XSB0178 : micromolar calcium activated neutral protease 1 [Bos taurus]
[ CaMP Format ]
This entry is computationally expanded from SB0020
* Basic Information
| Organism | Bos taurus (cattle) |
| Protein Names | Calpain-1 catalytic subunit; 3.4.22.52; Calpain-1 large subunit; Calcium-activated neutral proteinase 1; CANP 1; Calpain mu-type; muCANP; Micromolar-calpain; micromolar calcium activated neutral protease 1 |
| Gene Names | CAPN1 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 716 aa
Average Mass: 82.249 kDa
Monoisotopic Mass: 82.197 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
| --- cleavage 15 --- |
| Peptidase_C2 1. | 55 | 354 | 797.3 | 9.8e-237 |
| Calpain_III 1. | 365 | 524 | 371.3 | 1.8e-108 |
| efhand 1. | 548 | 575 | 0.1 | 20 |
| efhand 2. | 591 | 619 | 16.6 | 0.11 |
| efhand 3. | 621 | 649 | 15.4 | 0.23 |
| efhand 4. | 686 | 713 | 6.2 | 4.5 |
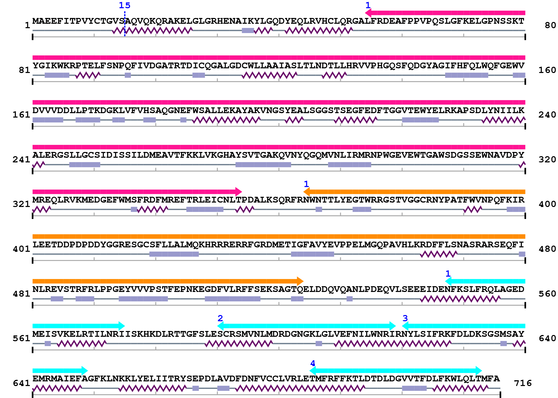
3. Sequence Information
Fasta Sequence: XSB0178.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] ITPVYCTGVS15-AQVQKQRAKE
Ser15  Ala
Ala
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| Ile6 | Thr7 | Pro8 | Val9 | Tyr10 | Cys11 | Thr12 | Gly13 | Val14 | Ser15 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Ala16 | Gln17 | Val18 | Gln19 | Lys20 | Gln21 | Arg22 | Ala23 | Lys24 | Glu25 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
MAEEFITPVYCTGVSAQVQKQRAKELGLGRHENAIKYLGQDYEQL
|
Summary
| # |
organism |
max score |
hits |
top seq |
| 1 |
Bos taurus |
94.40 |
6 |
micromolar calcium activated neutral protease 1 |
| 2 |
Macaca mulatta |
92.00 |
3 |
PREDICTED: calpain 1, large subunit isoform 4 |
| 3 |
Homo sapiens |
89.70 |
21 |
CAPN1 protein |
| 4 |
Pan troglodytes |
89.70 |
5 |
PREDICTED: hypothetical protein isoform 9 |
| 5 |
Canis familiaris |
89.70 |
4 |
PREDICTED: similar to Calpain-1 catalytic subunit |
| 6 |
synthetic construct |
89.70 |
1 |
Homo sapiens calpain 1, (mu/I) large subunit |
| 7 |
Pongo pygmaeus |
89.70 |
1 |
CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 8 |
N/A |
89.00 |
11 |
C |
| 9 |
Sus scrofa |
89.00 |
4 |
micromolar calcium-activated neutral protease 1 is |
| 10 |
Rattus norvegicus |
87.80 |
5 |
calpain 1 |
| 11 |
Mus musculus |
84.30 |
15 |
unnamed protein product |
| 12 |
Xenopus laevis |
72.40 |
7 |
MGC81785 protein |
| 13 |
Xenopus tropicalis |
71.60 |
3 |
CAPN1 protein |
| 14 |
Gallus gallus |
70.50 |
5 |
mu-calpain large subunit |
| 15 |
Danio rerio |
45.40 |
16 |
AF282675_1 calpain 1 |
| 16 |
Coturnix coturnix |
43.50 |
1 |
quail calpain |
| 17 |
Monodelphis domestica |
42.40 |
1 |
PREDICTED: similar to calpain 2 |
| 18 |
catalytic |
42.00 |
1 |
Calpain 2, large |
| 19 |
Stizostedion vitreum vitreum |
40.80 |
1 |
putative calpain-like protein |
| 20 |
Oncorhynchus mykiss |
39.30 |
3 |
calpain 1 catalytic subunit |
| 21 |
Tetraodon nigroviridis |
37.40 |
2 |
unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|
| Bos taurus |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|||||||||||||||#||||||||||||||||||||||||||||||
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|
| Macaca mulatta |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|||| ||||||||||#||||||||||||||||||||||||||||||
Sbjct 1 MAEEIITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|
| Homo sapiens |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|+|| ||||||||||#||||||||+|||||||||||||||||||||
Sbjct 1 MSEEIITPVYCTGVS#AQVQKQRARELGLGRHENAIKYLGQDYEQL 45
|
| Pan troglodytes |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|+|| ||||||||||#||||||||+|||||||||||||||||||||
Sbjct 1 MSEEIITPVYCTGVS#AQVQKQRARELGLGRHENAIKYLGQDYEQL 45
|
| Canis familiaris |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|||| ||||||||||#||||||||||||||||||||||+|||+|||
Sbjct 1 MAEEIITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYMGQDFEQL 45
|
| synthetic construct |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|+|| ||||||||||#||||||||+|||||||||||||||||||||
Sbjct 1 MSEEIITPVYCTGVS#AQVQKQRARELGLGRHENAIKYLGQDYEQL 45
|
| Pongo pygmaeus |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|+|| ||||||||||#||||||||+|||||||||||||||||||||
Sbjct 1 MSEEIITPVYCTGVS#AQVQKQRARELGLGRHENAIKYLGQDYEQL 45
|
| N/A |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|||| ||||||||||#||||| ||||||||||||||||||||||||
Sbjct 1 MAEEVITPVYCTGVS#AQVQKLRAKELGLGRHENAIKYLGQDYEQL 45
|
| Sus scrofa |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|||| ||||||||||#||||| ||||||||||||||||||||||||
Sbjct 1 MAEEVITPVYCTGVS#AQVQKLRAKELGLGRHENAIKYLGQDYEQL 45
|
| Rattus norvegicus |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|||| ||||||||||#||||||| |||||||||||||||||||| |
Sbjct 1 MAEELITPVYCTGVS#AQVQKQRDKELGLGRHENAIKYLGQDYENL 45
|
| Mus musculus |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
| || ||||||||||#|||||+| |||||||||||||||||||| |
Sbjct 1 MTEELITPVYCTGVS#AQVQKKRDKELGLGRHENAIKYLGQDYETL 45
|
| Xenopus laevis |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|||| | ||| ||+|#||+|||| |||+|+||||+|+|||||+++
Sbjct 1 MAEEVIVPVYATGIS#AQLQKQRDHELGVGKHENAVKFLGQDYDRI 45
|
| Xenopus tropicalis |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|||| | ||| +|+|#||+|||| +|||+|+||||+|+|||||+++
Sbjct 1 MAEEVIVPVYASGIS#AQLQKQRDQELGVGKHENAVKFLGQDYDRI 45
|
| Gallus gallus |
Query 1 MAEEFITPVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|||| + ||+|||||#||||+|||| ||||+|+||+++ |||| |
Sbjct 1 MAEEPVVPVFCTGVS#AQVQRQRAKALGLGQHQNAVRFRGQDYAAL 45
|
| Danio rerio |
Query 8 PVYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|+ ||++#|+++ | ++ |||++ ||+|+|||||| |
Sbjct 3 PICATGMA#ARLRSQWDRDAGLGQNHNAVKFLGQDYETL 40
|
| Coturnix coturnix |
Query 13 GVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|++#|++|+ | + |+| | ||+||| |||| |
Sbjct 6 GIA#ARLQRDRLRAEGVGEHNNAVKYLNQDYEAL 38
|
| Monodelphis domestica |
Query 12 TGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
+|++#|+| + | ||| || |+||| |||| |
Sbjct 2 SGIA#AKVVRDREAAAGLGTHEQAVKYLNQDYETL 35
|
| catalytic |
Query 13 GVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|++#|++ | | ||| || ||||| |||| |
Sbjct 32 GIA#AKLAKDREAAEGLGSHERAIKYLNQDYEAL 64
|
| Stizostedion vitreum vitreum |
Query 12 TGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
+|++#| +|++| || |+| |+||| |||| |
Sbjct 2 SGIA#ATLQRRREKEQGIGSCSQAVKYLNQDYEAL 35
|
| Oncorhynchus mykiss |
Query 9 VYCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
|| +|++#|+++ | ++ |||++ ||+|+ |||++ |
Sbjct 4 VYASGMA#AKLRSQWDRDEGLGQNHNAVKFQGQDFKSL 40
|
| Tetraodon nigroviridis |
Query 10 YCTGVS#AQVQKQRAKELGLGRHENAIKYLGQDYEQL 45
+ ||++# +++ | ++ |||++ |+|+|||||| |
Sbjct 7 HATGMA#YKLRSQWDRDEGLGQNHKAVKFLGQDYELL 42
|
* References
[PubMed ID: 11048924] Smith TP, Casas E, Rexroad CE 3rd, Kappes SM, Keele JW, Bovine CAPN1 maps to a region of BTA29 containing a quantitative trait locus for meat tenderness. J Anim Sci. 2000 Oct;78(10):2589-94.
 Ala
Ala

 Sequence conservation (by blast)
Sequence conservation (by blast)