XSB0244 : RecName: Full=Src substrate cortactin
[ CaMP Format ]
This entry is computationally expanded from SB0022
* Basic Information
| Organism | Mus musculus (house mouse) |
| Protein Names | Src substrate cortactin |
| Gene Names | Ems1 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
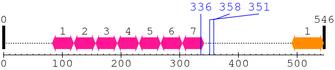
Length: 546 aa
Average Mass: 61.260 kDa
Monoisotopic Mass: 61.222 kDa
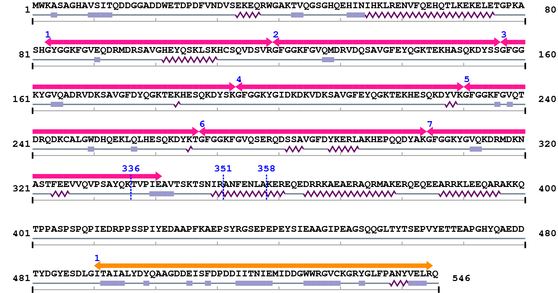
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| HS1_rep 1. | 83 | 119 | 67.0 | 7.3e-17 |
| HS1_rep 2. | 120 | 156 | 72.7 | 1.3e-18 |
| HS1_rep 3. | 157 | 193 | 74.7 | 3.4e-19 |
| HS1_rep 4. | 194 | 230 | 69.9 | 9.2e-18 |
| HS1_rep 5. | 231 | 267 | 68.5 | 2.5e-17 |
| HS1_rep 6. | 268 | 304 | 69.1 | 1.6e-17 |
| HS1_rep 7. | 305 | 341 | 26.6 | 0.0001 |
| --- cleavage 336 (inside HS1_rep 305..341) --- | ||||
| --- cleavage 358 --- | ||||
| --- cleavage 351 --- | ||||
| SH3_1 1. | 491 | 545 | 89.8 | 9.6e-24 |
3. Sequence Information
Fasta Sequence: XSB0244.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Known Structures in PDB: 2F9X (Model; -; A=1-546)
* Cleavage Information
3 [sites]
Cleavage sites (±10aa)
[Site 1] VVQVPSAYQK336-TVPIEAVTSK
Lys336  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Val327 | Val328 | Gln329 | Val330 | Pro331 | Ser332 | Ala333 | Tyr334 | Gln335 | Lys336 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr337 | Val338 | Pro339 | Ile340 | Glu341 | Ala342 | Val343 | Thr344 | Ser345 | Lys346 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| GGKYGVQKDRMDKNASTFEEVVQVPSAYQKTVPIEAVTSKTSNIRANFENLAKEREQEDR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 120.00 | 5 | cortactin |
| 2 | Rattus norvegicus | 120.00 | 4 | cortactin isoform B |
| 3 | N/A | 120.00 | 2 | S |
| 4 | mice, BALB/c 3T3 cells, Peptide, 546 aa | 120.00 | 1 | cortactin, p80/p85 |
| 5 | Homo sapiens | 113.00 | 10 | cortactin isoform a |
| 6 | synthetic construct | 113.00 | 3 | cortactin |
| 7 | Macaca mulatta | 113.00 | 2 | PREDICTED: similar to cortactin isoform a |
| 8 | Pongo pygmaeus | 111.00 | 1 | hypothetical protein |
| 9 | Canis familiaris | 108.00 | 5 | PREDICTED: similar to cortactin isoform a isoform |
| 10 | Bos taurus | 108.00 | 2 | cortactin |
| 11 | Gallus gallus | 94.40 | 2 | cortactin |
| 12 | Macaca fascicularis | 92.40 | 2 | unnamed protein product |
| 13 | Xenopus laevis | 90.10 | 3 | MGC80961 protein |
| 14 | Danio rerio | 84.30 | 10 | cortactin |
| 15 | Tetraodon nigroviridis | 79.70 | 3 | unnamed protein product |
| 16 | Strongylocentrotus purpuratus | 48.10 | 1 | SRC8 protein |
| 17 | Drosophila melanogaster | 47.00 | 2 | Cortactin |
| 18 | Drosophila pseudoobscura | 47.00 | 1 | GA17577-PA |
| 19 | Pan troglodytes | 43.50 | 5 | PREDICTED: cortactin isoform 3 |
| 20 | Tribolium castaneum | 42.40 | 1 | PREDICTED: similar to Xaa-Pro dipeptidase (X-Pro d |
| 21 | Monodelphis domestica | 39.30 | 1 | PREDICTED: hypothetical protein |
| 22 | Suberites domuncula | 35.40 | 1 | cortactin |
| 23 | Anopheles gambiae str. PEST | 33.50 | 1 | ENSANGP00000029554 |
| 24 | Xenopus tropicalis | 33.10 | 1 | cortactin |
| 25 | Aedes aegypti | 32.30 | 1 | cortactin |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |
| Rattus norvegicus | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |
| N/A | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |
| mice, BALB/c 3T3 cells, Peptide, 546 aa | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |
| Homo sapiens | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||||||||||||||||||+| || |||||#|||+||||||||||||||||||||+||||| Sbjct 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366 |
| synthetic construct | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||||||||||||||||||+| || |||||#|||+||||||||||||||||||||+||||| Sbjct 270 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 329 |
| Macaca mulatta | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||||||||||||||||||+| || |||||#|||+||||||||||||||||||||+||||| Sbjct 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366 |
| Pongo pygmaeus | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||||||||||||||||||+| || |||||#|||+||| ||||||||||||||||+||||| Sbjct 270 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVASKTSNIRANFENLAKEKEQEDR 329 |
| Canis familiaris | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||||||||||||||||||+| || ||||#|||+||| |+||||||||||||||+||||| Sbjct 307 GGKYGVQKDRMDKNASTFEDVAQVAPAYQK#TVPVEAVNSRTSNIRANFENLAKEKEQEDR 366 |
| Bos taurus | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||||||||||||||||||+| | |+|||#|||+||| ||||||||||||||||+||||| Sbjct 307 GGKYGVQKDRMDKNASTFEDVAAVSSSYQK#TVPVEAVNSKTSNIRANFENLAKEKEQEDR 366 |
| Gallus gallus | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||||||||||||||+|||++ + | |||#| |+| | +|||+|||| ||||||+||||| Sbjct 316 GGKYGVQKDRMDKNAATFEDIEKPTSTYQK#TKPVERVANKTSSIRANLENLAKEKEQEDR 375 |
| Macaca fascicularis | Query 317 MDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||||||||+| || |||||#|||+||||||||||||||||||||+||||| Sbjct 1 MDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 50 |
| Xenopus laevis | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 ||||||||||||| |++||+| +| |+|||#| |+| || |+||||||||||++||||| Sbjct 305 GGKYGVQKDRMDKAAASFEDVEKVSSSYQK#TRPVEVEGSKASSIRANFENLAKDKEQEDR 364 |
| Danio rerio | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||||||||||||+| ||||| + +||||#| |+|| +| |+|+| |||+||++|+|++ Sbjct 306 GGKYGVQKDRMDKSAGTFEEVQKPSAAYQK#TRPVEAASSSASSIKARFENIAKQKEEEEQ 365 |
| Tetraodon nigroviridis | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||+||||||||| | ||||| + +|||#| |+|| | +|+| |||+||++| ||| Sbjct 315 GGKFGVQKDRMDKAAGTFEEVEKPTPSYQK#TKPVEAAGSNAGSIKARFENMAKQKEDEDR 374 |
| Strongylocentrotus purpuratus | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||+|| |+ | | | ++ | |+|+|#| | | |+| || +|+ |+| | Sbjct 270 GGKFGVDKNAQDATAGGFGDMQGVSSSYKK#TRPQPPAKSGAGNMRNKFEQMAQAGEEESR 329 |
| Drosophila melanogaster | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVT----SKTSNIRANFENLAKERE 362 |||+|||+|| ||+| ++ + | + # | + +| ||+|| |||||| | Sbjct 197 GGKFGVQEDRKDKSAVGWDHK-EAPQKHAS#QVDHKVKPVIEGAKPSNLRAKFENLAKNSE 255 Query 363 QEDR 366 +| | Sbjct 256 EESR 259 |
| Drosophila pseudoobscura | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVT----SKTSNIRANFENLAKERE 362 |||+|||+|| ||+| ++ + | + # | + +| ||+|| |||||| | Sbjct 195 GGKFGVQEDRKDKSAVGWDHK-EAPQKHAS#QVDHKVKPVIEGAKPSNLRAKFENLAKNSE 253 Query 363 QEDR 366 +| | Sbjct 254 EESR 257 |
| Pan troglodytes | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKT 347 |||||||||||||++ + + + | ||# |+ |+ + | Sbjct 307 GGKYGVQKDRMDKSSPSLGQTIVESPAMQK#GFPVAAIVTTT 347 |
| Tribolium castaneum | Query 307 GGKYGVQKDRMDKNASTF-EEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENL 357 |||+|+| ||+||+|+ | +| +| + | |# | + +| ++|| |||+ Sbjct 736 GGKFGLQTDRVDKSAANFNDEPAKVGTNYTK#VKP-DIGDAKPKDLRAKFENM 786 |
| Monodelphis domestica | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTS---KTSNIRANFENLAKEREQ 363 ||||||||||||| | | | |#+ + | | |++ | | + | Sbjct 270 GGKYGVQKDRMDKRGGTAEPQAQSGRPQTK#SSLLTLATGVPCSISGIKSAFSNHQTPQAQ 329 Query 364 ED 365 | Sbjct 330 SD 331 |
| Suberites domuncula | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLA 358 ||||||++ | +| ++++ | | ++ # + +| +||+ |||+| Sbjct 271 GGKYGVEEGNQDSSAGGYDDMQAVKSDHRT#ERGVS--KGQTGSIRSKFENMA 320 |
| Anopheles gambiae str. PEST | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAV------TSKTSNIRANFENLAKE 360 |||+||| || ||+| ++ |+ | ++ # + + | ++| ||+|| ||| | Sbjct 195 GGKFGVQTDRKDKSAFGWDH-VEKPQMHES#QLDHKIVIKPDIGSAKPSNLRAKFENFAAT 253 Query 361 REQEDR 366 |+| | Sbjct 254 AEEEAR 259 Query 307 GGKYGVQKDRMDKNASTFEEVVQV 330 |||+||+||||||+| | + +| Sbjct 84 GGKFGVEKDRMDKSAVGHEHIEKV 107 Query 307 GGKYGVQKDRMDKNASTFEEVVQV 330 |||+||||||+||+| ++ | +| Sbjct 121 GGKFGVQKDRVDKSAHGWDHVEKV 144 |
| Xenopus tropicalis | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 ||++||| ||+|| || | |+ |||+|#| |+||+|| |+|+ |||+|+ |+|++ Sbjct 268 GGRFGVQTDRVDKCASGFGEIETPSSAYEK#TQPLEAMTSGAHNLRSRFENMARTAEEENK 327 Query 307 GGKYGVQKDRMDKNASTFEEVVQV 330 |||||||||| ||+| |+ +| Sbjct 120 GGKYGVQKDRTDKSAVGFDYKAKV 143 Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQE 364 |||||||||| ||+| ++ | ++# | ++ | | | + |+|+ + Sbjct 194 GGKYGVQKDRQDKSAHSW-------SHKEE#LQPHQSQTDYTQGFGGKF-GVQKDRQDK 243 |
| Aedes aegypti | Query 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366 |||+||| |||||+| |+| ++ + | |#| | + ++| ||||| ||| | |+| | Sbjct 231 GGKFGVQSDRMDKSALGFQEQDKIGTNYTK#TKP-DIGSAKPSNIRARFENFAMAAEEETR 289 Query 307 GGKYGVQKDRMDKNASTFEEVVQV 330 |||+||| ||+|| |+|+| +| Sbjct 120 GGKFGVQSDRVDKCAATWEHKEKV 143 |
[Site 2] NIRANFENLA358-KEREQEDRRK
Ala358  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn349 | Ile350 | Arg351 | Ala352 | Asn353 | Phe354 | Glu355 | Asn356 | Leu357 | Ala358 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys359 | Glu360 | Arg361 | Glu362 | Gln363 | Glu364 | Asp365 | Arg366 | Arg367 | Lys368 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| QVPSAYQKTVPIEAVTSKTSNIRANFENLAKEREQEDRRKAEAERAQRMAKERQEQEEAR |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 117.00 | 25 | cortactin |
| 2 | N/A | 117.00 | 4 | S |
| 3 | mice, BALB/c 3T3 cells, Peptide, 546 aa | 117.00 | 1 | cortactin, p80/p85 |
| 4 | Rattus norvegicus | 115.00 | 7 | cortactin isoform B |
| 5 | Homo sapiens | 112.00 | 44 | SRC8_HUMAN Src substrate cortactin (Amplaxin) (Onc |
| 6 | Macaca mulatta | 112.00 | 9 | PREDICTED: similar to cortactin isoform a |
| 7 | Macaca fascicularis | 112.00 | 3 | unnamed protein product |
| 8 | synthetic construct | 112.00 | 3 | cortactin |
| 9 | Pongo pygmaeus | 110.00 | 1 | hypothetical protein |
| 10 | Canis familiaris | 106.00 | 15 | PREDICTED: similar to cortactin isoform a isoform |
| 11 | Bos taurus | 103.00 | 5 | cortactin |
| 12 | Gallus gallus | 94.40 | 2 | cortactin |
| 13 | Xenopus laevis | 87.80 | 4 | cortactin |
| 14 | Tetraodon nigroviridis | 75.90 | 6 | unnamed protein product |
| 15 | Danio rerio | 75.50 | 17 | cortactin |
| 16 | Xenopus tropicalis | 65.90 | 1 | cortactin |
| 17 | Strongylocentrotus purpuratus | 53.10 | 4 | SRC8 protein |
| 18 | Aedes aegypti | 50.80 | 1 | cortactin |
| 19 | Drosophila melanogaster | 50.40 | 5 | Cortactin CG3637-PA |
| 20 | Drosophila pseudoobscura | 49.30 | 1 | GA17577-PA |
| 21 | Pan troglodytes | 48.10 | 18 | PREDICTED: hypothetical protein isoform 1 |
| 22 | Anopheles gambiae str. PEST | 42.40 | 2 | ENSANGP00000029554 |
| 23 | Parvibaculum lavamentivorans DS-1 | 41.20 | 1 | translation initiation factor IF-2 |
| 24 | Magnaporthe grisea 70-15 | 40.80 | 3 | hypothetical protein MGCH7_ch7g791 |
| 25 | Oryza sativa (japonica cultivar-group) | 37.70 | 1 | Os02g0602300 |
| 26 | Reinekea sp. MED297 | 37.00 | 1 | hypothetical protein MED297_01825 |
| 27 | Myxococcus xanthus DK 1622 | 36.60 | 1 | hypothetical protein MXAN_5743 |
| 28 | Trichomonas vaginalis G3 | 36.20 | 6 | eukaryotic initiation factor, putative |
| 29 | Aspergillus nidulans FGSC A4 | 36.20 | 3 | hypothetical protein AN7123.2 |
| 30 | Oryctolagus cuniculus | 36.20 | 2 | drebrin 1 isoform a |
| 31 | Felis catus | 36.20 | 1 | drebrin |
| 32 | Monodelphis domestica | 35.80 | 3 | PREDICTED: similar to myeloid/lymphoid or mixed-li |
| 33 | Entamoeba histolytica HM-1:IMSS | 35.80 | 2 | leucine rich repeat protein |
| 34 | Lodderomyces elongisporus NRRL YB-4239 | 35.80 | 1 | hypothetical protein LELG_02889 |
| 35 | Babesia bovis | 35.80 | 1 | anonymous antigen-2 |
| 36 | Bacillus cereus ATCC 14579 | 35.40 | 1 | hypothetical protein BC0683 |
| 37 | Coccidioides immitis RS | 35.40 | 1 | hypothetical protein CIMG_00823 |
| 38 | Myxococcus xanthus | 35.40 | 1 | unknown |
| 39 | Caenorhabditis elegans | 35.00 | 4 | DAF-12 Interacting Protein family member (din-1) |
| 40 | Dictyostelium discoideum AX4 | 35.00 | 3 | histone H3 domain-containing protein |
| 41 | Caenorhabditis briggsae | 35.00 | 3 | Hypothetical protein CBG04933 |
| 42 | Trypanosoma brucei TREU927 | 35.00 | 1 | hypothetical protein Tb11.01.7450 |
| 43 | Candida glabrata | 35.00 | 1 | unnamed protein product |
| 44 | Tetrahymena thermophila SB210 | 35.00 | 1 | hypothetical protein TTHERM_00043920 |
| 45 | Neosartorya fischeri NRRL 181 | 35.00 | 1 | mitochondrial translation initiation factor IF-2, |
| 46 | Candida albicans SC5314 | 35.00 | 1 | putative Sir2 family histone deacetylase |
| 47 | Trypanosoma cruzi strain CL Brener | 34.70 | 2 | hypothetical protein |
| 48 | Arabidopsis thaliana | 34.70 | 2 | unknown protein |
| 49 | Paramecium tetraurelia | 34.70 | 2 | hypothetical protein |
| 50 | Neurospora crassa OR74A | 34.70 | 1 | hypothetical protein |
| 51 | Stappia aggregata IAM 12614 | 34.70 | 1 | translation initiation factor IF-2 |
| 52 | Nitrobacter sp. Nb-311A | 34.70 | 1 | translation initiation factor IF-2 |
| 53 | Nitrobacter winogradskyi Nb-255 | 34.70 | 1 | translation initiation factor IF-2 |
| 54 | Aspergillus oryzae | 34.70 | 1 | unnamed protein product |
| 55 | Giardia lamblia ATCC 50803 | 34.70 | 1 | hypothetical protein GLP_384_18902_20299 |
| 56 | Sus scrofa | 34.70 | 1 | myosin VI |
| 57 | Aspergillus fumigatus Af293 | 34.30 | 2 | histone deacetylase complex subunit (Hos4), putati |
| 58 | Nitrobacter hamburgensis X14 | 34.30 | 1 | translation initiation factor IF-2 |
| 59 | Leishmania infantum JPCM5 | 34.30 | 1 | hypothetical protein LinJ30.2420 |
| 60 | Aspergillus clavatus NRRL 1 | 33.10 | 1 | ubiquitin-protein ligase (Tom1), putative |
| 61 | Apis mellifera | 32.70 | 1 | PREDICTED: similar to CG18497-PA, isoform A, parti |
| 62 | Borrelia burgdorferi B31 | 32.70 | 1 | ErpB |
| 63 | Alkalilimnicola ehrlichei MLHE-1 | 32.70 | 1 | TonB family protein |
| 64 | Borrelia burgdorferi | 32.70 | 1 | ErpB |
| 65 | Neurospora crassa | 32.70 | 1 | related to putative cytoplasmic structural protein |
| 66 | Coprinopsis cinerea okayama7#130 | 32.30 | 1 | predicted protein |
| 67 | Shewanella woodyi ATCC 51908 | 32.30 | 1 | TolA |
| 68 | Dichelobacter nodosus VCS1703A | 32.30 | 1 | TolA protein |
| 69 | Cucurbita maxima | 32.30 | 1 | preproMP73 |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| N/A | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| mice, BALB/c 3T3 cells, Peptide, 546 aa | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| Rattus norvegicus | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 ||||||||||||||||||||||||||||||#||||||||||||||||||||+||||||||| Sbjct 292 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAQERQEQEEAR 351 |
| Homo sapiens | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||||||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| Macaca mulatta | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||||||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 329 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 388 |
| Macaca fascicularis | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||||||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 13 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 72 |
| synthetic construct | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||||||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 292 QVSSAYQKTVPVEAVTSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 351 |
| Pongo pygmaeus | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 || ||||||||+||| ||||||||||||||#||+||||||||||||||||||||||||||| Sbjct 292 QVSSAYQKTVPVEAVASKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAKERQEQEEAR 351 |
| Canis familiaris | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 || |||||||+||| |+||||||||||||#||+|||||||||||+||||||||||||||| Sbjct 329 QVAPAYQKTVPVEAVNSRTSNIRANFENLA#KEKEQEDRRKAEAEKAQRMAKERQEQEEAR 388 |
| Bos taurus | Query 330 VPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | |+||||||+||| ||||||||||||||#||+|||||||||||||||||+|||||| || Sbjct 330 VSSSYQKTVPVEAVNSKTSNIRANFENLA#KEKEQEDRRKAEAERAQRMAQERQEQEAAR 388 |
| Gallus gallus | Query 332 SAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | |||| |+| | +|||+|||| ||||#||+|||||||||||||||||+|+||||||| Sbjct 341 STYQKTKPVERVANKTSSIRANLENLA#KEKEQEDRRKAEAERAQRMAREKQEQEEAR 397 |
| Xenopus laevis | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 +| |+|||| |+| || |+|||||||||#|++|||||+|||||||||+ +||+|||+|| Sbjct 327 KVSSSYQKTRPVEVEGSKASSIRANFENLA#KDKEQEDRKKAEAERAQRLERERREQEQAR 386 |
| Tetraodon nigroviridis | Query 333 AYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 +|||| |+|| | +|+| |||+|#|++| ||||+|| || +| ||||||||||+ Sbjct 341 SYQKTKPVEAAGSNAGSIKARFENMA#KQKEDEDRRRAEEERLRRQAKERQEQEEAQ 396 |
| Danio rerio | Query 329 QVPSA-YQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEA 387 | ||| |||| |+|| +| |+|+| |||+|#|++|+|++++ | ||++| |||+|||||| Sbjct 327 QKPSAAYQKTRPVEAASSSASSIKARFENIA#KQKEEEEQQRLEEERSRRQAKEKQEQEEA 386 Query 388 R 388 | Query 388 R 388 |
| Xenopus tropicalis | Query 332 SAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQE 385 |||+|| |+||+|| |+|+ |||+|#+ |+|+++||| || +| ||+||+| Sbjct 293 SAYEKTQPLEAMTSGAHNLRSRFENMA#RTAEEENKQKAEEERVRRQKKEKQEKE 346 |
| Strongylocentrotus purpuratus | Query 330 VPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQE 385 | |+|+|| | | |+| || +|#+ |+| ||||| || +| |+|++|+| Sbjct 293 VSSSYKKTRPQPPAKSGAGNMRNKFEQMA#QAGEEESRRKAEEERGRRQAREQKEKE 348 |
| Aedes aegypti | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEA 387 ++ + | || | + ++| ||||| ||| |# |+| |+||| ++ |+ |+++++||| Sbjct 253 KIGTNYTKTKP-DIGSAKPSNIRARFENFA#MAAEEETRKKAEEQKRLRLEKDKKDREEA 310 |
| Drosophila melanogaster | Query 345 SKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEA 387 +| ||+|| |||||#| |+| |++|| ++ | ||+++++||| Sbjct 238 AKPSNLRAKFENLA#KNSEEESRKRAEEQKRLREAKDKRDREEA 280 |
| Drosophila pseudoobscura | Query 345 SKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEA 387 +| ||+|| |||||#| |+| |+++| ++ | ||+++++||| Sbjct 236 AKPSNLRAKFENLA#KNSEEESRKRSEEQKRLREAKDKRDREEA 278 |
| Pan troglodytes | Query 332 SAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQE 385 +||+|| |||| +| ++| ||++|#+|+ |++ | |+||++|+ +||++ Sbjct 261 TAYKKTTPIEAASSGARGLKAKFESMA#EEK----RKREEEEKAQQVARRQQERK 310 |
| Anopheles gambiae str. PEST | Query 344 TSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEA 387 ++| ||+|| ||| |# |+| |++|+ ++ | |+| ++||| Sbjct 237 SAKPSNLRAKFENFA#ATAEEEARKRADEQKRLREEKDRCDREEA 280 |
| Parvibaculum lavamentivorans DS-1 | Query 331 PSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKE--RQEQEEA 387 || + | + +| + | || + |#+||| ||||+|| | |+| |+| |+| | | Sbjct 122 PSRSRSGVVLRTLTEEEKNARAQALDSA#REREGEDRRRAE-EEARRFAEEDARREAERA 179 |
| Magnaporthe grisea 70-15 | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQE 385 | | + + | | | | | || + #+ ||+| ||| || | + |+||+| | Sbjct 356 QAPGSSRPTAPPAPVGSPASGIRGPKMIMQ#ERREREQRRKDEATRREAEAREREELE 412 |
| Oryza sativa (japonica cultivar-group) | Query 336 KTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 || +|||+| + | + |+ +# | |+|+ + | | + +| ||+|| Sbjct 75 KTALMEAVSSGKLSAREHLESCS#NEEEEEEEEEEEEEEEEEEEEEEQEEEE 125 |
| Reinekea sp. MED297 | Query 355 ENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | #+|||+|+||+||||| ||+ +||| | | Sbjct 219 ERRR#REREEEERRQAEAER-QRLEQERQRQLE 249 |
| Myxococcus xanthus DK 1622 | Query 331 PSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQ-EDRRKAEAERAQRMAKERQEQEEAR 388 || | + + +| | + #++|++ |+||| | ||| +||+ ||| | Sbjct 531 PSTSNKRAQEDEARQREEELRKRREEVE#RQRKELEERRKQEEAAAQREQEERRRQEERR 589 |
| Trichomonas vaginalis G3 | Query 335 QKTVPIEAVTSKTSNIRANFENLA#KER-EQEDRRKAEAERAQRMAKERQEQEE 386 ++ ||| + + | |#+|+ |+| | ||| || +| |||+ |+|+ Sbjct 246 ERLAAIEAEKERQRRAKERAEQRA#REKAEREAREKAEKERLEREAKEKAEREK 298 |
| Aspergillus nidulans FGSC A4 | Query 345 SKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | + + +| | # ||+| + |+ +|| || |+||| | | | Sbjct 4 SSSPDYKALFLKAE#DERKQAEERQRQAEERQRQAEERQRQAEER 47 |
| Oryctolagus cuniculus | Query 330 VPSAYQKTVPIEAVTSKTSNIRANFENLA#KE---REQEDRRKAEAERA----QRMAKERQ 382 | + |||| || | | +| #|| |++|+|+|| || +|| +||| Sbjct 159 VGTTYQKTDA--AVEMKRINREQFWEQAK#KEEELRKEEERKKALDERLRFEQERMEQERQ 216 Query 383 EQEE 386 |||| Query 383 EQEE 386 |
| Felis catus | Query 330 VPSAYQKTVPIEAVTSKTSNIRANFENLA#KE---REQEDRRKAEAERA----QRMAKERQ 382 | + |||| || | | +| #|| |++|+|+|| || +|| +||| Sbjct 60 VGTTYQKTDA--AVEMKRINREQFWEQAK#KEEELRKEEERKKALDERLRFEQERMEQERQ 117 Query 383 EQEE 386 |||| Query 383 EQEE 386 |
| Monodelphis domestica | Query 359 KEREQEDRRKAEAERAQRMAKERQEQEE 386 + |++||||+ | |||+| |+|++ ||| Sbjct 1623 RARQEEDRRRQEEERAKRDAEEKRRQEE 1650 |
| Entamoeba histolytica HM-1:IMSS | Query 331 PSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | | || |+ | | #|++|+| |+| | ++ |+ |++||++|| | Sbjct 950 PRLEQNEVPKESEEEKRHR-----EEER#KQKEEEKRQKEEEDKKQKEAEKRQKEEEKR 1002 |
| Lodderomyces elongisporus NRRL YB-4239 | Query 341 EAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 || | + || | #+|+|+++ | | | + + ||++|+|| + Sbjct 879 EAFTIEISNPTKQQRELE#EEKEEKEEEKEEKEEKEELEKEKEEEEEEK 926 |
| Babesia bovis | Query 333 AYQKTVPIEAVTSKTSNIRANFEN-LA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | || + || + +| | | #||+| | +|+ | |+| |+| ||++||| Sbjct 455 ALQKRLAEEAAQKRLEQMRRQAEEKLR#KEKEAEMKRQEEEEQA-RIAAERKQQEE 508 |
| Bacillus cereus ATCC 14579 | Query 341 EAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 || + + ++ | | #|+| | +|||| || ||+|+|+++ |||| Sbjct 95 EAEKQRAAEVQRNAE-AE#KQRNAEAQRKAEEER-QRVAEEQRKAEEAR 140 |
| Coccidioides immitis RS | Query 331 PSAYQKTVPIE-AVTSKTSNIRAN-FENLA#KEREQEDRRKAE---AERAQRMAKERQEQE 385 | | || | || |+ +||++ | |# | |+ |+ | |+ +|| ||+||+| Sbjct 304 PKAKAKTTPASFAVESRARDIRSDRFSAAA#SARSQQRERQTEMIAAQLEERMRKEKQEEE 363 |
| Myxococcus xanthus | Query 331 PSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQ-EDRRKAEAERAQRMAKERQEQEEAR 388 || | + + +| | + #++|++ |+||| | ||| +||+ ||| | Sbjct 531 PSTSNKRAQEDEARQREEELRKRREEVE#RQRKELEERRKQEEAAAQREQEERRRQEERR 589 Query 329 QVPSAYQKTVPIE------AVTSK-----TSNIRANFENLA#KEREQEDR-RKAEAERAQR 376 + |+| ++ +|+ |+ || ||| || |+ |#++||+| | |+ | || ++ Sbjct 504 EAPAAEKEPLPMPPPPAQTALASKQKEPSTSNKRAQ-EDEA#RQREEELRKRREEVERQRK 562 Query 377 MAKERQEQEEA 387 +||++|||| Sbjct 563 ELEERRKQEEA 573 |
| Caenorhabditis elegans | Query 344 TSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | || +| |++ #|||+++++ | | || | +|+| +| | Sbjct 758 TPSTSTHQAMFKDKE#KERKKKEKEKEEREREARREMKRKETKEER 802 |
| Dictyostelium discoideum AX4 | Query 329 QVPSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | | +|| || + |+ + | + #+| |+|+ + | | + +| +|+|| Sbjct 203 QTPKIQRKTSQIEEESDDDSSSQNNSSDQE#EEEEEEEEEEEEEEEEEEEEEEEEEEEE 260 |
| Caenorhabditis briggsae | Query 350 IRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEA 387 +| ++ #| ||||| || + | |+| +|||+| || Sbjct 188 LRKREQDEI#KRREQEDIRKRDQEDARRRDQERQKQYEA 225 |
| Trypanosoma brucei TREU927 | Query 341 EAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | + + + + | |#|| |+|||++ + | +| |+|+|+|| | Sbjct 123 EVILKQKEEVEEDRELRA#KE-EEEDRKRKQREEEERKRKQREEEEEDR 169 |
| Candida glabrata | Query 355 ENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | #|++|+|++|| | | +| ||++|+|+ | Sbjct 121 EKKK#KQKEEEEKRKKELEEQERKKKEQEEEEKRR 154 |
| Tetrahymena thermophila SB210 | Query 339 PIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 ||++ +++ + + + | #||+|+ |+ | | | ++ ||++|| +|+ Sbjct 760 PIKSFSAEQAGGQTAMQKLK#KEKEENDKVKKEKEDQDKLKKEKEEQLKAQ 809 |
| Neosartorya fischeri NRRL 181 | Query 350 IRANFENLA#KEREQEDRR-----KAEAERAQRMAKERQEQEEAR 388 |+ | | #|+||+|+|| || || +| |+| +++|||| Sbjct 205 IQKQQEALR#KQREEEERRLAEEKAAEEERLRREAEEEKKREEAR 248 |
| Candida albicans SC5314 | Query 337 TVPIEAVTSKTS--NIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 ||| ||+| +| | + | | #+| |+|+ + | | + +| +|+|| Sbjct 61 TVPFNAVSSSSSSDNDQDNDEEEE#EEEEEEEEEEEEEEEEEEEEEEEEEEEE 112 |
| Trypanosoma cruzi strain CL Brener | Query 355 ENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | #|||| +|||+ | || +|| || +|+|| Sbjct 289 ERYR#KEREAKDRRE-ELERQERMQKEAKEREE 319 |
| Arabidopsis thaliana | Query 335 QKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 ++ + +| + |++ + + #|++++|||+| ||| | + |++++|| | Sbjct 1543 KEAMKLERAKQEQENLKK--QEIE#KKKKEEDRKKKEAEMAWKQEMEKKKKEEER 1594 |
| Paramecium tetraurelia | Query 336 KTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKER-QEQEEAR 388 | | + +| | +| | #||||+++| + ||| || +|| ++||| | Sbjct 4 KAPPPKKLTKKELKEKAEQEK--#KEREEQERIRKEAEEKQRQEEERLRKQEEER 55 |
| Neurospora crassa OR74A | Query 358 A#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 |#| ||++ || || ||+|+||| +|+ | Sbjct 112 A#KAREEKKRRLAEESRARRLAKEAEEERARR 142 |
| Stappia aggregata IAM 12614 | Query 340 IEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRM--AKERQEQEEAR 388 + +| + + || +|#| |++|||++ | | +| | +|| +|||| Sbjct 120 LRTLTKEEAAARAAALQMA#KVRDEEDRKRREEEDKRRAIEAAQRQAEEEAR 170 |
| Nitrobacter sp. Nb-311A | Query 338 VPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | + +| + ||+ |#| | +|+|| |||| |+| +|| || | Sbjct 95 VVLRKLTEDERSARASALAEA#KVRAEEERRIAEAEAARRNSKEGIEQAE 143 |
| Nitrobacter winogradskyi Nb-255 | Query 338 VPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | + +| + ||+ |#| | +|+|| |||| |+| +|| || | Sbjct 95 VVLRKLTEDERSARASALAEA#KVRAEEERRIAEAEAARRNSKEGIEQAE 143 |
| Aspergillus oryzae | Query 331 PSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQ 384 || +++| + |++ +|| |+#+| |+| +| ||||| || | + Sbjct 1934 PSLIPRSIPTRKGHPRVSSLAKHFEQLS#REFEKERQR----ERAQRAAKRSQSR 1983 |
| Giardia lamblia ATCC 50803 | Query 339 PIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | | | | + | ||#+|+ +++|++ | |+ +| |||+ ||+ | Sbjct 132 PRETVASPPQE---DLEALA#REKAEKERQRREQEQQERERKERERQEKER 178 |
| Sus scrofa | Query 332 SAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRM-----AKERQEQEE 386 || || | + |+ | #| ||++++|+ + | +|| || +||+|| Sbjct 912 SALQKKKQQEEEAERLRRIQEEMEKER#KRREEDEQRRRKEEEERRMKLEMEAKRKQEEEE 971 Query 387 AR 388 + Sbjct 972 RK 973 |
| Aspergillus fumigatus Af293 | Query 352 ANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | | ||# | |+ | | ||| |+|+|| |+|| + Sbjct 965 AEEERLA#AEAEKARLAKEEEERAARVARERAEEEERK 1001 |
| Nitrobacter hamburgensis X14 | Query 338 VPIEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | + +| ||+ |#| | +|+|| |||| |+| +|| || | Sbjct 92 VVLRTLTEDERTARASALAEA#KIRAEEERRIAEAEAARRNSKEGIEQAE 140 |
| Leishmania infantum JPCM5 | Query 353 NFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 +| | #++|+ |+||| ||+| | +++ |+|| + Sbjct 142 DFRKLR#EDRDMEERRKQRAEKAAREQQDKLEREEKK 177 |
| Aspergillus clavatus NRRL 1 | Query 351 RANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | | +|#|| |+++|++ | | | +|||+|| | Sbjct 2879 RERQERIA#KEEEEKERKRKEEEENARRQQERQQQEAER 2916 Query 357 LA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 + #+||++ + +|| || +|+||| +|+| | Sbjct 2865 ME#EERKRREEERAERERQERIAKEEEEKERKR 2896 |
| Apis mellifera | Query 351 RANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | | #+|||++++ +|| || +| +||+|+|| Sbjct 1402 RERQERER#REREEKEKERAERERREREERERREREE 1437 Query 355 ENLA#KEREQEDRR-KAEAERAQRMAKERQEQEEAR 388 | #+|||+ +|| + | || +| |+|+||+| || Sbjct 1421 ERER#REREERERREREERERKEREAREKQEKERAR 1455 Query 351 RANFENLA#KEREQEDRRKAEAERAQRMAKERQEQE 385 | | #+|||+ +| + | |||+| +||+|+| Sbjct 1397 RERLERER#QERERREREEKEKERAERERREREERE 1431 Query 351 RANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | | | #+||++ +||+ | + +| +||+|+|| Sbjct 1394 RRERERLE#RERQERERREREEKEKERAERERREREE 1429 Query 351 RANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | | #|| |+ +| + | || +| +||+|+|+ | Sbjct 1382 REKEERRE#KEEERRERERLERERQERERREREEKEKER 1419 Query 355 ENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | #||||+++||+ | +| ||| |+| | Sbjct 1452 ERAR#KEREEQERREKEERERRREEKERVERERKR 1485 |
| Borrelia burgdorferi B31 | Query 329 QVPSAYQKTVPIEAVTSKTSNIRA-NFENLA#KEREQEDRRKAEAERAQR-MAKERQEQEE 386 +| + +| | | | + ||#|+ |++ +|||| |+ +| +|||++|| Sbjct 121 KVKKVEESEAKVEGKEEKQENTEERNKQELA#KQEEEQQKRKAEQEKQKREEEQERQKREE 180 Query 355 ENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | #++||+| |||+||+ + |||+||| Sbjct 172 EQER#QKREEEQERKAKAEKEAKEKAERQKQEE 203 |
| Alkalilimnicola ehrlichei MLHE-1 | Query 351 RANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | | | #++|++|+||+ | || +||+ ||| Sbjct 179 RRRQEELE#RQRQEEERRRQEEAERQRQEEERRRQEE 214 Query 351 RANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | | #+ +|+|+||+ | || ++ +||| ||| | Sbjct 142 RLRREEEQ#RRQEEEERRRQEEERRRQEEEERQRQEEER 179 Query 355 ENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | #+ |++|+||+ | | || +||+ ||| Sbjct 153 EEEE#RRRQEEERRRQEEEERQRQEEERRRQEE 184 Query 351 RANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEEA 387 | | #++|++|+||+ | || +||+ |||| Sbjct 164 RRRQEEEE#RQRQEEERRRQEELERQRQEEERRRQEEA 200 |
| Borrelia burgdorferi | Query 329 QVPSAYQKTVPIEAVTSKTSNIRA-NFENLA#KEREQEDRRKAEAERAQR-MAKERQEQEE 386 +| + +| | | | + ||#|+ |++ +|||| |+ +| +|||++|| Sbjct 121 KVKKVEESEAKVEGKEEKQENTEERNKQELA#KQEEEQQKRKAEQEKQKREEEQERQKREE 180 Query 355 ENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | #++||+| |||+||+ + |||+||| Sbjct 172 EQER#QKREEEQERKAKAEKEAKEKAERQKQEE 203 |
| Neurospora crassa | Query 330 VPSAYQKTVPIEAV----------TSKTSNIRANFENLA#KER-EQEDRRKAEAERAQR-- 376 || +|+|| +| | + + +| | +|#+|+ |+ +| ||| |+|+| Sbjct 1534 VPPQHQETVDTKAQEDQLKAQKVETERLEHEKAEQERVA#REKAERAEREKAEREKAEREQ 1593 Query 377 ----MAKERQEQEEA 387 |+|+ |||+| Sbjct 1594 VALEKAREKAEQEKA 1608 Query 333 AYQKTVPIEAVTSKTSNIRANFENLA#K---EREQEDRRKAEAERAQRMAKERQEQEEAR 388 | |+ | | | +| | +|#+ |+|+ +||||+ |+|+| || |||| Sbjct 1719 AEQERVEREKAQEKALQEKAEQERIA#RKKAEQEKAERRKADLEKAER---ERVALEEAR 1774 |
| Coprinopsis cinerea okayama7#130 | Query 355 ENLA#KEREQEDRRKAEAERAQRMAKERQEQE 385 | #+|||+|+|++ | || +| |+||+|+| Sbjct 1434 ERER#EEREREERKREEREREERDAREREERE 1464 Query 351 RANFENLA#KER-EQEDRRKAEAERAQRMAKERQEQEE 386 | | | #||| |+|+||| | || + +|++|+|| Sbjct 1494 RLEQERLE#KERLEEEERRKEEEERQKAEEQEKKEREE 1530 Query 356 NLA#KEREQEDRRKAEAERAQRMAKERQEQEEA 387 | |#| +|+ | +|+ | |||+||| ++ | Sbjct 1181 NRA#KTQEENDELRAKTEEAQRLAKEASDKHRA 1212 |
| Shewanella woodyi ATCC 51908 | Query 341 EAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | | + | | |#|+ |+| +||||||| ++ +| + |+| Sbjct 150 EEAARKAAEKRKAEEAAA#KKAEEERKRKAEAERKRKAEEEARRQQE 195 Query 331 PSAYQKTVPIEAVTSKTSNIRANFENLA#KEREQEDR----RKAEAERAQRMAKERQEQEE 386 | ++|| | + + | | # |+++ +| |+ || | | |++ +|+|+ Sbjct 35 PQVQASAPAVQAVVVDQSKVAQHVERLK#AEKKEAERKEKARQDEANRQVREARKEREREQ 94 Query 387 AR 388 || Query 387 AR 388 |
| Dichelobacter nodosus VCS1703A | Query 345 SKTSNIRANFEN--LA#KEREQEDRRKAEAER-----AQRMAKERQEQEE 386 ++ + ||| | ||# |||+ + ||| || |+|+|||| |||| Sbjct 83 ARLAKIRAEEEEKRLA#AERERLAKEKAEQERRARLEAERLAKERAEQEE 131 Query 340 IEAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQE 385 +||+ +| | ||#| | +|+ ++ ||| +|+|||+ ||| Sbjct 70 LEAIAAKKEAIEQ--ARLA#KIRAEEEEKRLAAER-ERLAKEKAEQE 112 |
| Cucurbita maxima | Query 341 EAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 || + | | |#++||+|+ || | | |++ +| +|+|| Sbjct 562 EARKREEEEARKREEEEA#RKREEEEARKREKEEARKREEEEREREE 607 Query 355 ENLA#KEREQEDRRKAEAERAQRMAKERQEQEEAR 388 | #+|||+|+ || | || + +||+ +|| | Sbjct 500 EEEE#REREEEEARKREEEREREEEEERRREEEER 533 Query 341 EAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKERQ-EQEEAR 388 | + | | |#++||+|+| + | | +| +||+ |+|||| Sbjct 464 EEARKREEEEREREEEEA#RKREEEEREREEEEARKREEEEREREEEEAR 512 Query 351 RANFENLA#KEREQEDRRKAEAERAQRMAKERQ-EQEEAR 388 | | |#++||+|+| + | | +| +||+ |+|||| Sbjct 459 REREEEEA#RKREEEEREREEEEARKREEEEREREEEEAR 497 Query 355 ENLA#KEREQEDRRKAEAERAQRMAKERQEQEE 386 | #+|||+|+ || | | |++ +| +|+|| Sbjct 529 EEEE#REREEEEARKREEEEARKREEEEREREE 560 Query 341 EAVTSKTSNIRANFENLA#KEREQEDRRKAEAERAQRMAKE---RQEQEEAR 388 | + | | |#++||+|+ || | | |++ +| ++|+|||| Sbjct 546 EEARKREEEEREREEEEA#RKREEEEARKREEEEARKREEEEARKREKEEAR 596 |
[Site 3] AVTSKTSNIR351-ANFENLAKER
Arg351  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala342 | Val343 | Thr344 | Ser345 | Lys346 | Thr347 | Ser348 | Asn349 | Ile350 | Arg351 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala352 | Asn353 | Phe354 | Glu355 | Asn356 | Leu357 | Ala358 | Lys359 | Glu360 | Arg361 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| STFEEVVQVPSAYQKTVPIEAVTSKTSNIRANFENLAKEREQEDRRKAEAERAQRMAKER |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 117.00 | 7 | cortactin |
| 2 | N/A | 117.00 | 2 | S |
| 3 | mice, BALB/c 3T3 cells, Peptide, 546 aa | 117.00 | 1 | cortactin, p80/p85 |
| 4 | Rattus norvegicus | 115.00 | 6 | cortactin isoform B |
| 5 | Homo sapiens | 109.00 | 24 | SRC8_HUMAN Src substrate cortactin (Amplaxin) (Onc |
| 6 | synthetic construct | 109.00 | 3 | cortactin |
| 7 | Macaca fascicularis | 109.00 | 2 | unnamed protein product |
| 8 | Macaca mulatta | 109.00 | 2 | PREDICTED: similar to cortactin isoform a |
| 9 | Pongo pygmaeus | 107.00 | 1 | hypothetical protein |
| 10 | Canis familiaris | 103.00 | 6 | PREDICTED: similar to cortactin isoform a isoform |
| 11 | Bos taurus | 102.00 | 3 | cortactin |
| 12 | Gallus gallus | 88.20 | 2 | cortactin |
| 13 | Xenopus laevis | 84.00 | 4 | cortactin |
| 14 | Tetraodon nigroviridis | 72.00 | 5 | unnamed protein product |
| 15 | Danio rerio | 70.10 | 10 | cortactin |
| 16 | Xenopus tropicalis | 62.80 | 1 | cortactin |
| 17 | Strongylocentrotus purpuratus | 49.70 | 1 | SRC8 protein |
| 18 | Aedes aegypti | 47.00 | 1 | cortactin |
| 19 | Pan troglodytes | 45.80 | 9 | PREDICTED: hypothetical protein isoform 1 |
| 20 | Drosophila melanogaster | 43.50 | 2 | Cortactin CG3637-PA |
| 21 | Drosophila pseudoobscura | 42.40 | 1 | GA17577-PA |
| 22 | Parvibaculum lavamentivorans DS-1 | 39.30 | 1 | translation initiation factor IF-2 |
| 23 | Anopheles gambiae str. PEST | 37.00 | 2 | ENSANGP00000029554 |
| 24 | Magnaporthe grisea 70-15 | 37.00 | 2 | hypothetical protein MGCH7_ch7g791 |
| 25 | Monodelphis domestica | 35.00 | 1 | PREDICTED: similar to nexilin (F actin binding pro |
| 26 | Aspergillus fumigatus Af293 | 34.30 | 1 | 1-phosphatidylinositol-3-phosphate 5-kinase (Fab1) |
| 27 | Neosartorya fischeri NRRL 181 | 33.50 | 1 | 1-phosphatidylinositol-3-phosphate 5-kinase (Fab1) |
| 28 | Neurospora crassa | 33.50 | 1 | related to putative cytoplasmic structural protein |
| 29 | Neurospora crassa OR74A | 33.50 | 1 | hypothetical protein |
| 30 | Dictyostelium discoideum AX4 | 33.50 | 1 | hypothetical protein DDBDRAFT_0206214 |
| 31 | Arabidopsis thaliana | 33.10 | 2 | unnamed protein product |
| 32 | Aspergillus oryzae | 32.70 | 1 | unnamed protein product |
| 33 | Paramecium tetraurelia | 32.70 | 1 | hypothetical protein |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 |
| N/A | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 |
| mice, BALB/c 3T3 cells, Peptide, 546 aa | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 |
| Rattus norvegicus | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||+|| Sbjct 285 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAQER 344 |
| Homo sapiens | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |
| synthetic construct | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 285 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 344 |
| Macaca fascicularis | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 6 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 65 |
| Macaca mulatta | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+|||||||||||#|||||||||+|||||||||||||||||||| Sbjct 322 STFEDVTQVSSAYQKTVPVEAVTSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 381 |
| Pongo pygmaeus | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||+| || ||||||||+||| |||||||#|||||||||+|||||||||||||||||||| Sbjct 285 STFEDVTQVSSAYQKTVPVEAVASKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAKER 344 |
| Canis familiaris | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||+| || |||||||+||| |+|||||#|||||||||+|||||||||||+|||||||| Sbjct 322 STFEDVAQVAPAYQKTVPVEAVNSRTSNIR#ANFENLAKEKEQEDRRKAEAEKAQRMAKER 381 |
| Bos taurus | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||+| | |+||||||+||| |||||||#|||||||||+|||||||||||||||||+|| Sbjct 322 STFEDVAAVSSSYQKTVPVEAVNSKTSNIR#ANFENLAKEKEQEDRRKAEAERAQRMAQER 381 |
| Gallus gallus | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 +|||++ + | |||| |+| | +|||+||#|| ||||||+|||||||||||||||||+|+ Sbjct 331 ATFEDIEKPTSTYQKTKPVERVANKTSSIR#ANLENLAKEKEQEDRRKAEAERAQRMAREK 390 |
| Xenopus laevis | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ++||+| +| |+|||| |+| || |+||#||||||||++|||||+|||||||||+ +|| Sbjct 320 ASFEDVEKVSSSYQKTRPVEVEGSKASSIR#ANFENLAKDKEQEDRKKAEAERAQRLERER 379 |
| Tetraodon nigroviridis | Query 323 TFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||| + +|||| |+|| | +|+#| |||+||++| ||||+|| || +| |||| Sbjct 331 TFEEVEKPTPSYQKTKPVEAAGSNAGSIK#ARFENMAKQKEDEDRRRAEEERLRRQAKER 389 |
| Danio rerio | Query 323 TFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ||||| + +||||| |+|| +| |+|+#| |||+||++|+|++++ | ||++| |||+ Sbjct 322 TFEEVQKPSAAYQKTRPVEAASSSASSIK#ARFENIAKQKEEEEQQRLEEERSRRQAKEK 380 |
| Xenopus tropicalis | Query 322 STFEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 | | |+ |||+|| |+||+|| |+|#+ |||+|+ |+|+++||| || +| ||+ Sbjct 283 SGFGEIETPSSAYEKTQPLEAMTSGAHNLR#SRFENMARTAEEENKQKAEEERVRRQKKEK 342 |
| Strongylocentrotus purpuratus | Query 324 FEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 | ++ | |+|+|| | | |+|# || +|+ |+| ||||| || +| |+|+ Sbjct 287 FGDMQGVSSSYKKTRPQPPAKSGAGNMR#NKFEQMAQAGEEESRRKAEEERGRRQAREQ 344 |
| Aedes aegypti | Query 324 FEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 |+| ++ + | || | + ++| ||||#| ||| | |+| |+||| ++ |+ |++ Sbjct 248 FQEQDKIGTNYTKTKP-DIGSAKPSNIR#ARFENFAMAAEEETRKKAEEQKRLRLEKDK 304 |
| Pan troglodytes | Query 324 FEEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 | |+ +||+|| |||| +| ++#| ||++|+|+ |++ | |+||++|+ + Sbjct 253 FNEMEAPTTAYKKTTPIEAASSGARGLK#AKFESMAEEK----RKREEEEKAQQVARRQ 306 |
| Drosophila melanogaster | Query 345 SKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 +| ||+|#| |||||| |+| |++|| ++ | ||++ Sbjct 238 AKPSNLR#AKFENLAKNSEEESRKRAEEQKRLREAKDK 274 |
| Drosophila pseudoobscura | Query 345 SKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 +| ||+|#| |||||| |+| |+++| ++ | ||++ Sbjct 236 AKPSNLR#AKFENLAKNSEEESRKRSEEQKRLREAKDK 272 |
| Parvibaculum lavamentivorans DS-1 | Query 327 VVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKE 380 | + || + | + +| + | |#| + |+||| ||||+|| | |+| |+| Sbjct 118 VEKKPSRSRSGVVLRTLTEEEKNAR#AQALDSAREREGEDRRRAE-EEARRFAEE 170 |
| Anopheles gambiae str. PEST | Query 344 TSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 ++| ||+|#| ||| | |+| |++|+ ++ | |+| Sbjct 237 SAKPSNLR#AKFENFAATAEEEARKRADEQKRLREEKDR 274 |
| Magnaporthe grisea 70-15 | Query 329 QVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 | | + + | | | | | ||# + + ||+| ||| || | + |+|| Sbjct 356 QAPGSSRPTAPPAPVGSPASGIR#GPKMIMQERREREQRRKDEATRREAEARER 408 |
| Monodelphis domestica | Query 341 EAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAKER 381 || + |++#| || +|| ||+|++|+ | ++ || |+ Sbjct 429 EAPFTHKVNMK#ARFEQMAKAREEEEQRRIEQQKLLRMQFEQ 469 |
| Aspergillus fumigatus Af293 | Query 331 PSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAK 379 || ++|| ++ ||+ # +|| |++| |+| +| || || || Sbjct 1905 PSLIPRSVPSRKNNTRVSNLA#KHFEQLSREFEKERQR----ERQQRAAK 1949 |
| Neosartorya fischeri NRRL 181 | Query 331 PSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAK 379 || ++|| ++ ||+ # +|| |++| |+| +| || || || Sbjct 1904 PSLIPRSVPGRKNNTRVSNLA#KHFEQLSREFEKERQR----ERQQRAAK 1948 |
| Neurospora crassa | Query 325 EEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDR-RKAEAERAQRMAKER 381 || + || +|+|| +| + +# | | |+ +++| + +||||+| || Sbjct 1529 EETLYVPPQHQETVDTKAQEDQLKAQK#VETERLEHEKAEQERVAREKAERAEREKAER 1586 |
| Neurospora crassa OR74A | Query 325 EEVVQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDR-RKAEAERAQRMAKER 381 || + || +|+|| +| + +# | | |+ +++| + +||||+| || Sbjct 1452 EETLYVPPQHQETVDTKAQEDQLKAQK#VETERLEHEKAEQERVAREKAERAEREKAER 1509 |
| Dictyostelium discoideum AX4 | Query 328 VQVPSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMA 378 ++ |++ | +| + | ++ # | + |++|+| +|| | | |||+| Sbjct 374 IEEPTSAPKDDVVEVMDSWDNDDY#ETVEEIQKKKEEEAKRKEEEEEAQRLA 424 |
| Arabidopsis thaliana | Query 342 AVTSKTSNIR#ANFENLAKEREQEDRRKAEA---ERAQRMAKE 380 | | | +#|+ + + ||+|+|||++ || || +|+| + Sbjct 736 ARADKLRNFK#ADLQKMKKEKEEEDRKRIEALKIERQKRIASK 777 |
| Aspergillus oryzae | Query 331 PSAYQKTVPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRMAK 379 || +++| + |++ # +|| |++| |+| +| ||||| || Sbjct 1934 PSLIPRSIPTRKGHPRVSSLA#KHFEQLSREFEKERQR----ERAQRAAK 1978 |
| Paramecium tetraurelia | Query 325 EEVVQVPSAYQKT--VPIEAVTSKTSNIR#ANFENLAKEREQEDRRKAEAERAQRM 377 |+++++ |||| +|+ + + #| |++||| ++| ||+++|++ Sbjct 213 EQILEMAPAYQKDEDLPVSKKVKRQKKLM#AKKFETNKKQEQESQQKIEAQQSQQL 267 |
* References
[PubMed ID: 7516062] Miglarese MR, Mannion-Henderson J, Wu H, Parsons JT, Bender TP, The protein tyrosine kinase substrate cortactin is differentially expressed in murine B lymphoid tumors. Oncogene. 1994 Jul;9(7):1989-97.
[PubMed ID: 7693700] ... Zhan X, Hu X, Hampton B, Burgess WH, Friesel R, Maciag T, Murine cortactin is phosphorylated in response to fibroblast growth factor-1 on tyrosine residues late in the G1 phase of the BALB/c 3T3 cell cycle. J Biol Chem. 1993 Nov 15;268(32):24427-31.
[PubMed ID: 12913069] ... Hou P, Estrada L, Kinley AW, Parsons JT, Vojtek AB, Gorski JL, Fgd1, the Cdc42 GEF responsible for Faciogenital Dysplasia, directly interacts with cortactin and mAbp1 to modulate cell shape. Hum Mol Genet. 2003 Aug 15;12(16):1981-93.
[PubMed ID: 17114649] ... Munton RP, Tweedie-Cullen R, Livingstone-Zatchej M, Weinandy F, Waidelich M, Longo D, Gehrig P, Potthast F, Rutishauser D, Gerrits B, Panse C, Schlapbach R, Mansuy IM, Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol Cell Proteomics. 2007 Feb;6(2):283-93. Epub 2006 Nov 17.
[PubMed ID: 17242355] ... Villen J, Beausoleil SA, Gerber SA, Gygi SP, Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007 Jan 30;104(5):1488-93. Epub 2007 Jan 22.