XSB0172 : coatomer protein complex, subunit beta 1 [Rattus norvegicus]
[ CaMP Format ]
This entry is computationally expanded from SB0044
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | Coatomer subunit beta; Beta-coat protein; Beta-COP; coatomer protein complex; subunit beta 1; receptor for activated C-kinase-2 |
| Gene Names | Copb1; Copb; coatomer protein complex, subunit beta 1 |
| Gene Locus | 1q34; chromosome 1 |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| NP_542959 | NM_080781 | 114023 | P23514 | N/A | N/A | N/A | rno:114023 |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 953 aa
Average Mass: 107.011 kDa
Monoisotopic Mass: 106.943 kDa
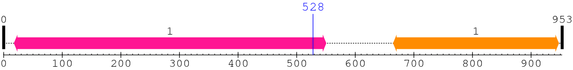
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Adaptin_N 1. | 18 | 550 | 532.0 | 7.3e-157 |
| --- cleavage 528 (inside Adaptin_N 18..550) --- | ||||
| Coatamer_beta_C 1. | 665 | 947 | 563.1 | 3.2e-166 |
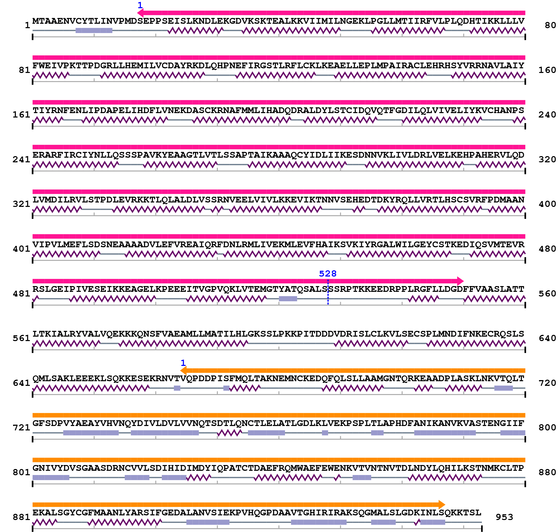
3. Sequence Information
Fasta Sequence: XSB0172.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] GTYATQSALS528-SSRPTKKEED
Ser528  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Gly519 | Thr520 | Tyr521 | Ala522 | Thr523 | Gln524 | Ser525 | Ala526 | Leu527 | Ser528 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser529 | Ser530 | Arg531 | Pro532 | Thr533 | Lys534 | Lys535 | Glu536 | Glu537 | Asp538 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ELKPEEEITVGPVQKLVTEMGTYATQSALSSSRPTKKEEDRPPLRGFLLDGDFFVAASLA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 120.00 | 3 | coatomer protein complex, subunit beta 1 |
| 2 | Pan troglodytes | 120.00 | 3 | PREDICTED: coatomer protein complex, subunit beta |
| 3 | Homo sapiens | 120.00 | 2 | beta-cop homolog |
| 4 | Pongo pygmaeus | 120.00 | 1 | hypothetical protein |
| 5 | Rattus norvegicus | 120.00 | 1 | coatomer protein complex, subunit beta 1 |
| 6 | Gallus gallus | 119.00 | 1 | coatomer protein complex, subunit beta |
| 7 | Bos taurus | 118.00 | 1 | hypothetical protein LOC535533 |
| 8 | Canis familiaris | 118.00 | 1 | PREDICTED: similar to coatomer protein complex, su |
| 9 | Monodelphis domestica | 115.00 | 1 | PREDICTED: hypothetical protein |
| 10 | Xenopus laevis | 111.00 | 2 | hypothetical protein LOC735158 |
| 11 | Danio rerio | 104.00 | 2 | coatomer protein complex, subunit beta 1 |
| 12 | Tetraodon nigroviridis | 103.00 | 1 | unnamed protein product |
| 13 | Strongylocentrotus purpuratus | 67.80 | 1 | PREDICTED: similar to MGC80934 protein, partial |
| 14 | Caenorhabditis elegans | 61.60 | 1 | Y25C1A.5 |
| 15 | Apis mellifera | 57.00 | 1 | PREDICTED: similar to coatomer protein complex, su |
| 16 | Anopheles gambiae str. PEST | 56.20 | 1 | ENSANGP00000016931 |
| 17 | Aedes aegypti | 55.10 | 1 | coatomer beta subunit |
| 18 | Drosophila melanogaster | 54.70 | 1 | beta-coatomer protein CG6223-PA |
| 19 | N/A | 53.90 | 4 | - |
| 20 | Drosophila pseudoobscura | 53.50 | 1 | GA19453-PA |
| 21 | Caenorhabditis briggsae | 53.50 | 1 | Hypothetical protein CBG19635 |
| 22 | Kluyveromyces lactis | 50.40 | 1 | unnamed protein product |
| 23 | Debaryomyces hansenii CBS767 | 48.50 | 1 | hypothetical protein DEHA0F20548g |
| 24 | Pichia stipitis CBS 6054 | 46.20 | 1 | predicted protein |
| 25 | Candida glabrata | 45.10 | 1 | unnamed protein product |
| 26 | Phaeosphaeria nodorum SN15 | 43.50 | 1 | hypothetical protein SNOG_15084 |
| 27 | Ashbya gossypii ATCC 10895 | 42.70 | 1 | ABL112Wp |
| 28 | Lodderomyces elongisporus NRRL YB-4239 | 40.80 | 1 | coatomer beta subunit |
| 29 | Chaetomium globosum CBS 148.51 | 40.80 | 1 | conserved hypothetical protein |
| 30 | Aspergillus terreus NIH2624 | 40.40 | 1 | coatomer beta subunit |
| 31 | Aspergillus nidulans FGSC A4 | 40.00 | 1 | hypothetical protein AN1177.2 |
| 32 | Pichia guilliermondii ATCC 6260 | 39.70 | 1 | hypothetical protein PGUG_00251 |
| 33 | Neurospora crassa OR74A | 39.70 | 1 | hypothetical protein |
| 34 | Aspergillus oryzae | 39.30 | 1 | unnamed protein product |
| 35 | Aspergillus niger | 39.30 | 1 | hypothetical protein An08g03270 |
| 36 | Aspergillus fumigatus Af293 | 39.30 | 1 | Coatomer subunit beta, putative |
| 37 | Aspergillus clavatus NRRL 1 | 39.30 | 1 | Coatomer subunit beta, putative |
| 38 | Neosartorya fischeri NRRL 181 | 39.30 | 1 | Coatomer subunit beta, putative |
| 39 | Magnaporthe grisea 70-15 | 38.90 | 1 | conserved hypothetical protein |
| 40 | Candida albicans SC5314 | 38.50 | 1 | hypothetical protein CaO19_8161 |
| 41 | Gibberella zeae PH-1 | 38.10 | 1 | conserved hypothetical protein |
| 42 | Schizosaccharomyces pombe 972h- | 37.70 | 1 | coatomer subunit beta |
| 43 | Cryptococcus neoformans var. neoformans JEC21 | 37.00 | 1 | ER to Golgi transport-related protein |
| 44 | Coprinopsis cinerea okayama7#130 | 36.60 | 1 | hypothetical protein CC1G_00705 |
| 45 | Ustilago maydis 521 | 36.20 | 1 | hypothetical protein UM03361.1 |
| 46 | Yarrowia lipolytica | 36.20 | 1 | hypothetical protein |
| 47 | Coccidioides immitis RS | 35.00 | 1 | hypothetical protein CIMG_06935 |
| 48 | Arabidopsis thaliana | 34.70 | 3 | coatomer beta subunit, putative / beta-coat protei |
| 49 | Ostreococcus lucimarinus CCE9901 | 34.70 | 1 | predicted protein |
| 50 | Ostreococcus tauri | 34.30 | 1 | coatomer subunit beta (ISS) |
| 51 | Oryza sativa (japonica cultivar-group) | 33.10 | 6 | COPB1_ORYSJ Coatomer subunit beta-1 (Beta-coat pro |
| 52 | Oryza sativa (indica cultivar-group) | 33.10 | 2 | hypothetical protein OsI_001379 |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |
| Pan troglodytes | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |
| Homo sapiens | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |
| Pongo pygmaeus | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |
| Rattus norvegicus | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |
| Gallus gallus | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |||||||+||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 499 ELKPEEEVTVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |
| Bos taurus | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 ||||||||||||||||||||||||||||||#|||||||||+|||||||||||||||||||| Sbjct 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEERPPLRGFLLDGDFFVAASLA 558 |
| Canis familiaris | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 ||||||||||||||||||||||||||||||#|||||||||+|||||||||||||||||||| Sbjct 600 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEERPPLRGFLLDGDFFVAASLA 659 |
| Monodelphis domestica | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |||||||| |||||||||||||||||||||#||||||||||||||| |||||||||||||| Sbjct 499 ELKPEEEINVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRAFLLDGDFFVAASLA 558 |
| Xenopus laevis | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |||||+++|||| |||||||||||||||||#|||| ||||+|||||||||||||+|||||| Sbjct 499 ELKPEDDVTVGPAQKLVTEMGTYATQSALS#SSRPAKKEEERPPLRGFLLDGDFYVAASLA 558 |
| Danio rerio | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |+|||||+| | ||||||||| ||||||#+|||+|||||||||||||+||||+|||||| Sbjct 499 EVKPEEEVTAAPAPKLVTEMGTYVTQSALS#TSRPSKKEEDRPPLRGFLMDGDFYVAASLA 558 |
| Tetraodon nigroviridis | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |+|||+|++ | |||||||||| ||||||#||||+|||||||||||||+||||+||| || Sbjct 525 EVKPEDEVSAAPAQKLVTEMGTYVTQSALS#SSRPSKKEEDRPPLRGFLMDGDFYVAACLA 584 |
| Strongylocentrotus purpuratus | Query 500 LKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 | ||+ | ++|||||||||||||||# ||+|| || |||||+|||||| | || Sbjct 276 LTPEK---VSASKRLVTEMGTYATQSALS#VVS-TKQEEQRPALRGFLMDGDFFVGACLA 330 |
| Caenorhabditis elegans | Query 499 ELKPEEEITVGPVQKL-----VTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFV 553 | ||||+ | |+ || ||||||+|||#|+ | |+ ++|||| |||||+||+ Sbjct 497 EQKPEEQ---QPEQQQKQKPKVTADGTYATQTALS#STVTTSKKNEKPPLRRFLLDGEFFL 553 Query 554 AASLA 558 |||| Sbjct 554 GASLA 558 |
| Apis mellifera | Query 508 VGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 | | | ||| |||||||| |#++ |||| || | ++++||||+ |||| Sbjct 500 VTPAQ-LVTSDGTYATQSAFS#ATSARKKEEKRPALVQYMMEGDFFIGASLA 549 |
| Anopheles gambiae str. PEST | Query 505 EITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 | | | || |||||||| |# + | |+ +||||| +++||||||||+|| Sbjct 510 ETTANSTNK-VTSDGTYATQSAFS#VA-PVAKKVERPPLRQYMMDGDFFVAATLA 561 |
| Aedes aegypti | Query 515 VTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 || |||||||| |# + | |+ +||||| +|+|||||+ |+|| Sbjct 517 VTSDGTYATQSAFS#VA-PVSKKVERPPLRQYLMDGDFFIGATLA 559 |
| Drosophila melanogaster | Query 509 GPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 | || |||||||| |# + | | | ||||| +|+|||||+ |+|+ Sbjct 522 GNASNKVTSDGTYATQSAYS#LA-PVAKAEKRPPLRQYLMDGDFFIGAALS 570 |
| N/A | Query 509 GPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 | || |||||||| |# | | | ||||| +|+|||||+ |+|+ Sbjct 521 GNASNKVTSDGTYATQSAYS#LP-PVAKAEKRPPLRQYLMDGDFFIGAALS 569 |
| Drosophila pseudoobscura | Query 515 VTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 || |||||||| |# + | | | ||||| +|+|||||+ |+|+ Sbjct 528 VTSDGTYATQSAYS#LA-PVAKVEKRPPLRQYLMDGDFFIGAALS 570 |
| Caenorhabditis briggsae | Query 503 EEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 ++++ || || ||||||+|||#++ +|| | +|||| |||+|+||+ |||| Sbjct 505 QKKVPSGPK---VTADGTYATQTALS#TTVVSKKNE-KPPLRRFLLNGEFFLGASLA 556 |
| Kluyveromyces lactis | Query 506 ITVGPVQKLVTEMGTYATQSALS#SSRPTK-----KEEDRPPLRGFLLDGDFFVAASLA 558 ++ ||| | |||||+||||# | | ++| ||||| |+|+|||| |+ || Sbjct 510 VSTGPV---VLPDGTYATESALS#VSEKVKSTSNEEKEARPPLRRFVLNGDFFTASVLA 564 |
| Debaryomyces hansenii CBS767 | Query 503 EEEITVGPVQK-----LVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASL 557 +|+ | | |+| +| |||||++||+#+ | +|++||+| +|+|||++ + | Sbjct 498 DEDETNGSVEKPKKRPVVLPDGTYATENALT#TEIKTSVDEEKPPIRRLILEGDFYLGSVL 557 Query 558 A 558 | Query 558 A 558 |
| Pichia stipitis CBS 6054 | Query 504 EEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 | ||| | |||||+|||+#+ +++||+| +|||||++|| |+ Sbjct 506 EHTKKGPV---VLPDGTYATESALT#AEVKDTSNDEKPPVRKHILDGDFYLAAVLS 557 |
| Candida glabrata | Query 503 EEEIT--VGPVQKLVTEMGTYATQSALS#SSRPTKKE-----EDRPPLRGFLLDGDFFVAA 555 || | ||| + |||||++| #|| |+ |+||||| |+| |||+ |+ Sbjct 509 EESATKQAGPV---ILPDGTYATENAFG#SSNNDNKKKLVENENRPPLRRFVLGGDFYTAS 565 Query 556 SLA 558 || Sbjct 566 ILA 568 |
| Phaeosphaeria nodorum SN15 | Query 519 GTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVAASLA 558 |||||+|||+#|| | | +|||| +||||+++| |+ Sbjct 525 GTYATESALT#SSAAAKAKLEAVKNSQKPPLRQLVLDGDYYLATVLS 570 |
| Ashbya gossypii ATCC 10895 | Query 502 PEEEITVGPVQKLVTEMGTYATQSAL---S#SSRPTKKEED-RPPLRGFLLDGDFFVAASL 557 ||+ + || | ||| |+||| |#+ + +|+++| ||||| |+| | |+ + | Sbjct 507 PEDNVPKGPT---VLPDGTYVTESALTVQS#TKQLSKEDKDSRPPLRRFILSGYFYAGSVL 563 Query 558 A 558 | Query 558 A 558 |
| Lodderomyces elongisporus NRRL YB-4239 | Query 519 GTYATQSALS#S-SRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 |||||++| |#| | ++ | +|||| || |||++ | |+ Sbjct 178 GTYATETAFS#SESYESQDSESKPPLRKHLLAGDFYLGAVLS 218 |
| Chaetomium globosum CBS 148.51 | Query 507 TVGPVQKLVTEMGTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVAASLA 558 | | +|++ + |||||++||+#| | +|||| +||||+++| || Sbjct 515 TPGGSRKVLAD-GTYATETALT#SQSAAAARLEAVKASSKPPLRQLILDGDYYLATVLA 571 |
| Aspergillus terreus NIH2624 | Query 519 GTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVAASLA 558 |||||+|||+#| | +|||| +|||||++| |+ Sbjct 525 GTYATESALT#SESAAAARLAAVKAAQKPPLRQLILDGDFYLATVLS 570 |
| Aspergillus nidulans FGSC A4 | Query 502 PEEEITV-GPVQKLVTEMGTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVA 554 |+|+| | | |||||+|||+#| | +|||| +||||+++| Sbjct 502 PKEQINGHGKSAPKVLADGTYATESALT#SQSAAAARLQAVKAAQKPPLRQLILDGDYYLA 561 Query 555 ASLA 558 |+ Sbjct 562 TVLS 565 |
| Pichia guilliermondii ATCC 6260 | Query 503 EEEITVGPVQK--LVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVA 554 +|| + | + | +| |||||++|++# +||++||+| || |||++| Sbjct 550 QEEESSGKLTKKPVVLPDGTYATENAMT#VD-VVHQEEEKPPVRKLLLGGDFYLA 602 |
| Neurospora crassa OR74A | Query 519 GTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVAASLA 558 |||||++||+#| | +|||| +||||+++| || Sbjct 524 GTYATETALT#SQSTAAARLEAVKASSKPPLRQLILDGDYYLATVLA 569 |
| Aspergillus oryzae | Query 519 GTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVAASLA 558 |||||+|||+#| | +|||| +||||+++| |+ Sbjct 525 GTYATESALT#SQSAAAARLEAVKAAQKPPLRQLILDGDYYLATVLS 570 |
| Aspergillus niger | Query 519 GTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVAASLA 558 |||||+|||+#| | +|||| +||||+++| |+ Sbjct 525 GTYATESALT#SQSAAAARLEAVKAAQKPPLRQLILDGDYYLATVLS 570 |
| Aspergillus fumigatus Af293 | Query 519 GTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVAASLA 558 |||||+|||+#| | +|||| +||||+++| |+ Sbjct 525 GTYATESALT#SESAAAARLEAVKAAQKPPLRQLILDGDYYLATVLS 570 |
| Aspergillus clavatus NRRL 1 | Query 519 GTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVAASLA 558 |||||+|||+#| | +|||| +||||+++| |+ Sbjct 525 GTYATESALT#SESAAAAKLAAVKAAQKPPLRQLILDGDYYLATVLS 570 |
| Neosartorya fischeri NRRL 181 | Query 519 GTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVAASLA 558 |||||+|||+#| | +|||| +||||+++| |+ Sbjct 525 GTYATESALT#SESAAAARLEAVKAAQKPPLRQLILDGDYYLATVLS 570 |
| Magnaporthe grisea 70-15 | Query 519 GTYATQSALS#S--SRPTK----KEEDRPPLRGFLLDGDFFVAASLA 558 |||||++||+#| |+ | | +|||| +||||+++| |+ Sbjct 525 GTYATETALT#SQSSKAAKLEAVKAAQKPPLRQLILDGDYYLATVLS 570 |
| Candida albicans SC5314 | Query 509 GPVQKLVTEMGTYATQSALS#SSRPTKKEED-RPPLRGFLLDGDFFVAASLA 558 ||| | |||||+|||+#| | | + |+| +| |||++ | || Sbjct 513 GPV---VLPDGTYATESALT#SETTDSLESDSKTPIRKQILAGDFYLGAVLA 560 |
| Gibberella zeae PH-1 | Query 519 GTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVAASLA 558 |||||++||+#| | +|||| +||||+++| |+ Sbjct 525 GTYATETALT#SQSSAAARLEAVKTAQKPPLRQLILDGDYYLATVLS 570 |
| Schizosaccharomyces pombe 972h- | Query 502 PEEEITV-----GPVQKLVTEMGTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGD 550 ||+++ + + | |||||+||++#| + | +|||| +| || Sbjct 495 PEDDLLIDISAPASTSRKVLPDGTYATESAVT#SEALSAARLEAVKASKKPPLRTQILSGD 554 Query 551 FFVAASLA 558 +++|| || Sbjct 555 YYLAAVLA 562 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 519 GTYATQSALS#SSRPTKKEE-----DRPPLRGFLLDGDFFVAASLA 558 |||||++ +#|| + | +|||| +| |||| + || Sbjct 520 GTYATETVYT#SSASAARLEAVRSASKPPLRSLILGGDFFTGSVLA 564 |
| Coprinopsis cinerea okayama7#130 | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEED-----RPPLRGFLLDGDFFV 553 | |+| + | |||||++| |#+| + | +|||| +| |||| Sbjct 446 EESEEKEKPAVSTRPRVLADGTYATETAFS#TSSANARLEAVKAAAKPPLRTLILGGDFFT 505 Query 554 AASLA 558 + |+ Sbjct 506 GSVLS 510 |
| Ustilago maydis 521 | Query 519 GTYATQSALS#SSR------PTKKEEDRPPLRGFLLDGDFFVAASLA 558 |||||++| |#| | +|||| || |||+ + || Sbjct 537 GTYATETAFS#SETNQAARLEAVKNASKPPLRSLLLLGDFYTGSVLA 582 |
| Yarrowia lipolytica | Query 503 EEEITVG------PVQKLVTEMGTYATQSALS#SSRPTKKEE-DRPPLRGFLL-DGDFFVA 554 ||| || | +|++ + |||||+|||+#++ + +| + ||| +| ||++||| Sbjct 498 EEEPDVGETATHKPSRKVLAD-GTYATESALT#TTVKAEVDEFEGQPLRSIILKDGNYFVA 556 Query 555 ASL 557 | Sbjct 557 GVL 559 |
| Coccidioides immitis RS | Query 519 GTYATQSALS#SSRPTK------KEEDRPPLRGFLLDGDFFVAASLA 558 |||| ++||+#| | +|||| +||||+++| |+ Sbjct 523 GTYAQETALT#SQSAAAAKLEAIKAARKPPLRQLILDGDYYLATVLS 568 |
| Arabidopsis thaliana | Query 499 ELKPEEEITVGPVQKLVTEMGTYATQSALS#S---SRPTKKEEDRPP--LRGFLLDGDFFV 553 +++| | + ++ |||||||| |# | || + || || ||||+ Sbjct 498 KIQPTSSAMVSSRKPVILADGTYATQSAAS#ETTFSSPTVVQGSLTSGNLRALLLTGDFFL 557 Query 554 AASLA 558 | +| Sbjct 558 GAVVA 562 |
| Ostreococcus lucimarinus CCE9901 | Query 503 EEEITVGPVQKLVTEMGTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 +|+ | + | ||||||+| |#+| + | || || || |++| +| Sbjct 499 DEDTTETTTRPAVLADGTYATQAAYS#TSAAISQV---PNLREMLLKGDSFLSAVIA 551 |
| Ostreococcus tauri | Query 519 GTYATQSALS#SSRPTKKEEDRPPLRGFLLDGDFFVAASLA 558 ||||||+| +#+|+ | || || ||||+|+ +| Sbjct 507 GTYATQAAFT#ASKAIPV---LPNLRELLLKGDFFLASVIA 543 |
| Oryza sativa (japonica cultivar-group) | Query 506 ITVGPVQKLVTEMGTYATQSALS#SSRPTKKE------EDRPPLRGFLLDGDFFVAASLA 558 +|| + +| |||||||| +# + + || +| ||||+|| +| Sbjct 508 VTVSSRRPVVLADGTYATQSAAT#ETAISSPAVAPGSLSSTQNLRSLILSGDFFLAAVVA 566 |
| Oryza sativa (indica cultivar-group) | Query 506 ITVGPVQKLVTEMGTYATQSALS#SSRPTKKE-----EDRPPLRGFLLDGDFFVAASLA 558 +|| + +| |||||||| +# + | || +| ||||+|| ++ Sbjct 507 VTVSSRRPVVLADGTYATQSAAT#EAISTPSVAPGSLSSTLNLRSLILSGDFFLAAVIS 564 |
* References
[PubMed ID: 15039458] Huang X, Walker JW, Myofilament anchoring of protein kinase C-epsilon in cardiac myocytes. J Cell Sci. 2004 Apr 15;117(Pt 10):1971-8. Epub 2004 Mar 23.
[PubMed ID: 9114004] ... Zhao L, Helms JB, Brugger B, Harter C, Martoglio B, Graf R, Brunner J, Wieland FT, Direct and GTP-dependent interaction of ADP ribosylation factor 1 with coatomer subunit beta. Proc Natl Acad Sci U S A. 1997 Apr 29;94(9):4418-23.
[PubMed ID: 8376457] ... Peter F, Plutner H, Zhu H, Kreis TE, Balch WE, Beta-COP is essential for transport of protein from the endoplasmic reticulum to the Golgi in vitro. J Cell Biol. 1993 Sep;122(6):1155-67.
[PubMed ID: 1840503] ... Duden R, Griffiths G, Frank R, Argos P, Kreis TE, Beta-COP, a 110 kd protein associated with non-clathrin-coated vesicles and the Golgi complex, shows homology to beta-adaptin. Cell. 1991 Feb 8;64(3):649-65.