Peptide Partial, 680 aa | 33.50 | 1 | beta 3 integrin, GPIIIA |
| 50 | Equus caballus | 33.50 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
Top-ranked sequences
| organism | matching |
|---|---|
| Sigmodon hispidus | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||||||||||||||||||||||||||||||#||||||| Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| N/A | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |||| |||||||||||||||||||||||||#||||||| Sbjct 735 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 771 |
| Homo sapiens | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |||| |||||||||||||||||||||||||#||||||| Sbjct 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Mus musculus | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |||| |||||||||||||||||||||||||#||||||| Sbjct 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Pan troglodytes | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |||| |||||||||||||||||||||||||#||||||| Sbjct 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| synthetic construct | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |||| |||||||||||||||||||||||||#||||||| Sbjct 733 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Rattus norvegicus | Query 734 EYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||| |||||||||||||||||||||||||#||||||| Sbjct 733 EYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 768 |
| Bos taurus | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||| |||||||||||||||||||||||||#||||||| Sbjct 733 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Ovis aries | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||| |||||||||||||||||||||||||#||||||| Sbjct 734 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Capra hircus | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||| |||||||||||||||||||||||||#||||||| Sbjct 734 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Ovis canadensis | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||| |||||||||||||||||||||||||#||||||| Sbjct 734 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Bubalus bubalis | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||| |||||||||||||||||||||||||#||||||| Sbjct 733 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Sus scrofa | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAE 768 |||+ |||||||||||||||||||||||||#|||||| Sbjct 733 REYKRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAE 768 |
| Canis familiaris | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||++ ||||||+||||||||||||||||||#||+|||| Sbjct 728 REFKRFEKEKLRSQWNNDNPLFKSATTTVM#NPRFAES 764 |
| Gallus gallus | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 |||| ||||| |++|| |||||||||||||#||+| Sbjct 735 REYRRFEKEKSKAKWNEADNPLFKSATTTVM#NPRF 769 |
| Monodelphis domestica | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NP 764 |||+ |||||||||||||||||| |||++# | Sbjct 1199 REYKRFEKEKLKSQWNNDNPLFKKKTTTII#IP 1230 |
| Danio rerio | Query 733 REYRHFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 766 ||+ |||||+ ++|+ +||++||| |||+#|||+ Sbjct 758 REFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKY 792 |
| Tetraodon nigroviridis | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 ||+ |||||+ ++|+ +||++||| |||+#|||+ Sbjct 754 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 788 |
| Xenopus laevis | Query 734 EYRHFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 767 ||+ ||||++ |+|| |||+ +||||| #|| |+ Sbjct 736 EYQRFEKERMNSKWNPGHNPLYHNATTTVQ#NPNFS 770 |
| Ictalurus punctatus | Query 733 REYRHFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 766 ||+ |||||+ ++|+ +||++||| |||+#|||+ Sbjct 770 REFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKY 804 |
| Oryctolagus cuniculus | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 ||+ |||||+ ++|+ +||++||| |||+#|||+ Sbjct 26 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 60 |
| Xenopus tropicalis | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 ||+ |||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Pongo pygmaeus | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 ||+ |||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Felis catus | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 ||+ |||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Mus sp. | Query 733 REYRHFEKEKLKSQWNNDN-PLFKSATTTVM#NPKF 766 |||| ||||+ + | || ||+||| || +#||+| Sbjct 755 REYRRFEKEQQQLNWKQDNNPLYKSAITTTV#NPRF 789 |
| Macaca mulatta | Query 733 REYRHFEKEKLKSQWNND-NPLFKSATTTVM#NPKFAES 769 ||| ||||+ + | | |||+||| || +#||+| |+ Sbjct 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTTI#NPRFQEA 669 |
| Oncorhynchus mykiss | Query 733 REYRHFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 766 +|+| || |+ | +|+ ||||| +||||| #|| | Sbjct 740 KEFRRFENEQKKGKWSPGDNPLFMNATTTVA#NPTF 774 |
| Pacifastacus leniusculus | Query 733 REYRHFEKE-KLKSQWNNDNPLFKSATTTVM#NPKFAES 769 ||| |||| || | +|||+||| || #|| ||+| Sbjct 763 REYAKFEKERKLPSGKRAENPLYKSAKTTFQ#NPAFAQS 800 |
| Anopheles gambiae | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ ||||++ ++|+ +||++| |||| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYA 835 |
| Anopheles gambiae str. PEST | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ ||||++ ++|+ +||++| |||| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYA 835 |
| Aedes aegypti | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ ||||++ ++|+ +||++| |||| #|| +| Sbjct 798 REFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYA 833 |
| Tribolium castaneum | Query 733 REYRHFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 766 +|| || |+ | ||+ |||||++ ||+| #|| + Sbjct 717 QEYAKFENERQKMQWHRNDNPLYRQATSTFK#NPVY 751 |
| Cyprinus carpio | Query 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFA 767 ||++ |||++ + + ||||++||||| #|| |+ Sbjct 730 REWKRFEKDRKHEKTSGTNPLFQNATTTVQ#NPTFS 764 |
| Drosophila melanogaster | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ ||||++ ++|+ +||++| ||+| #|| +| Sbjct 809 REFARFEKERMNAKWDTGENPIYKQATSTFK#NPMYA 844 |
| Pseudoplusia includens | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ ||||++ ++|+ +||++| ||+| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYA 835 |
| Ostrinia furnacalis | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 767 ||+ ||||++ ++|+ +||++| ||+| #|| +| Sbjct 794 REFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYA 829 |
| Strongylocentrotus purpuratus | Query 733 REYRHFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFAE 768 ||| +| + |+||+ +|||++||+||| #|| + + Sbjct 766 REYAQWENDCKKAQWDQSDNPIYKSSTTTFK#NPTYGK 802 |
| Caenorhabditis briggsae | Query 734 EYRHFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 767 || | |+| ++|+ |+||++| |||| #|| +| Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTFK#NPVYA 806 |
| Apis mellifera | Query 733 REYRHFEKEK-LKSQWNN-DNPLFKSATTTVM#NPKF 766 ||++ ||+|+ | ++|| ||||+| ||+| #|| | Sbjct 778 REFKKFERERILANKWNRRDNPLYKEATSTFK#NPTF 813 |
| Caenorhabditis elegans | Query 734 EYRHFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 767 || | |+| ++|+ |+||++| |||| #|| +| Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTFK#NPVYA 805 |
| Acropora millepora | Query 734 EYRHFEKEKLKSQWNND-NPLFKSATTTVM#NPKFA 767 ||+ ||+|++ |+| + |||+++| || #|| +| Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTFE#NPTYA 788 |
| Crassostrea gigas | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFAE 768 || |||| + ++|+ +||++| ||+| +#|| + + Sbjct 763 RELARFEKEAMNARWDTGENPIYKQATSTFV#NPTYRQ 799 |
| Biomphalaria glabrata | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 ||+ ||||+ ++|+ +||++| ||+| #|| + Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATSTFK#NPTY 785 |
| Podocoryne carnea | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 ||+ |||++ +|++ +||++| ||+| #|| + Sbjct 744 REFAKFEKDRQNPKWDSGENPIYKKATSTFQ#NPMY 778 |
| Lytechinus variegatus | Query 733 REYRHFEKEKLKSQW-NNDNPLFKSATTTVM#NPKF 766 ||+++||||+ + | +||++| +|+ #|| + Sbjct 769 REFQNFEKERANATWEGGENPIYKPSTSVFK#NPTY 803 |
| Schistosoma japonicum | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFAES 769 || +|+++ +| +||+|+| || |+#|| | |+ Sbjct 249 RELANFKQQGENMRWEMAENPIFESPTTNVL#NPTFEEN 286 |
| rats, Peptide Partial, 723 aa | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#N 763 +|+ ||+|+ +++|+ +|||+| ||+| #| Sbjct 686 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 717 |
| Canis lupus familiaris | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#N 763 +|+ ||+|+ +++|+ +|||+| ||+| #| Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 778 |
| mice, Peptide Partial, 680 aa | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#N 763 +|+ ||+|+ +++|+ +|||+| ||+| #| Sbjct 647 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 678 |
| Equus caballus | Query 733 REYRHFEKEKLKSQWNN-DNPLFKSATTTVM#N 763 +|+ ||+|+ +++|+ +|||+| ||+| #| Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 778 |
[Site 2] NPLFKSATTT760-VMNPKFAES
Thr760  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn751 | Pro752 | Leu753 | Phe754 | Lys755 | Ser756 | Ala757 | Thr758 | Thr759 | Thr760 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val761 | Met762 | Asn763 | Pro764 | Lys765 | Phe766 | Ala767 | Glu768 | Ser769 | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DLREYRHFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Sigmodon hispidus | 85.50 | 1 | AF445415_1 integrin beta-2 precursor |
| 2 | N/A | 82.40 | 20 | I |
| 3 | Homo sapiens | 82.40 | 14 | integrin beta chain, beta 2 precursor variant |
| 4 | Mus musculus | 82.40 | 10 | integrin beta 2 |
| 5 | Pan troglodytes | 82.40 | 6 | integrin, beta 2 |
| 6 | synthetic construct | 82.40 | 3 | integrin, beta 2 (antigen CD18 (p95), lymphocyte f |
| 7 | Rattus norvegicus | 80.50 | 8 | PREDICTED: similar to Integrin beta-2 precursor (C |
| 8 | Bos taurus | 80.50 | 6 | integrin, beta 2 |
| 9 | Capra hircus | 80.50 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 10 | Ovis aries | 80.50 | 1 | antigen CD18 |
| 11 | Bubalus bubalis | 80.50 | 1 | integrin beta-2 |
| 12 | Ovis canadensis | 80.50 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 13 | Sus scrofa | 79.70 | 4 | CD18 leukocyte adhesion molecule |
| 14 | Canis familiaris | 77.40 | 4 | AF181965_1 B-2 integrin |
| 15 | Monodelphis domestica | 63.20 | 4 | PREDICTED: similar to Stromal interaction molecule |
| 16 | Gallus gallus | 62.00 | 5 | integrin, beta 2 |
| 17 | Felis catus | 48.90 | 1 | integrin beta 1 |
| 18 | Danio rerio | 48.50 | 12 | integrin, beta 1b |
| 19 | Tetraodon nigroviridis | 48.50 | 6 | unnamed protein product |
| 20 | Xenopus laevis | 48.50 | 5 | hypothetical protein LOC379708 |
| 21 | Ictalurus punctatus | 48.50 | 2 | beta-1 integrin |
| 22 | Oryctolagus cuniculus | 48.50 | 2 | integrin beta 1 |
| 23 | Xenopus tropicalis | 48.50 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 24 | Pongo pygmaeus | 48.50 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 25 | Mus sp. | 48.50 | 1 | beta 7 integrin |
| 26 | Macaca mulatta | 48.10 | 2 | PREDICTED: integrin, beta 7 |
| 27 | Oncorhynchus mykiss | 47.80 | 1 | CD18 protein |
| 28 | Cyprinus carpio | 45.80 | 2 | CD18 type 2 |
| 29 | Pacifastacus leniusculus | 44.70 | 1 | integrin |
| 30 | Anopheles gambiae str. PEST | 44.30 | 1 | ENSANGP00000021373 |
| 31 | Anopheles gambiae | 44.30 | 1 | integrin beta subunit |
| 32 | Aedes aegypti | 44.30 | 1 | integrin beta subunit |
| 33 | Tribolium castaneum | 43.50 | 3 | PREDICTED: similar to CG1560-PA |
| 34 | Drosophila melanogaster | 43.10 | 1 | myospheroid CG1560-PA |
| 35 | Ostrinia furnacalis | 42.70 | 1 | integrin beta 1 precursor |
| 36 | Pseudoplusia includens | 42.70 | 1 | integrin beta 1 |
| 37 | Strongylocentrotus purpuratus | 42.40 | 5 | integrin beta L subunit |
| 38 | Apis mellifera | 42.00 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 39 | Caenorhabditis briggsae | 42.00 | 1 | Hypothetical protein CBG03601 |
| 40 | Caenorhabditis elegans | 41.60 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 41 | Biomphalaria glabrata | 40.00 | 1 | beta integrin subunit |
| 42 | Crassostrea gigas | 40.00 | 1 | integrin beta cgh |
| 43 | Acropora millepora | 39.70 | 1 | integrin subunit betaCn1 |
| 44 | Podocoryne carnea | 38.50 | 1 | AF308652_1 integrin beta chain |
| 45 | Lytechinus variegatus | 37.00 | 1 | beta-C integrin subunit |
| 46 | Schistosoma japonicum | 35.80 | 1 | SJCHGC06221 protein |
| 47 | Canis lupus familiaris | 35.00 | 1 | integrin beta chain, beta 3 |
| 48 | rats, Peptide Partial, 723 aa | 35.00 | 1 | beta 3 integrin, GPIIIA |
| 49 | Equus caballus | 35.00 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 50 | mice, Peptide Partial, 680 aa | 35.00 | 1 | beta 3 integrin, GPIIIA |
Top-ranked sequences
| organism | matching |
|---|---|
| Sigmodon hispidus | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||| Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| N/A | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |||||| |||||||||||||||||||||||#||||||||| Sbjct 733 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 771 |
| Homo sapiens | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |||||| |||||||||||||||||||||||#||||||||| Sbjct 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Mus musculus | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |||||| |||||||||||||||||||||||#||||||||| Sbjct 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Pan troglodytes | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |||||| |||||||||||||||||||||||#||||||||| Sbjct 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| synthetic construct | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |||||| |||||||||||||||||||||||#||||||||| Sbjct 731 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Rattus norvegicus | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 || ||| |||||||||||||||||||||||#||||||||| Sbjct 730 DLNEYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 768 |
| Bos taurus | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||| |||||||||||||||||||||||#||||||||| Sbjct 731 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Capra hircus | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||| |||||||||||||||||||||||#||||||||| Sbjct 732 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Ovis aries | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||| |||||||||||||||||||||||#||||||||| Sbjct 732 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Bubalus bubalis | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||| |||||||||||||||||||||||#||||||||| Sbjct 731 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Ovis canadensis | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||| |||||||||||||||||||||||#||||||||| Sbjct 732 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Sus scrofa | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAE 768 |||||+ |||||||||||||||||||||||#|||||||| Sbjct 731 DLREYKRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAE 768 |
| Canis familiaris | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 ||||++ ||||||+||||||||||||||||#||||+|||| Sbjct 726 DLREFKRFEKEKLRSQWNNDNPLFKSATTT#VMNPRFAES 764 |
| Monodelphis domestica | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNP 764 |||||+ |||||||||||||||||| |||#++ | Sbjct 1197 DLREYKRFEKEKLKSQWNNDNPLFKKKTTT#IIIP 1230 |
| Gallus gallus | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | |||| ||||| |++|| |||||||||||#||||+| Sbjct 733 DRREYRRFEKEKSKAKWNEADNPLFKSATTT#VMNPRF 769 |
| Felis catus | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ |||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DTREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Danio rerio | Query 731 DLREYRHFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 766 | ||+ |||||+ ++|+ +||++||| ||#|+|||+ Sbjct 756 DRREFAKFEKEKMNAKWDAGENPIYKSAVTT#VVNPKY 792 |
| Tetraodon nigroviridis | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ |||||+ ++|+ +||++||| ||#|+|||+ Sbjct 752 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 788 |
| Xenopus laevis | Query 731 DLREYRHFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 767 | ||+ ||||++ |+|| |||+ +||||#| || |+ Sbjct 733 DRNEYQRFEKERMNSKWNPGHNPLYHNATTT#VQNPNFS 770 |
| Ictalurus punctatus | Query 731 DLREYRHFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 766 | ||+ |||||+ ++|+ +||++||| ||#|+|||+ Sbjct 768 DRREFAKFEKEKMNAKWDAGENPIYKSAVTT#VVNPKY 804 |
| Oryctolagus cuniculus | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ |||||+ ++|+ +||++||| ||#|+|||+ Sbjct 24 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 60 |
| Xenopus tropicalis | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ |||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Pongo pygmaeus | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ |||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Mus sp. | Query 731 DLREYRHFEKEKLKSQWNND-NPLFKSATTT#VMNPKF 766 | |||| ||||+ + | | |||+||| ||# +||+| Sbjct 753 DRREYRRFEKEQQQLNWKQDNNPLYKSAITT#TVNPRF 789 |
| Macaca mulatta | Query 731 DLREYRHFEKEKLKSQWNND-NPLFKSATTT#VMNPKFAES 769 | ||| ||||+ + | | |||+||| ||# +||+| |+ Sbjct 630 DRREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEA 669 |
| Oncorhynchus mykiss | Query 731 DLREYRHFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 766 |++|+| || |+ | +|+ ||||| +||||#| || | Sbjct 738 DVKEFRRFENEQKKGKWSPGDNPLFMNATTT#VANPTF 774 |
| Cyprinus carpio | Query 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFA 767 ||||++ |||++ + + ||||++||||#| || |+ Sbjct 728 DLREWKRFEKDRKHEKTSGTNPLFQNATTT#VQNPTFS 764 |
| Pacifastacus leniusculus | Query 731 DLREYRHFEKE-KLKSQWNNDNPLFKSATTT#VMNPKFAES 769 | ||| |||| || | +|||+||| ||# || ||+| Sbjct 761 DRREYAKFEKERKLPSGKRAENPLYKSAKTT#FQNPAFAQS 800 |
| Anopheles gambiae str. PEST | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ ||||++ ++|+ +||++| ||||# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYA 835 |
| Anopheles gambiae | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ ||||++ ++|+ +||++| ||||# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYA 835 |
| Aedes aegypti | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ ||||++ ++|+ +||++| ||||# || +| Sbjct 796 DRREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYA 833 |
| Tribolium castaneum | Query 731 DLREYRHFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 766 | +|| || |+ | ||+ |||||++ ||+|# || + Sbjct 715 DRQEYAKFENERQKMQWHRNDNPLYRQATST#FKNPVY 751 |
| Drosophila melanogaster | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ ||||++ ++|+ +||++| ||+|# || +| Sbjct 807 DRREFARFEKERMNAKWDTGENPIYKQATST#FKNPMYA 844 |
| Ostrinia furnacalis | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ ||||++ ++|+ +||++| ||+|# || +| Sbjct 792 DRREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYA 829 |
| Pseudoplusia includens | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 767 | ||+ ||||++ ++|+ +||++| ||+|# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYA 835 |
| Strongylocentrotus purpuratus | Query 731 DLREYRHFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFAE 768 | ||| +| + |+||+ +|||++||+|||# || + + Sbjct 764 DKREYAQWENDCKKAQWDQSDNPIYKSSTTT#FKNPTYGK 802 |
| Apis mellifera | Query 731 DLREYRHFEKEK-LKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||++ ||+|+ | ++|| ||||+| ||+|# || | Sbjct 776 DNREFKKFERERILANKWNRRDNPLYKEATST#FKNPTF 813 |
| Caenorhabditis briggsae | Query 731 DLREYRHFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 767 | || | |+| ++|+ |+||++| ||||# || +| Sbjct 769 DRAEYAKFNNERLMAKWDTNENPIYKQATTT#FKNPVYA 806 |
| Caenorhabditis elegans | Query 731 DLREYRHFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 767 | || | |+| ++|+ |+||++| ||||# || +| Sbjct 768 DRSEYATFNNERLMAKWDTNENPIYKQATTT#FKNPVYA 805 |
| Biomphalaria glabrata | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ ||||+ ++|+ +||++| ||+|# || + Sbjct 749 DTREFAKFEKERQNAKWDTGENPIYKQATST#FKNPTY 785 |
| Crassostrea gigas | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFAE 768 | || |||| + ++|+ +||++| ||+|# +|| + + Sbjct 761 DKRELARFEKEAMNARWDTGENPIYKQATST#FVNPTYRQ 799 |
| Acropora millepora | Query 731 DLREYRHFEKEKLKSQWNND-NPLFKSATTT#VMNPKFA 767 | ||+ ||+|++ |+| + |||+++| ||# || +| Sbjct 751 DRIEYQKFERERMHSKWTREKNPLYQAAKTT#FENPTYA 788 |
| Podocoryne carnea | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 | ||+ |||++ +|++ +||++| ||+|# || + Sbjct 742 DRREFAKFEKDRQNPKWDSGENPIYKKATST#FQNPMY 778 |
| Lytechinus variegatus | Query 731 DLREYRHFEKEKLKSQW-NNDNPLFKSATTT#VMNPKF 766 | ||+++||||+ + | +||++| +|+ # || + Sbjct 767 DRREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTY 803 |
| Schistosoma japonicum | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFAES 769 | || +|+++ +| +||+|+| || #|+|| | |+ Sbjct 247 DRRELANFKQQGENMRWEMAENPIFESPTTN#VLNPTFEEN 286 |
| Canis lupus familiaris | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMN 763 | +|+ ||+|+ +++|+ +|||+| ||+|# | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 778 |
| rats, Peptide Partial, 723 aa | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMN 763 | +|+ ||+|+ +++|+ +|||+| ||+|# | Sbjct 684 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 717 |
| Equus caballus | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMN 763 | +|+ ||+|+ +++|+ +|||+| ||+|# | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 778 |
| mice, Peptide Partial, 680 aa | Query 731 DLREYRHFEKEKLKSQWNN-DNPLFKSATTT#VMN 763 | +|+ ||+|+ +++|+ +|||+| ||+|# | Sbjct 645 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 678 |
* References
None.