XSB0193 : protein kinase C, beta isoform 2 [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0039
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Protein kinase C beta type; PKC-beta; PKC-B; 2.7.11.13; protein kinase C; beta isoform 2; beta 1 polypeptide |
| Gene Names | PRKCB; PKCB, PRKCB1; PKCB; PRKCB1; protein kinase C, beta |
| Gene Locus | 16p11.2; chromosome 16 |
| GO Function | Not available |
* Information From OMIM
Description: PRKCB1 is a member of the protein kinase C (PKC) gene family (see Coussens et al., 1986 and PRKCA; OMIM:176960).
Function: The 66-kD isoform of the growth factor adaptor SHC, p66(SHC) (OMIM:600560), translates oxidative damage into cell death by acting as a reactive oxygen species producer within mitochondria. Pinton et al. (2007) demonstrated that protein kinase C-beta, activated by oxidative conditions in the cell, induces phosphorylation of p66(SHC) and triggers mitochondrial accumulation of the protein after it is recognized by the prolyl isomerase PIN1 (OMIM:601052). Once imported, p66(SHC) causes alterations of mitochondrial calcium ion responses and 3-dimensional structure, thus causing apoptosis. Pinton et al. (2007) concluded that their data identified a signaling route that activates an apoptotic inducer shortening the life span.
* Structure Information
1. Primary Information
Length: 673 aa
Average Mass: 77.012 kDa
Monoisotopic Mass: 76.962 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| C1_1 1. | 37 | 89 | 80.1 | 8.2e-21 |
| C1_1 2. | 102 | 154 | 97.3 | 5.4e-26 |
| C2 1. | 173 | 260 | 126.7 | 7.5e-35 |
| --- cleavage 311 --- | ||||
| Pkinase 1. | 342 | 600 | 262.2 | 1.2e-75 |
| Pkinase_C 1. | 620 | 665 | 55.6 | 1.9e-13 |
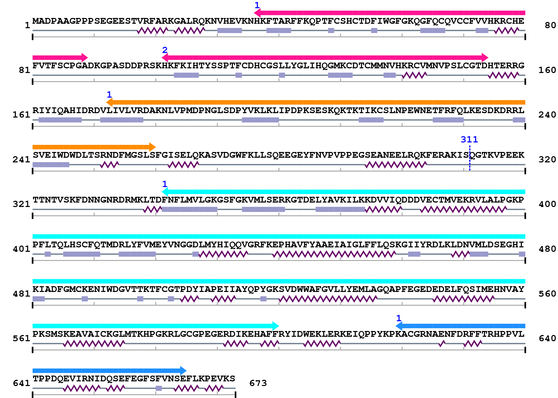
3. Sequence Information
Fasta Sequence: XSB0193.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 2I0E (X-ray; 260 A; A/B=321-661)
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] RQKFERAKIS311-QGTKVPEEKT
Ser311  Gln
Gln
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg302 | Gln303 | Lys304 | Phe305 | Glu306 | Arg307 | Ala308 | Lys309 | Ile310 | Ser311 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gln312 | Gly313 | Thr314 | Lys315 | Val316 | Pro317 | Glu318 | Glu319 | Lys320 | Thr321 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EGEYFNVPVPPEGSEANEELRQKFERAKISQGTKVPEEKTTNTVSKFDNNGNRDRMKLTD |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 125.00 | 9 | protein kinase C, beta isoform 2 |
| 2 | Macaca mulatta | 124.00 | 6 | PREDICTED: protein kinase C, beta 1 isoform 5 |
| 3 | Oryctolagus cuniculus | 120.00 | 3 | unnamed protein product |
| 4 | Canis familiaris | 119.00 | 2 | PREDICTED: similar to Protein kinase C, beta type |
| 5 | N/A | 118.00 | 10 | - |
| 6 | Rattus norvegicus | 118.00 | 4 | protein kinase C, beta |
| 7 | Mus musculus | 118.00 | 4 | protein kinase C, beta 1 |
| 8 | Bos taurus | 115.00 | 3 | protein kinase C, beta |
| 9 | Monodelphis domestica | 111.00 | 3 | PREDICTED: similar to Protein kinase C beta type ( |
| 10 | Gallus gallus | 105.00 | 3 | PREDICTED: similar to protein kinase C beta-1 |
| 11 | Latimeria chalumnae | 95.90 | 2 | protein kinase C beta 1 |
| 12 | Protopterus dolloi | 92.80 | 2 | protein kinase C beta 1 |
| 13 | Danio rerio | 89.70 | 7 | protein kinase C, beta 1 |
| 14 | Tetraodon nigroviridis | 89.00 | 3 | unnamed protein product |
| 15 | Scyliorhinus canicula | 83.20 | 2 | protein kinase C |
| 16 | Petromyzon marinus | 55.50 | 1 | protein kinase C |
| 17 | Takifugu rubripes | 53.90 | 1 | protein kinase C, alpha type |
| 18 | synthetic construct | 52.40 | 1 | protein kinase C alpha |
| 19 | Xenopus tropicalis | 50.10 | 1 | hypothetical protein LOC780209 |
| 20 | Xenopus laevis | 46.20 | 1 | protein kinase C, alpha |
| 21 | Myxine glutinosa | 44.70 | 1 | protein kinase C |
| 22 | Caenorhabditis briggsae | 39.70 | 1 | Hypothetical protein CBG16150 |
| 23 | Aplysia californica | 38.10 | 1 | KPC1_APLCA Calcium-dependent protein kinase C (APL |
| 24 | Aedes aegypti | 35.80 | 1 | protein kinase c |
| 25 | Apis mellifera | 34.30 | 1 | PREDICTED: similar to Protein C kinase 53E CG6622- |
| 26 | Trichomonas vaginalis G3 | 32.30 | 1 | SMC family, C-terminal domain containing protein |
| 27 | Anopheles gambiae str. PEST | 32.30 | 1 | ENSANGP00000009078 |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 |
| Macaca mulatta | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||| ||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 |
| Oryctolagus cuniculus | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||| ||||||||||||| #|||| ||||||||+|||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKTPEEKTTNTISKFDNNGNRDRMKLTD 341 |
| Canis familiaris | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||| ||||||||||||||#|||| ||||||||+|| ||||||||||||| Sbjct 274 EGEYFNVPVPPEGSEGNEELRQKFERAKIS#QGTKAPEEKTTNTISKLDNNGNRDRMKLTD 333 |
| N/A | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||| ||||||||||||| #|||| ||||| ||+|||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKAPEEKTANTISKFDNNGNRDRMKLTD 341 |
| Rattus norvegicus | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||| ||||||||||||| #|||| ||||| ||+|||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKAPEEKTANTISKFDNNGNRDRMKLTD 341 |
| Mus musculus | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||| ||||||||||||| #|||| ||||| ||+|||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#QGTKAPEEKTANTISKFDNNGNRDRMKLTD 341 |
| Bos taurus | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||||| ||||||||||||| # | | ||||||||+|||||||||||||||| Sbjct 282 EGEYFNVPVPPEGSEGNEELRQKFERAKIG#PGPKTPEEKTTNTISKFDNNGNRDRMKLTD 341 |
| Monodelphis domestica | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| | ||||||||||||| #||||| |||| | +|| ||||||||||||| Sbjct 279 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#QGTKVTEEKTANAISKIDNNGNRDRMKLTD 338 |
| Gallus gallus | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| | ||||||||||||| # |+| +||||| +|||||||+||||||+| Sbjct 281 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#TGSKAADEKTTNAISKFDNNGSRDRMKLSD 340 |
| Latimeria chalumnae | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| | |||||+||||||| # |+| |+| |+ ||| |||||||+||+| Sbjct 204 EGEYFNVPVPPEGEEGNEELRKKFERAKIG#PGSKATEDKVKNSSSKFGNNGNRDRIKLSD 263 |
| Protopterus dolloi | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| | ||||||||||||| # | + |+| | +||||||||||+||+| Sbjct 213 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#PGAEAAEDKIAN-AAKFDNNGNRDRVKLSD 271 |
| Danio rerio | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| | ||||||||||||| # | + ++| +||||+|||||||||+| Sbjct 281 EGEYFNVPVPPEGEEGNEELRQKFERAKI-#-GPSKTDGSSSNAISKFDSNGNRDRMKLSD 338 |
| Tetraodon nigroviridis | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||||||||||| | ||||||||||||| # | | + |+ | |+||+|||+||||| | Sbjct 218 EGEYFNVPVPPEGEEGNEELRQKFERAKIG#PG-KNTDGKSANAASRFDSNGNQDRMKLAD 276 |
| Scyliorhinus canicula | Query 282 EGEYFNVPVPPE-GSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLT 340 ||||+||||||| || |||||||||||+| # |+| ||| +++ || | ||||||||| Sbjct 202 EGEYYNVPVPPEEDSEGNEELRQKFERARIG#PGSKAAEEKRSSSWSKID-NGNRDRMKLA 260 Query 341 D 341 | Query 341 D 341 |
| Petromyzon marinus | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTK--VPEEKTTNTVSKFDNNGNRDRMKL 339 ||||+|||+ |+ + + |+|+|||+|++ # | +|+ |++| || | ||+| Sbjct 212 EGEYYNVPIAPDIEDGDTEMRRKFEKARLG#PSVKPRASDERRANSLSAILNNANVDRVKA 271 Query 340 TD 341 | Sbjct 272 DD 273 |
| Takifugu rubripes | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||+|||+| | + | |||||||+||+ #|| || | +|| ||++| | Sbjct 280 EGEYYNVPIP-EVDDVNLELRQKFEKAKLG#QGKKVITPSDHRRFSL--PSGNMDRVRLND 336 |
| synthetic construct | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS-#QGTKVPEEKTTNTVSKFDNNGNRDRMKLT 340 ||||+|||+| | | |||||||+||+ # | || + + + + | ||+||| Sbjct 282 EGEYYNVPIPEGDEEGNMELRQKFEKAKLGP#AGNKV----ISPSEDRKQPSNNLDRVKLT 337 Query 341 D 341 | Query 341 D 341 |
| Xenopus tropicalis | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS-#QGTKVPEEKTTNTVSKFDNNGNRDRMKLT 340 ||||+|||+| | + | |||||||+||+ # | || |+ + + | | ++|| Sbjct 286 EGEYYNVPIPEEEDDGNIELRQKFEKAKLGP#AGNKV--ISPTDERRPYVPSNNIDSVRLT 343 Query 341 D 341 | Query 341 D 341 |
| Xenopus laevis | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS-#QGTKVPEEKTTNTVSKFDNNGNRDRMKLT 340 ||||+|||+| | + | |||||||+||+ # | || | + + | | ++|| Sbjct 286 EGEYYNVPIP-EADDGNLELRQKFEKAKLGP#AGNKV--INPTGERRPYIPSNNIDSIRLT 342 Query 341 D 341 | Query 341 D 341 |
| Myxine glutinosa | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||+|||| ||| | ||||+ +|++|+#+ | + + | + ||+ | | Sbjct 203 EGEYYNVPVAPEGEE-GLELRQRLQRSQIA#RSDKNKQSPAKDQPSDSASLSGLDRVNLED 261 |
| Caenorhabditis briggsae | Query 282 EGEYFNVPVPPEGSEANEELRQKF-ERAKIS#QGTKV--PEEKTTNTVSKFDNNGNRDRMK 338 |||++|+ + || | |++|+| + |||#+ + | | | | | ||| +| Sbjct 352 EGEFYNINITPEYDEDMEKVRKKMNDNFKIS#RDSSAGKPRESTPRATSTSLTNTNRDVIK 411 Query 339 LTD 341 +| Sbjct 412 ASD 414 |
| Aplysia californica | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFD 329 |||++ ||| + +|+ +|++ | |+ ||# + || +|| | Sbjct 266 EGEFYGVPVTDDITESIQEIKSKMHRSSIS#SEKRYPEPDKVQNMSKQD 313 |
| Aedes aegypti | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNGNRDRMKLTD 341 ||||+||||| ||++ +|+ + + ||#+ | +| | | +| ++ || Sbjct 230 EGEYYNVPVPEEGTDL-VQLKSQMRKTSIS#KKAPVLCDKDVP-----HNMGKKDVIRATD 283 |
| Apis mellifera | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKF--DNNGNRDRMKL 339 ||||+||||| || + ||+ | | |# +||| | | | | | ++ Sbjct 299 EGEYYNVPVPEEGVDL-AELKMKPSPQKTS#-----VTKKTTTTQDKEVPHNMGKSDLIRA 352 Query 340 TD 341 +| Sbjct 353 SD 354 |
| Trichomonas vaginalis G3 | Query 293 EGSEANEELRQKFERAKIS#QGTKVPEEKTTNTVSKFDNNG-NRDRMKLTD 341 | + ||+| |+ |+|| # |+ |||| | | + | + || | Sbjct 436 ENKKLNEKLNQELEKAKSD#LSQKIEEEKTLNGKSDIEEKTLNSLKEKLED 485 |
| Anopheles gambiae str. PEST | Query 282 EGEYFNVPVPPEGSEANEELRQKFERAKIS#QGTKVP 317 ||||+||||| ||++ ++ | + | |# |+| Sbjct 222 EGEYYNVPVPEEGADL---VQLKSQMRKTS#ITKKIP 254 |
* References
[PubMed ID: 19168795] Holler C, Pinon JD, Denk U, Heyder C, Hofbauer S, Greil R, Egle A, PKCbeta is essential for the development of chronic lymphocytic leukemia in the TCL1 transgenic mouse model: validation of PKCbeta as a therapeutic target in chronic lymphocytic leukemia. Blood. 2009 Mar 19;113(12):2791-4. Epub 2009 Jan 23.
[PubMed ID: 19091746] ... Gould CM, Kannan N, Taylor SS, Newton AC, The chaperones Hsp90 and Cdc37 mediate the maturation and stabilization of protein kinase C through a conserved PXXP motif in the C-terminal tail. J Biol Chem. 2009 Feb 20;284(8):4921-35. Epub 2008 Dec 17.
[PubMed ID: 19112601] ... Guo K, Li Y, Kang X, Sun L, Cui J, Gao D, Liu Y, Role of PKCbeta in hepatocellular carcinoma cells migration and invasion in vitro: a potential therapeutic target. Clin Exp Metastasis. 2009;26(3):189-95. Epub 2008 Dec 27.
[PubMed ID: 18981301] ... Osto E, Kouroedov A, Mocharla P, Akhmedov A, Besler C, Rohrer L, von Eckardstein A, Iliceto S, Volpe M, Luscher TF, Cosentino F, Inhibition of protein kinase Cbeta prevents foam cell formation by reducing scavenger receptor A expression in human macrophages. Circulation. 2008 Nov 18;118(21):2174-82. Epub 2008 Nov 3.
[PubMed ID: 18937679] ... Pettigrew KA, McKnight AJ, Martin RJ, Patterson CC, Kilner J, Sadlier D, Maxwell AP, Savage DA, No support for association of protein kinase C, beta 1 (PRKCB1) gene promoter polymorphisms c.-1504C>T and c.-546C>G with diabetic nephropathy in Type 1 diabetes. Diabet Med. 2008 Sep;25(9):1127-9.
[PubMed ID: 7961692] ... Orr JW, Newton AC, Requirement for negative charge on "activation loop" of protein kinase C. J Biol Chem. 1994 Nov 4;269(44):27715-8.
[PubMed ID: 8034726] ... Zhang J, Wang L, Schwartz J, Bond RW, Bishop WR, Phosphorylation of Thr642 is an early event in the processing of newly synthesized protein kinase C beta 1 and is essential for its activation. J Biol Chem. 1994 Jul 29;269(30):19578-84.
[PubMed ID: 8327493] ... Zhang J, Wang L, Petrin J, Bishop WR, Bond RW, Characterization of site-specific mutants altered at protein kinase C beta 1 isozyme autophosphorylation sites. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6130-4.
[PubMed ID: 1400396] ... Obeid LM, Blobe GC, Karolak LA, Hannun YA, Cloning and characterization of the major promoter of the human protein kinase C beta gene. Regulation by phorbol esters. J Biol Chem. 1992 Oct 15;267(29):20804-10.
[PubMed ID: 1324914] ... Goode N, Hughes K, Woodgett JR, Parker PJ, Differential regulation of glycogen synthase kinase-3 beta by protein kinase C isotypes. J Biol Chem. 1992 Aug 25;267(24):16878-82.
[PubMed ID: 1321150] ... Moss SJ, Doherty CA, Huganir RL, Identification of the cAMP-dependent protein kinase and protein kinase C phosphorylation sites within the major intracellular domains of the beta 1, gamma 2S, and gamma 2L subunits of the gamma-aminobutyric acid type A receptor. J Biol Chem. 1992 Jul 15;267(20):14470-6.
[PubMed ID: 1611098] ... Devalia V, Thomas NS, Roberts PJ, Jones HM, Linch DC, Down-regulation of human protein kinase C alpha is associated with terminal neutrophil differentiation. Blood. 1992 Jul 1;80(1):68-76.
[PubMed ID: 1556124] ... Niino YS, Ohno S, Suzuki K, Positive and negative regulation of the transcription of the human protein kinase C beta gene. J Biol Chem. 1992 Mar 25;267(9):6158-63.