XSB0250 : unnamed protein product [Mus musculus]
[ CaMP Format ]
This entry is computationally expanded from SB0063
* Basic Information
| Organism | Mus musculus (house mouse) |
| Protein Names | Integrin beta-1; Fibronectin receptor subunit beta; Integrin VLA-4 subunit beta |
| Gene Names | Itgb1 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
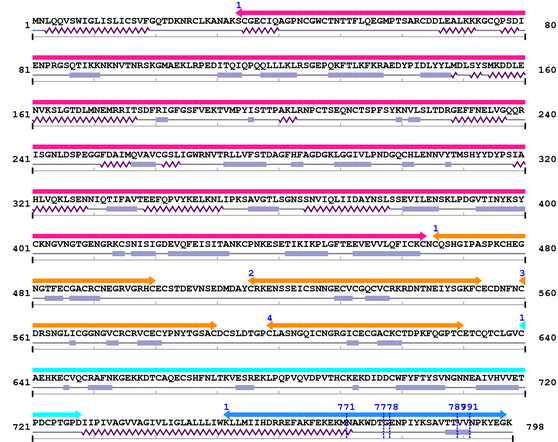
Length: 798 aa
Average Mass: 88.246 kDa
Monoisotopic Mass: 88.188 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Integrin_beta 1. | 34 | 464 | 1128.7 | 0 |
| EGF_2 1. | 466 | 500 | 11.0 | 0.16 |
| EGF_2 2. | 516 | 553 | 21.3 | 0.0041 |
| EGF_2 3. | 560 | 590 | 23.8 | 0.0007 |
| EGF_2 4. | 599 | 630 | 36.0 | 1.5e-07 |
| Integrin_B_tail 1. | 640 | 728 | 155.2 | 1.9e-43 |
| Integrin_b_cyt 1. | 752 | 797 | 112.3 | 1.6e-30 |
| --- cleavage 791 (inside Integrin_b_cyt 752..797) --- | ||||
| --- cleavage 789 (inside Integrin_b_cyt 752..797) --- | ||||
| --- cleavage 778 (inside Integrin_b_cyt 752..797) --- | ||||
| --- cleavage 771 (inside Integrin_b_cyt 752..797) --- | ||||
| --- cleavage 777 (inside Integrin_b_cyt 752..797) --- | ||||
3. Sequence Information
Fasta Sequence: XSB0250.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
5 [sites]
Cleavage sites (±10aa)
[Site 1] IYKSAVTTVV791-NPKYEGK
Val791  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ile782 | Tyr783 | Lys784 | Ser785 | Ala786 | Val787 | Thr788 | Thr789 | Val790 | Val791 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn792 | Pro793 | Lys794 | Tyr795 | Glu796 | Gly797 | Lys798 | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| EFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKYEGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 81.30 | 28 | I |
| 2 | Homo sapiens | 81.30 | 22 | integrin beta 1 isoform 1A precursor |
| 3 | Mus musculus | 81.30 | 14 | Fn receptor beta prechain |
| 4 | Bos taurus | 81.30 | 13 | integrin beta 1 |
| 5 | Pan troglodytes | 81.30 | 12 | PREDICTED: hypothetical protein |
| 6 | Rattus norvegicus | 81.30 | 10 | Integrin beta 1 (fibronectin receptor beta) |
| 7 | Gallus gallus | 81.30 | 6 | integrin, beta 1 (fibronectin receptor, beta polyp |
| 8 | Monodelphis domestica | 81.30 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 9 | Sus scrofa | 81.30 | 6 | integrin beta-1 subunit |
| 10 | Tetraodon nigroviridis | 81.30 | 5 | unnamed protein product |
| 11 | Xenopus laevis | 81.30 | 5 | integrin beta 1 |
| 12 | synthetic construct | 81.30 | 5 | hypothetical protein |
| 13 | Oryctolagus cuniculus | 81.30 | 2 | integrin beta 1 |
| 14 | Pongo pygmaeus | 81.30 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Xenopus tropicalis | 81.30 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 16 | Felis catus | 81.30 | 1 | integrin beta 1 |
| 17 | Danio rerio | 80.90 | 12 | integrin beta1 subunit-like protein 1 |
| 18 | Ictalurus punctatus | 79.30 | 2 | beta-1 integrin |
| 19 | Drosophila melanogaster | 65.50 | 1 | myospheroid CG1560-PA |
| 20 | Anopheles gambiae str. PEST | 63.90 | 1 | ENSANGP00000021373 |
| 21 | Anopheles gambiae | 63.90 | 1 | integrin beta subunit |
| 22 | Aedes aegypti | 63.90 | 1 | integrin beta subunit |
| 23 | Pseudoplusia includens | 62.40 | 1 | integrin beta 1 |
| 24 | Ostrinia furnacalis | 62.40 | 1 | integrin beta 1 precursor |
| 25 | Tribolium castaneum | 61.20 | 2 | PREDICTED: similar to CG1560-PA |
| 26 | Biomphalaria glabrata | 60.80 | 1 | beta integrin subunit |
| 27 | Canis familiaris | 60.10 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 28 | Crassostrea gigas | 59.30 | 1 | integrin beta cgh |
| 29 | Podocoryne carnea | 57.40 | 1 | AF308652_1 integrin beta chain |
| 30 | Caenorhabditis briggsae | 56.20 | 1 | Hypothetical protein CBG03601 |
| 31 | Caenorhabditis elegans | 53.90 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Macaca mulatta | 50.80 | 12 | PREDICTED: similar to integrin beta chain, beta 3 |
| 33 | rats, Peptide Partial, 723 aa | 50.80 | 1 | beta 3 integrin, GPIIIA |
| 34 | Canis lupus familiaris | 50.80 | 1 | integrin beta chain, beta 3 |
| 35 | Equus caballus | 50.80 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 36 | Mus sp. | 49.30 | 1 | beta 7 integrin |
| 37 | mice, Peptide Partial, 680 aa | 47.40 | 1 | beta 3 integrin, GPIIIA |
| 38 | Strongylocentrotus purpuratus | 47.00 | 5 | integrin beta-C subunit |
| 39 | Lytechinus variegatus | 47.00 | 1 | beta-C integrin subunit |
| 40 | Acropora millepora | 46.60 | 1 | integrin subunit betaCn1 |
| 41 | Oncorhynchus mykiss | 46.20 | 1 | CD18 protein |
| 42 | Ovis aries | 45.80 | 2 | antigen CD18 |
| 43 | Ovis canadensis | 45.80 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 44 | Bubalus bubalis | 45.80 | 1 | integrin beta-2 |
| 45 | Capra hircus | 45.80 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 46 | Sigmodon hispidus | 45.10 | 1 | AF445415_1 integrin beta-2 precursor |
| 47 | Schistosoma japonicum | 40.40 | 1 | SJCHGC06221 protein |
| 48 | Pacifastacus leniusculus | 39.70 | 1 | integrin |
| 49 | Apis mellifera | 39.30 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 50 | Halocynthia roretzi | 38.10 | 1 | integrin beta Hr2 precursor |
| 51 | Ophlitaspongia tenuis | 38.10 | 1 | integrin subunit betaPo1 |
| 52 | Cyprinus carpio | 35.80 | 2 | CD18 type 2 |
| 53 | Geodia cydonium | 34.30 | 1 | integrin beta subunit |
| 54 | Suberites domuncula | 33.50 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Homo sapiens | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Mus musculus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Bos taurus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Pan troglodytes | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Rattus norvegicus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Gallus gallus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Monodelphis domestica | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Sus scrofa | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Tetraodon nigroviridis | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Xenopus laevis | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| synthetic construct | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Oryctolagus cuniculus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Pongo pygmaeus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Xenopus tropicalis | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Felis catus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||||||||||||||||||#||||||| Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |
| Danio rerio | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |||||||||||||||||||||||||||||+#||||||| Sbjct 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVI#NPKYEGK 798 |
| Ictalurus punctatus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 ||||||||||||||| ||||||||||||||#||||||| Sbjct 771 EFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKYEGK 807 |
| Drosophila melanogaster | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |||+||||+|||||||||||||| | +| #|| | || Sbjct 810 EFARFEKERMNAKWDTGENPIYKQATSTFK#NPMYAGK 846 |
| Anopheles gambiae str. PEST | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |||+||||+| |||||||||||| | || #|| | || Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYAGK 837 |
| Anopheles gambiae | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |||+||||+| |||||||||||| | || #|| | || Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYAGK 837 |
| Aedes aegypti | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |||+||||+| |||||||||||| | || #|| | || Sbjct 799 EFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYAGK 835 |
| Pseudoplusia includens | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |||+||||+| |||||||||||| | +| #|| | || Sbjct 801 EFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYAGK 837 |
| Ostrinia furnacalis | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |||+||||+| |||||||||||| | +| #|| | || Sbjct 795 EFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYAGK 831 |
| Tribolium castaneum | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |||||||| | |+|||||||||| | +| #|| | || Sbjct 793 EFAKFEKEAMMARWDTGENPIYKQATSTFK#NPTYAGK 829 |
| Biomphalaria glabrata | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 ||||||||+ ||||||||||||| | +| #|| | Sbjct 752 EFAKFEKERQNAKWDTGENPIYKQATSTFK#NPTY 785 |
| Canis familiaris | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |||||||||||||||| ||||||| + #|| | | Sbjct 785 EFAKFEKEKMNAKWDTQENPIYKSPINNFK#NPNYGRK 821 |
| Crassostrea gigas | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 | |+|||| |||+|||||||||| | +| |#|| | Sbjct 764 ELARFEKEAMNARWDTGENPIYKQATSTFV#NPTY 797 |
| Podocoryne carnea | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |||||||++ | |||+||||||| | +| #|| | | Sbjct 745 EFAKFEKDRQNPKWDSGENPIYKKATSTFQ#NPMYGNK 781 |
| Caenorhabditis briggsae | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |+||| |++ ||||| |||||| | || #|| | || Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTFK#NPVYAGK 808 |
| Caenorhabditis elegans | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 |+| | |++ ||||| |||||| | || #|| | || Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTFK#NPVYAGK 807 |
| Macaca mulatta | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 797 ||||||+|+ ||||| ||+|| | +| #| | | Sbjct 752 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITYRG 787 |
| rats, Peptide Partial, 723 aa | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 797 ||||||+|+ ||||| ||+|| | +| #| | | Sbjct 687 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITYRG 722 |
| Canis lupus familiaris | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 797 ||||||+|+ ||||| ||+|| | +| #| | | Sbjct 748 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITYRG 783 |
| Equus caballus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 797 ||||||+|+ ||||| ||+|| | +| #| | | Sbjct 748 EFAKFEEERARAKWDTANNPLYKEATSTFT#NITYRG 783 |
| Mus sp. | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 797 |+ +||||+ | ||+||||+|| |#||+++| Sbjct 756 EYRRFEKEQQQLNWKQDNNPLYKSAITTTV#NPRFQG 791 |
| mice, Peptide Partial, 680 aa | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#N 792 ||||||+|+ ||||| ||+|| | +| #| Sbjct 648 EFAKFEEERARAKWDTANNPLYKEATSTFT#N 678 |
| Strongylocentrotus purpuratus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 || ||||+ || |+ ||||||| + + #|| | | Sbjct 770 EFQNFEKERANATWEGGENPIYKPSTSVFK#NPTYNIK 806 |
| Lytechinus variegatus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 || ||||+ || |+ ||||||| + + #|| | | Sbjct 770 EFQNFEKERANATWEGGENPIYKPSTSVFK#NPTYNIK 806 |
| Acropora millepora | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 797 |+ |||+|+|++|| +||+|++| || #|| | | Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTFE#NPTYAG 789 |
| Oncorhynchus mykiss | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEGK 798 || +|| |+ || |+||++ +| ||| #|| + |+ Sbjct 741 EFRRFENEQKKGKWSPGDNPLFMNATTTVA#NPTFTGE 777 |
| Ovis aries | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 735 EYHRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 |
| Ovis canadensis | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 735 EYHRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 |
| Bubalus bubalis | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 734 EYHRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 |
| Capra hircus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 735 EYHRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 |
| Sigmodon hispidus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |+ |||||+ ++|+ +||++||| |||+#|||+ Sbjct 734 EYRHFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 766 |
| Schistosoma japonicum | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYE 796 | | |+++ | +|+ ||||++| | |+#|| +| Sbjct 250 ELANFKQQGENMRWEMAENPIFESPTTNVL#NPTFE 284 |
| Pacifastacus leniusculus | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |+||||||+ |||+|||| || #|| + Sbjct 764 EYAKFEKERKLPSGKRAENPLYKSAKTTFQ#NPAF 797 |
| Apis mellifera | Query 762 EFAKFEKEKMNA-KWDTGENPIYKSAVTTVV#NPKY 795 || |||+|++ | ||+ +||+|| | +| #|| + Sbjct 779 EFKKFERERILANKWNRRDNPLYKEATSTFK#NPTF 813 |
| Halocynthia roretzi | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 797 |+ ||+||+ || ||||+| +| + #|| + | Sbjct 798 EYEKFKKEEDLRKWTKGENPVYVNASSKFD#NPMFGG 833 |
| Ophlitaspongia tenuis | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 | |||+| |||+ |||+|+|| #|| | Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATKDYQ#NPLY 836 |
| Cyprinus carpio | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKYEG 797 |+ +|||++ + | +| ||++++| ||| #|| + | Sbjct 731 EWKRFEKDRKHEK-TSGTNPLFQNATTTVQ#NPTFSG 765 |
| Geodia cydonium | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 | ||| | |+ |||+| || | #|| | Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTEHK#NPIY 876 |
| Suberites domuncula | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 | ||||| |+ +||+|+|| #|| | Sbjct 729 EVKKFEKELAKTKYPKHQNPLYRSAAKDYQ#NPIY 762 |
[Site 2] NPIYKSAVTT789-VVNPKYEGK
Thr789  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn780 | Pro781 | Ile782 | Tyr783 | Lys784 | Ser785 | Ala786 | Val787 | Thr788 | Thr789 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val790 | Val791 | Asn792 | Pro793 | Lys794 | Tyr795 | Glu796 | Gly797 | Lys798 | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| RREFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKYEGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 85.10 | 31 | I |
| 2 | Homo sapiens | 85.10 | 25 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 3 | Mus musculus | 85.10 | 14 | integrin beta 1 (fibronectin receptor beta) |
| 4 | Pan troglodytes | 85.10 | 14 | PREDICTED: hypothetical protein |
| 5 | Bos taurus | 85.10 | 13 | integrin beta 1 |
| 6 | Rattus norvegicus | 85.10 | 10 | Integrin beta 1 (fibronectin receptor beta) |
| 7 | Gallus gallus | 85.10 | 7 | integrin, beta 1 (fibronectin receptor, beta polyp |
| 8 | Monodelphis domestica | 85.10 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 9 | Sus scrofa | 85.10 | 6 | integrin beta-1 subunit |
| 10 | Xenopus laevis | 85.10 | 6 | integrin beta 1 |
| 11 | Tetraodon nigroviridis | 85.10 | 5 | unnamed protein product |
| 12 | synthetic construct | 85.10 | 5 | hypothetical protein |
| 13 | Oryctolagus cuniculus | 85.10 | 2 | integrin beta 1 |
| 14 | Pongo pygmaeus | 85.10 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Xenopus tropicalis | 85.10 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 16 | Danio rerio | 84.70 | 14 | integrin beta1 subunit-like protein 1 |
| 17 | Ictalurus punctatus | 83.20 | 2 | beta-1 integrin |
| 18 | Felis catus | 83.20 | 1 | integrin beta 1 |
| 19 | Drosophila melanogaster | 69.30 | 1 | myospheroid CG1560-PA |
| 20 | Anopheles gambiae str. PEST | 67.80 | 1 | ENSANGP00000021373 |
| 21 | Anopheles gambiae | 67.80 | 1 | integrin beta subunit |
| 22 | Aedes aegypti | 67.80 | 1 | integrin beta subunit |
| 23 | Pseudoplusia includens | 66.20 | 1 | integrin beta 1 |
| 24 | Ostrinia furnacalis | 66.20 | 1 | integrin beta 1 precursor |
| 25 | Tribolium castaneum | 65.10 | 2 | PREDICTED: similar to CG1560-PA |
| 26 | Canis familiaris | 63.90 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 27 | Biomphalaria glabrata | 62.80 | 1 | beta integrin subunit |
| 28 | Crassostrea gigas | 62.00 | 1 | integrin beta cgh |
| 29 | Podocoryne carnea | 61.20 | 1 | AF308652_1 integrin beta chain |
| 30 | Caenorhabditis briggsae | 57.80 | 1 | Hypothetical protein CBG03601 |
| 31 | Caenorhabditis elegans | 55.50 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Macaca mulatta | 53.50 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 33 | Equus caballus | 53.50 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 34 | Canis lupus familiaris | 53.50 | 1 | integrin beta chain, beta 3 |
| 35 | rats, Peptide Partial, 723 aa | 53.50 | 1 | beta 3 integrin, GPIIIA |
| 36 | Mus sp. | 53.10 | 1 | beta 7 integrin |
| 37 | Strongylocentrotus purpuratus | 50.80 | 5 | integrin beta-C subunit |
| 38 | Lytechinus variegatus | 50.80 | 1 | beta-C integrin subunit |
| 39 | mice, Peptide Partial, 680 aa | 50.10 | 1 | beta 3 integrin, GPIIIA |
| 40 | Ovis aries | 47.80 | 2 | antigen CD18 |
| 41 | Bubalus bubalis | 47.80 | 1 | integrin beta-2 |
| 42 | Capra hircus | 47.80 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 43 | Ovis canadensis | 47.80 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 44 | Acropora millepora | 47.40 | 1 | integrin subunit betaCn1 |
| 45 | Sigmodon hispidus | 47.00 | 1 | AF445415_1 integrin beta-2 precursor |
| 46 | Oncorhynchus mykiss | 47.00 | 1 | CD18 protein |
| 47 | Schistosoma japonicum | 44.30 | 1 | SJCHGC06221 protein |
| 48 | Pacifastacus leniusculus | 43.50 | 1 | integrin |
| 49 | Apis mellifera | 41.20 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 50 | Halocynthia roretzi | 40.80 | 1 | integrin beta Hr2 precursor |
| 51 | Ophlitaspongia tenuis | 38.10 | 1 | integrin subunit betaPo1 |
| 52 | Cyprinus carpio | 37.70 | 2 | CD18 type 2 |
| 53 | Geodia cydonium | 34.30 | 1 | integrin beta subunit |
| 54 | Manduca sexta | 33.90 | 1 | plasmatocyte-specific integrin beta 1 |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Homo sapiens | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Mus musculus | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Pan troglodytes | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Bos taurus | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Rattus norvegicus | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Gallus gallus | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Monodelphis domestica | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Sus scrofa | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Xenopus laevis | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Tetraodon nigroviridis | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| synthetic construct | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Oryctolagus cuniculus | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Pongo pygmaeus | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Xenopus tropicalis | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||| Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Danio rerio | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||||||||||||||||#|+||||||| Sbjct 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VINPKYEGK 798 |
| Ictalurus punctatus | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 ||||||||||||||||| ||||||||||||#||||||||| Sbjct 769 RREFAKFEKEKMNAKWDAGENPIYKSAVTT#VVNPKYEGK 807 |
| Felis catus | Query 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||||||||||||||||||||||||||||#||||||||| Query 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |
| Drosophila melanogaster | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||||+||||+|||||||||||||| | +|# || | || Sbjct 808 RREFARFEKERMNAKWDTGENPIYKQATST#FKNPMYAGK 846 |
| Anopheles gambiae str. PEST | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||||+||||+| |||||||||||| | ||# || | || Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYAGK 837 |
| Anopheles gambiae | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||||+||||+| |||||||||||| | ||# || | || Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYAGK 837 |
| Aedes aegypti | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||||+||||+| |||||||||||| | ||# || | || Sbjct 797 RREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYAGK 835 |
| Pseudoplusia includens | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||||+||||+| |||||||||||| | +|# || | || Sbjct 799 RREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYAGK 837 |
| Ostrinia furnacalis | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||||+||||+| |||||||||||| | +|# || | || Sbjct 793 RREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYAGK 831 |
| Tribolium castaneum | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||||||||| | |+|||||||||| | +|# || | || Sbjct 791 RREFAKFEKEAMMARWDTGENPIYKQATST#FKNPTYAGK 829 |
| Canis familiaris | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||||||||||||||||| ||||||| + # || | | Sbjct 783 RREFAKFEKEKMNAKWDTQENPIYKSPINN#FKNPNYGRK 821 |
| Biomphalaria glabrata | Query 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |||||||||+ ||||||||||||| | +|# || | Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATST#FKNPTY 785 |
| Crassostrea gigas | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 +|| |+|||| |||+|||||||||| | +|# ||| | Sbjct 762 KRELARFEKEAMNARWDTGENPIYKQATST#FVNPTY 797 |
| Podocoryne carnea | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||||||||++ | |||+||||||| | +|# || | | Sbjct 743 RREFAKFEKDRQNPKWDSGENPIYKKATST#FQNPMYGNK 781 |
| Caenorhabditis briggsae | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 | |+||| |++ ||||| |||||| | ||# || | || Sbjct 770 RAEYAKFNNERLMAKWDTNENPIYKQATTT#FKNPVYAGK 808 |
| Caenorhabditis elegans | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 | |+| | |++ ||||| |||||| | ||# || | || Sbjct 769 RSEYATFNNERLMAKWDTNENPIYKQATTT#FKNPVYAGK 807 |
| Macaca mulatta | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 797 |+||||||+|+ ||||| ||+|| | +|# | | | Sbjct 750 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITYRG 787 |
| Equus caballus | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 797 |+||||||+|+ ||||| ||+|| | +|# | | | Sbjct 746 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITYRG 783 |
| Canis lupus familiaris | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 797 |+||||||+|+ ||||| ||+|| | +|# | | | Sbjct 746 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITYRG 783 |
| rats, Peptide Partial, 723 aa | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 797 |+||||||+|+ ||||| ||+|| | +|# | | | Sbjct 685 RKEFAKFEEERARAKWDTANNPLYKEATST#FTNITYRG 722 |
| Mus sp. | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 797 |||+ +||||+ | ||+||||+||# |||+++| Sbjct 754 RREYRRFEKEQQQLNWKQDNNPLYKSAITT#TVNPRFQG 791 |
| Strongylocentrotus purpuratus | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||| ||||+ || |+ ||||||| + + # || | | Sbjct 768 RREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTYNIK 806 |
| Lytechinus variegatus | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 |||| ||||+ || |+ ||||||| + + # || | | Sbjct 768 RREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTYNIK 806 |
| mice, Peptide Partial, 680 aa | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVN 792 |+||||||+|+ ||||| ||+|| | +|# | Sbjct 646 RKEFAKFEEERARAKWDTANNPLYKEATST#FTN 678 |
| Ovis aries | Query 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 734 REYHRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 |
| Bubalus bubalis | Query 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 733 REYHRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 |
| Capra hircus | Query 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 734 REYHRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 |
| Ovis canadensis | Query 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 734 REYHRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 |
| Acropora millepora | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 797 | |+ |||+|+|++|| +||+|++| ||# || | | Sbjct 752 RIEYQKFERERMHSKWTREKNPLYQAAKTT#FENPTYAG 789 |
| Sigmodon hispidus | Query 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 ||+ |||||+ ++|+ +||++||| ||#|+|||+ Sbjct 733 REYRHFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 766 |
| Oncorhynchus mykiss | Query 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEGK 798 +|| +|| |+ || |+||++ +| ||#| || + |+ Sbjct 740 KEFRRFENEQKKGKWSPGDNPLFMNATTT#VANPTFTGE 777 |
| Schistosoma japonicum | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYE 796 ||| | |+++ | +|+ ||||++| | #|+|| +| Sbjct 248 RRELANFKQQGENMRWEMAENPIFESPTTN#VLNPTFE 284 |
| Pacifastacus leniusculus | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |||+||||||+ |||+|||| ||# || + Sbjct 762 RREYAKFEKERKLPSGKRAENPLYKSAKTT#FQNPAF 797 |
| Apis mellifera | Query 761 REFAKFEKEKMNA-KWDTGENPIYKSAVTT#VVNPKY 795 ||| |||+|++ | ||+ +||+|| | +|# || + Sbjct 778 REFKKFERERILANKWNRRDNPLYKEATST#FKNPTF 813 |
| Halocynthia roretzi | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 797 +||+ ||+||+ || ||||+| +| + # || + | Sbjct 796 KREYEKFKKEEDLRKWTKGENPVYVNASSK#FDNPMFGG 833 |
| Ophlitaspongia tenuis | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 | |||+| |||+ |||+|+|| # || | Sbjct 803 EVRKFEREIKNAKYTKNENPLYRSATKD#YQNPLY 836 |
| Cyprinus carpio | Query 761 REFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKYEG 797 ||+ +|||++ + | +| ||++++| ||#| || + | Sbjct 730 REWKRFEKDRKHEK-TSGTNPLFQNATTT#VQNPTFSG 765 |
| Geodia cydonium | Query 762 EFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 | ||| | |+ |||+| || | # || | Sbjct 843 ELKKFENELAKVKYTKNENPLYHSATTE#HKNPIY 876 |
| Manduca sexta | Query 760 RREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 +||+||||+| + +| ||+|+ # || | Sbjct 726 KREYAKFEEESRSRGFDVSLNPLYQEPEIN#FSNPVY 761 |
[Site 3] EKMNAKWDTG778-ENPIYKSAVT
Gly778  Glu
Glu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu769 | Lys770 | Met771 | Asn772 | Ala773 | Lys774 | Trp775 | Asp776 | Thr777 | Gly778 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Glu779 | Asn780 | Pro781 | Ile782 | Tyr783 | Lys784 | Ser785 | Ala786 | Val787 | Thr788 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LIWKLLMIIHDRREFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKYEGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 107.00 | 33 | I |
| 2 | Homo sapiens | 107.00 | 25 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 3 | Mus musculus | 107.00 | 18 | integrin beta 1 (fibronectin receptor beta) |
| 4 | Pan troglodytes | 107.00 | 15 | PREDICTED: hypothetical protein |
| 5 | Danio rerio | 107.00 | 14 | integrin beta1 subunit-like protein 1 |
| 6 | Bos taurus | 107.00 | 13 | integrin beta 1 |
| 7 | Rattus norvegicus | 107.00 | 10 | Integrin beta 1 (fibronectin receptor beta) |
| 8 | Tetraodon nigroviridis | 107.00 | 8 | unnamed protein product |
| 9 | Gallus gallus | 107.00 | 7 | integrin, beta 1 (fibronectin receptor, beta polyp |
| 10 | Sus scrofa | 107.00 | 6 | integrin beta-1 subunit |
| 11 | Monodelphis domestica | 107.00 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 12 | Xenopus laevis | 107.00 | 6 | integrin beta 1 |
| 13 | synthetic construct | 107.00 | 5 | hypothetical protein |
| 14 | Oryctolagus cuniculus | 107.00 | 2 | integrin beta 1 |
| 15 | Pongo pygmaeus | 107.00 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 16 | Xenopus tropicalis | 107.00 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 17 | Felis catus | 105.00 | 1 | integrin beta 1 |
| 18 | Ictalurus punctatus | 104.00 | 2 | beta-1 integrin |
| 19 | Drosophila melanogaster | 87.00 | 1 | myospheroid CG1560-PA |
| 20 | Canis familiaris | 86.70 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 21 | Aedes aegypti | 85.50 | 1 | integrin beta subunit |
| 22 | Anopheles gambiae | 84.00 | 1 | integrin beta subunit |
| 23 | Anopheles gambiae str. PEST | 84.00 | 1 | ENSANGP00000021373 |
| 24 | Tribolium castaneum | 81.30 | 3 | PREDICTED: similar to CG1560-PA |
| 25 | Ostrinia furnacalis | 81.30 | 1 | integrin beta 1 precursor |
| 26 | Biomphalaria glabrata | 80.50 | 1 | beta integrin subunit |
| 27 | Pseudoplusia includens | 78.20 | 1 | integrin beta 1 |
| 28 | Podocoryne carnea | 76.60 | 1 | AF308652_1 integrin beta chain |
| 29 | Caenorhabditis briggsae | 76.30 | 1 | Hypothetical protein CBG03601 |
| 30 | Crassostrea gigas | 75.50 | 1 | integrin beta cgh |
| 31 | Caenorhabditis elegans | 73.90 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Macaca mulatta | 72.80 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 33 | Equus caballus | 72.80 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 34 | Canis lupus familiaris | 72.80 | 1 | integrin beta chain, beta 3 |
| 35 | rats, Peptide Partial, 723 aa | 72.80 | 1 | beta 3 integrin, GPIIIA |
| 36 | mice, Peptide Partial, 680 aa | 69.30 | 1 | beta 3 integrin, GPIIIA |
| 37 | Strongylocentrotus purpuratus | 68.20 | 5 | integrin beta-C subunit |
| 38 | Lytechinus variegatus | 68.20 | 1 | beta-C integrin subunit |
| 39 | Ovis aries | 62.00 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 40 | Pacifastacus leniusculus | 61.20 | 1 | integrin |
| 41 | Mus sp. | 60.50 | 1 | beta 7 integrin |
| 42 | Bubalus bubalis | 57.40 | 1 | integrin beta-2 |
| 43 | Capra hircus | 57.40 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 44 | Ovis canadensis | 57.00 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 45 | Sigmodon hispidus | 56.20 | 1 | AF445415_1 integrin beta-2 precursor |
| 46 | Schistosoma japonicum | 55.50 | 1 | SJCHGC06221 protein |
| 47 | Apis mellifera | 54.70 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 48 | Halocynthia roretzi | 53.10 | 2 | integrin beta Hr2 precursor |
| 49 | Oncorhynchus mykiss | 53.10 | 1 | CD18 protein |
| 50 | Acropora millepora | 51.20 | 1 | integrin subunit betaCn1 |
| 51 | Manduca sexta | 48.10 | 1 | plasmatocyte-specific integrin beta 1 |
| 52 | Spodoptera frugiperda | 47.00 | 1 | integrin beta 1 |
| 53 | Ophlitaspongia tenuis | 43.10 | 1 | integrin subunit betaPo1 |
| 54 | Cyprinus carpio | 42.70 | 2 | CD18 type 2 |
| 55 | Geodia cydonium | 41.60 | 1 | integrin beta subunit |
| 56 | Suberites domuncula | 39.70 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Homo sapiens | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Mus musculus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Pan troglodytes | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Danio rerio | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||+||||||| Sbjct 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVINPKYEGK 798 |
| Bos taurus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Rattus norvegicus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Tetraodon nigroviridis | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Gallus gallus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Sus scrofa | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Monodelphis domestica | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Xenopus laevis | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| synthetic construct | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Oryctolagus cuniculus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Pongo pygmaeus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Xenopus tropicalis | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||||||||| Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Felis catus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||| ||||||||||||||||||#|||||||||||||||||||| Sbjct 749 LIWKLLMIIHDTREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |
| Ictalurus punctatus | Query 750 IWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||| |#|||||||||||||||||||| Sbjct 759 IWKLLMIIHDRREFAKFEKEKMNAKWDAG#ENPIYKSAVTTVVNPKYEGK 807 |
| Drosophila melanogaster | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |+|||| ||||||||+||||+||||||||#|||||| | +| || | || Sbjct 797 LLWKLLTTIHDRREFARFEKERMNAKWDTG#ENPIYKQATSTFKNPMYAGK 846 |
| Canis familiaris | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||| #||||||| + || | | Sbjct 772 LIWKLLMIIHDRREFAKFEKEKMNAKWDTQ#ENPIYKSPINNFKNPNYGRK 821 |
| Aedes aegypti | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |+|||| ||||||||+||||+| ||||||#|||||| | || || | || Sbjct 786 LLWKLLTTIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTYAGK 835 |
| Anopheles gambiae | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |+||+| ||||||||+||||+| ||||||#|||||| | || || | || Sbjct 788 LLWKVLTSIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |+||+| ||||||||+||||+| ||||||#|||||| | || || | || Sbjct 788 LLWKVLTSIHDRREFARFEKERMMAKWDTG#ENPIYKQATTTFKNPTYAGK 837 |
| Tribolium castaneum | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |+||| ||||||||||||| | |+||||#|||||| | +| || | || Sbjct 780 LLWKLFTTIHDRREFAKFEKEAMMARWDTG#ENPIYKQATSTFKNPTYAGK 829 |
| Ostrinia furnacalis | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ++||++ ||||||||+||||+| ||||||#|||||| | +| || | || Sbjct 782 MLWKMVTTIHDRREFARFEKERMMAKWDTG#ENPIYKQATSTFKNPTYAGK 831 |
| Biomphalaria glabrata | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 795 |+|||| ||| |||||||||+ |||||||#|||||| | +| || | Sbjct 739 LLWKLLTFIHDTREFAKFEKERQNAKWDTG#ENPIYKQATSTFKNPTY 785 |
| Pseudoplusia includens | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 ++||+ |||||||+||||+| ||||||#|||||| | +| || | || Sbjct 788 MLWKMATTSHDRREFARFEKERMMAKWDTG#ENPIYKQATSTFKNPTYAGK 837 |
| Podocoryne carnea | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |||||| | ||||||||||++ | |||+|#|||||| | +| || | | Sbjct 732 LIWKLLATIQDRREFAKFEKDRQNPKWDSG#ENPIYKKATSTFQNPMYGNK 781 |
| Caenorhabditis briggsae | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |+|||| ++||| |+||| |++ ||||| #|||||| | || || | || Sbjct 759 LLWKLLTVLHDRAEYAKFNNERLMAKWDTN#ENPIYKQATTTFKNPVYAGK 808 |
| Crassostrea gigas | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 795 ||||| | |+|| |+|||| |||+||||#|||||| | +| ||| | Sbjct 751 LIWKLFTTISDKRELARFEKEAMNARWDTG#ENPIYKQATSTFVNPTY 797 |
| Caenorhabditis elegans | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |+|||| ++||| |+| | |++ ||||| #|||||| | || || | || Sbjct 758 LLWKLLTVLHDRSEYATFNNERLMAKWDTN#ENPIYKQATTTFKNPVYAGK 807 |
| Macaca mulatta | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 797 ||||||+ ||||+||||||+|+ ||||| # ||+|| | +| | | | Sbjct 739 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITYRG 787 |
| Equus caballus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 797 ||||||+ ||||+||||||+|+ ||||| # ||+|| | +| | | | Sbjct 735 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITYRG 783 |
| Canis lupus familiaris | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 797 ||||||+ ||||+||||||+|+ ||||| # ||+|| | +| | | | Sbjct 735 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITYRG 783 |
| rats, Peptide Partial, 723 aa | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 797 ||||||+ ||||+||||||+|+ ||||| # ||+|| | +| | | | Sbjct 674 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTNITYRG 722 |
| mice, Peptide Partial, 680 aa | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVN 792 ||||||+ ||||+||||||+|+ ||||| # ||+|| | +| | Sbjct 635 LIWKLLITIHDRKEFAKFEEERARAKWDTA#NNPLYKEATSTFTN 678 |
| Strongylocentrotus purpuratus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |||+|| ||||||| ||||+ || |+ |#|||||| + + || | | Sbjct 757 LIWRLLTYIHDRREFQNFEKERANATWEGG#ENPIYKPSTSVFKNPTYNIK 806 |
| Lytechinus variegatus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |||+|| ||||||| ||||+ || |+ |#|||||| + + || | | Sbjct 757 LIWRLLTYIHDRREFQNFEKERANATWEGG#ENPIYKPSTSVFKNPTYNIK 806 |
| Ovis aries | Query 750 IWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 |||||+ |||+| |||| |+ ||| ||# ||+|+ + +| | |+ | Sbjct 728 IWKLLVSFHDRKEVAKFEAERSKAKWQTG#TNPLYRGSTSTFKNVTYKHK 776 |
| Pacifastacus leniusculus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 795 |||||| +|||||+||||||+ #|||+|||| || || + Sbjct 751 LIWKLLTTLHDRREYAKFEKERKLPSGKRA#ENPLYKSAKTTFQNPAF 797 |
| Mus sp. | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 797 | ++| + |+||||+ +||||+ | # ||+||||+|| |||+++| Sbjct 743 LAYRLSVEIYDRREYRRFEKEQQQLNWKQD#NNPLYKSAITTTVNPRFQG 791 |
| Bubalus bubalis | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 795 +||| | + | ||+ +|||||+ ++|+ #+||++||| |||+|||+ Sbjct 721 VIWKALTHLSDLREYHRFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 766 |
| Capra hircus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 795 +||| | + | ||+ +|||||+ ++|+ #+||++||| |||+|||+ Sbjct 722 VIWKALTHLSDLREYHRFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 767 |
| Ovis canadensis | Query 750 IWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 795 ||| | + | ||+ +|||||+ ++|+ #+||++||| |||+|||+ Sbjct 723 IWKALTHLSDLREYHRFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 767 |
| Sigmodon hispidus | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 795 +||| | + | ||+ |||||+ ++|+ #+||++||| |||+|||+ Sbjct 721 VIWKALTHLTDLREYRHFEKEKLKSQWN-N#DNPLFKSATTTVMNPKF 766 |
| Schistosoma japonicum | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYE 796 ||+||++ | |||| | |+++ | +|+ #||||++| | |+|| +| Sbjct 237 LIYKLVITIDDRRELANFKQQGENMRWEMA#ENPIFESPTTNVLNPTFE 284 |
| Apis mellifera | Query 749 LIWKLLMIIHDRREFAKFEKEKMNA-KWDTG#ENPIYKSAVTTVVNPKY 795 | ||+| +| | ||| |||+|++ | ||+ #+||+|| | +| || + Sbjct 766 LTWKILTMIRDNREFKKFERERILANKWNRR#DNPLYKEATSTFKNPTF 813 |
| Halocynthia roretzi | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 797 +|||++ + |+||+ ||+||+ || |#|||+| +| + || + | Sbjct 785 IIWKVIQTLRDKREYEKFKKEEDLRKWTKG#ENPVYVNASSKFDNPMFGG 833 |
| Oncorhynchus mykiss | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 || | ++ + | +|| +|| |+ || |#+||++ +| ||| || + |+ Sbjct 728 LIIKAILYMRDVKEFRRFENEQKKGKWSPG#DNPLFMNATTTVANPTFTGE 777 |
| Acropora millepora | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 797 ++ | | + || |+ |||+|+|++|| #+||+|++| || || | | Sbjct 741 IVIKGLFTMVDRIEYQKFERERMHSKWTRE#KNPLYQAAKTTFENPTYAG 789 |
| Manduca sexta | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 795 + ||+|+ +||+||+||||+| + +| # ||+|+ || | Sbjct 715 IAWKILVDLHDKREYAKFEEESRSRGFDVS#LNPLYQEPEINFSNPVY 761 |
| Spodoptera frugiperda | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEGK 798 +|||+|+ +||++|+ ||+ | + | +| # ||+|+ || | + Sbjct 730 IIWKILVDMHDKKEYKKFQDEALAAGYDVS#LNPLYQDPSINFSNPVYNNQ 779 |
| Ophlitaspongia tenuis | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 795 |+ | |+++ | | |||+| |||+ #|||+|+|| || | Sbjct 790 LLLKGLLLLWDVVEVRKFEREIKNAKYTKN#ENPLYRSATKDYQNPLY 836 |
| Cyprinus carpio | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKYEG 797 || | ++ | ||+ +|||++ + | +|# ||++++| ||| || + | Sbjct 718 LIIKAVLYASDLREWKRFEKDRKHEK-TSG#TNPLFQNATTTVQNPTFSG 765 |
| Geodia cydonium | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 795 ++ |+|+++ | | ||| | |+ #|||+| || | || | Sbjct 830 ILLKILLMVLDYTELKKFENELAKVKYTKN#ENPLYHSATTEHKNPIY 876 |
| Suberites domuncula | Query 749 LIWKLLMIIHDRREFAKFEKEKMNAKWDTG#ENPIYKSAVTTVVNPKY 795 |+ || + | | | ||||| |+ #+||+|+|| || | Sbjct 716 LLLKLFLFIIDVIEVKKFEKELAKTKYPKH#QNPLYRSAAKDYQNPIY 762 |
[Site 4] EFAKFEKEKM771-NAKWDTGENP
Met771  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu762 | Phe763 | Ala764 | Lys765 | Phe766 | Glu767 | Lys768 | Glu769 | Lys770 | Met771 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn772 | Ala773 | Lys774 | Trp775 | Asp776 | Thr777 | Gly778 | Glu779 | Asn780 | Pro781 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LIGLALLLIWKLLMIIHDRREFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKYEGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 90.50 | 33 | I |
| 2 | Homo sapiens | 90.50 | 25 | integrin beta 1 isoform 1A precursor |
| 3 | Mus musculus | 90.50 | 16 | unnamed protein product |
| 4 | Pan troglodytes | 90.50 | 14 | PREDICTED: hypothetical protein |
| 5 | Bos taurus | 90.50 | 13 | integrin beta 1 |
| 6 | Rattus norvegicus | 90.50 | 10 | Integrin beta 1 (fibronectin receptor beta) |
| 7 | Gallus gallus | 90.50 | 7 | integrin, beta 1 (fibronectin receptor, beta polyp |
| 8 | Monodelphis domestica | 90.50 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 9 | Sus scrofa | 90.50 | 6 | integrin beta-1 subunit |
| 10 | Xenopus laevis | 90.50 | 6 | integrin beta 1 |
| 11 | Tetraodon nigroviridis | 90.50 | 5 | unnamed protein product |
| 12 | synthetic construct | 90.50 | 5 | hypothetical protein |
| 13 | Oryctolagus cuniculus | 90.50 | 2 | integrin beta 1 |
| 14 | Pongo pygmaeus | 90.50 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Xenopus tropicalis | 90.50 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 16 | Danio rerio | 90.10 | 14 | integrin beta1 subunit-like protein 1 |
| 17 | Ictalurus punctatus | 88.60 | 2 | beta-1 integrin |
| 18 | Felis catus | 88.20 | 1 | integrin beta 1 |
| 19 | Drosophila melanogaster | 74.70 | 1 | myospheroid CG1560-PA |
| 20 | Anopheles gambiae str. PEST | 73.20 | 1 | ENSANGP00000021373 |
| 21 | Anopheles gambiae | 73.20 | 1 | integrin beta subunit |
| 22 | Aedes aegypti | 73.20 | 1 | integrin beta subunit |
| 23 | Pseudoplusia includens | 71.60 | 1 | integrin beta 1 |
| 24 | Ostrinia furnacalis | 71.60 | 1 | integrin beta 1 precursor |
| 25 | Tribolium castaneum | 70.50 | 3 | PREDICTED: similar to CG1560-PA |
| 26 | Canis familiaris | 69.30 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 27 | Biomphalaria glabrata | 67.80 | 1 | beta integrin subunit |
| 28 | Crassostrea gigas | 64.30 | 1 | integrin beta cgh |
| 29 | Podocoryne carnea | 63.50 | 1 | AF308652_1 integrin beta chain |
| 30 | Caenorhabditis briggsae | 63.20 | 1 | Hypothetical protein CBG03601 |
| 31 | Caenorhabditis elegans | 60.80 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Macaca mulatta | 58.90 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 33 | Equus caballus | 58.90 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 34 | rats, Peptide Partial, 723 aa | 58.90 | 1 | beta 3 integrin, GPIIIA |
| 35 | Canis lupus familiaris | 58.90 | 1 | integrin beta chain, beta 3 |
| 36 | Strongylocentrotus purpuratus | 56.20 | 5 | integrin beta-C subunit |
| 37 | Lytechinus variegatus | 56.20 | 1 | beta-C integrin subunit |
| 38 | Mus sp. | 56.20 | 1 | beta 7 integrin |
| 39 | mice, Peptide Partial, 680 aa | 55.50 | 1 | beta 3 integrin, GPIIIA |
| 40 | Ovis aries | 51.60 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 41 | Acropora millepora | 49.70 | 1 | integrin subunit betaCn1 |
| 42 | Bubalus bubalis | 49.30 | 1 | integrin beta-2 |
| 43 | Ovis canadensis | 49.30 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 44 | Capra hircus | 49.30 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 45 | Pacifastacus leniusculus | 48.90 | 1 | integrin |
| 46 | Sigmodon hispidus | 48.50 | 1 | AF445415_1 integrin beta-2 precursor |
| 47 | Oncorhynchus mykiss | 48.10 | 1 | CD18 protein |
| 48 | Schistosoma japonicum | 46.60 | 1 | SJCHGC06221 protein |
| 49 | Apis mellifera | 43.50 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 50 | Halocynthia roretzi | 43.10 | 2 | integrin beta Hr2 precursor |
| 51 | Cyprinus carpio | 39.30 | 2 | CD18 type 2 |
| 52 | Manduca sexta | 39.30 | 1 | plasmatocyte-specific integrin beta 1 |
| 53 | Ophlitaspongia tenuis | 38.10 | 1 | integrin subunit betaPo1 |
| 54 | Spodoptera frugiperda | 36.60 | 1 | integrin beta 1 |
| 55 | Geodia cydonium | 35.40 | 1 | integrin beta subunit |
| 56 | Suberites domuncula | 33.50 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Homo sapiens | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Mus musculus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Pan troglodytes | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Bos taurus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Rattus norvegicus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Gallus gallus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Monodelphis domestica | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Sus scrofa | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Xenopus laevis | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Tetraodon nigroviridis | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| synthetic construct | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Oryctolagus cuniculus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Pongo pygmaeus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Xenopus tropicalis | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||||||||||||||||||||||||| Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Danio rerio | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#|||||||||||||||||||+||||||| Sbjct 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVINPKYEGK 798 |
| Ictalurus punctatus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#||||| ||||||||||||||||||||| Sbjct 767 HDRREFAKFEKEKM#NAKWDAGENPIYKSAVTTVVNPKYEGK 807 |
| Felis catus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 || |||||||||||#||||||||||||||||||||||||||| Sbjct 758 HDTREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |
| Drosophila melanogaster | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |||||||+||||+|#||||||||||||| | +| || | || Sbjct 806 HDRREFARFEKERM#NAKWDTGENPIYKQATSTFKNPMYAGK 846 |
| Anopheles gambiae str. PEST | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |||||||+||||+|# |||||||||||| | || || | || Sbjct 797 HDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAGK 837 |
| Anopheles gambiae | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |||||||+||||+|# |||||||||||| | || || | || Sbjct 797 HDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAGK 837 |
| Aedes aegypti | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |||||||+||||+|# |||||||||||| | || || | || Sbjct 795 HDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAGK 835 |
| Pseudoplusia includens | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |||||||+||||+|# |||||||||||| | +| || | || Sbjct 797 HDRREFARFEKERM#MAKWDTGENPIYKQATSTFKNPTYAGK 837 |
| Ostrinia furnacalis | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |||||||+||||+|# |||||||||||| | +| || | || Sbjct 791 HDRREFARFEKERM#MAKWDTGENPIYKQATSTFKNPTYAGK 831 |
| Tribolium castaneum | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |||||||||||| |# |+|||||||||| | +| || | || Sbjct 789 HDRREFAKFEKEAM#MARWDTGENPIYKQATSTFKNPTYAGK 829 |
| Canis familiaris | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||#|||||| ||||||| + || | | Sbjct 781 HDRREFAKFEKEKM#NAKWDTQENPIYKSPINNFKNPNYGRK 821 |
| Biomphalaria glabrata | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 795 || |||||||||+ #||||||||||||| | +| || | Sbjct 748 HDTREFAKFEKERQ#NAKWDTGENPIYKQATSTFKNPTY 785 |
| Crassostrea gigas | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 795 |+|| |+|||| |#||+|||||||||| | +| ||| | Sbjct 761 DKRELARFEKEAM#NARWDTGENPIYKQATSTFVNPTY 797 |
| Podocoryne carnea | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||||||||||++ #| |||+||||||| | +| || | | Sbjct 742 DRREFAKFEKDRQ#NPKWDSGENPIYKKATSTFQNPMYGNK 781 |
| Caenorhabditis briggsae | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||| |+||| |++# ||||| |||||| | || || | || Sbjct 768 HDRAEYAKFNNERL#MAKWDTNENPIYKQATTTFKNPVYAGK 808 |
| Caenorhabditis elegans | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||| |+| | |++# ||||| |||||| | || || | || Sbjct 767 HDRSEYATFNNERL#MAKWDTNENPIYKQATTTFKNPVYAGK 807 |
| Macaca mulatta | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |||+||||||+|+ # ||||| ||+|| | +| | | | Sbjct 748 HDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 787 |
| Equus caballus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |||+||||||+|+ # ||||| ||+|| | +| | | | Sbjct 744 HDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 783 |
| rats, Peptide Partial, 723 aa | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |||+||||||+|+ # ||||| ||+|| | +| | | | Sbjct 683 HDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 722 |
| Canis lupus familiaris | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |||+||||||+|+ # ||||| ||+|| | +| | | | Sbjct 744 HDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRG 783 |
| Strongylocentrotus purpuratus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |||||| ||||+ #|| |+ ||||||| + + || | | Sbjct 766 HDRREFQNFEKERA#NATWEGGENPIYKPSTSVFKNPTYNIK 806 |
| Lytechinus variegatus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |||||| ||||+ #|| |+ ||||||| + + || | | Sbjct 766 HDRREFQNFEKERA#NATWEGGENPIYKPSTSVFKNPTYNIK 806 |
| Mus sp. | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 +||||+ +||||+ # | ||+||||+|| |||+++| Sbjct 752 YDRREYRRFEKEQQ#QLNWKQDNNPLYKSAITTTVNPRFQG 791 |
| mice, Peptide Partial, 680 aa | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVN 792 |||+||||||+|+ # ||||| ||+|| | +| | Sbjct 644 HDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTN 678 |
| Ovis aries | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 |||+| |||| |+ # ||| || ||+|+ + +| | |+ | Sbjct 736 HDRKEVAKFEAERS#KAKWQTGTNPLYRGSTSTFKNVTYKHK 776 |
| Acropora millepora | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 || |+ |||+|+|#++|| +||+|++| || || | | Sbjct 751 DRIEYQKFERERM#HSKWTREKNPLYQAAKTTFENPTYAG 789 |
| Bubalus bubalis | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 795 | ||+ +|||||+# ++|+ +||++||| |||+|||+ Sbjct 731 DLREYHRFEKEKL#KSQWNN-DNPLFKSATTTVMNPKF 766 |
| Ovis canadensis | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 795 | ||+ +|||||+# ++|+ +||++||| |||+|||+ Sbjct 732 DLREYHRFEKEKL#KSQWNN-DNPLFKSATTTVMNPKF 767 |
| Capra hircus | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 795 | ||+ +|||||+# ++|+ +||++||| |||+|||+ Sbjct 732 DLREYHRFEKEKL#KSQWNN-DNPLFKSATTTVMNPKF 767 |
| Pacifastacus leniusculus | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 795 |||||+||||||+ # |||+|||| || || + Sbjct 760 HDRREYAKFEKERK#LPSGKRAENPLYKSAKTTFQNPAF 797 |
| Sigmodon hispidus | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 795 | ||+ |||||+# ++|+ +||++||| |||+|||+ Sbjct 731 DLREYRHFEKEKL#KSQWNN-DNPLFKSATTTVMNPKF 766 |
| Oncorhynchus mykiss | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 | +|| +|| |+ # || |+||++ +| ||| || + |+ Sbjct 738 DVKEFRRFENEQK#KGKWSPGDNPLFMNATTTVANPTFTGE 777 |
| Schistosoma japonicum | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYE 796 |||| | |+++ #| +|+ ||||++| | |+|| +| Sbjct 247 DRRELANFKQQGE#NMRWEMAENPIFESPTTNVLNPTFE 284 |
| Apis mellifera | Query 759 DRREFAKFEKEKM#NA-KWDTGENPIYKSAVTTVVNPKY 795 | ||| |||+|++# | ||+ +||+|| | +| || + Sbjct 776 DNREFKKFERERI#LANKWNRRDNPLYKEATSTFKNPTF 813 |
| Halocynthia roretzi | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |+||+ ||+||+ # || ||||+| +| + || + | Sbjct 795 DKREYEKFKKEED#LRKWTKGENPVYVNASSKFDNPMFGG 833 |
| Cyprinus carpio | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 | ||+ +|||++ #+ | +| ||++++| ||| || + | Sbjct 728 DLREWKRFEKDRK#HEK-TSGTNPLFQNATTTVQNPTFSG 765 |
| Manduca sexta | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 795 ||+||+||||+| #+ +| ||+|+ || | Sbjct 724 HDKREYAKFEEESR#SRGFDVSLNPLYQEPEINFSNPVY 761 |
| Ophlitaspongia tenuis | Query 762 EFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 795 | |||+| #|||+ |||+|+|| || | Sbjct 803 EVRKFEREIK#NAKYTKNENPLYRSATKDYQNPLY 836 |
| Spodoptera frugiperda | Query 758 HDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEGK 798 ||++|+ ||+ | +# | +| ||+|+ || | + Sbjct 739 HDKKEYKKFQDEAL#AAGYDVSLNPLYQDPSINFSNPVYNNQ 779 |
| Geodia cydonium | Query 759 DRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 795 | | ||| | # |+ |||+| || | || | Sbjct 840 DYTELKKFENELA#KVKYTKNENPLYHSATTEHKNPIY 876 |
| Suberites domuncula | Query 762 EFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKY 795 | ||||| # |+ +||+|+|| || | Sbjct 729 EVKKFEKELA#KTKYPKHQNPLYRSAAKDYQNPIY 762 |
[Site 5] KEKMNAKWDT777-GENPIYKSAV
Thr777  Gly
Gly
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys768 | Glu769 | Lys770 | Met771 | Asn772 | Ala773 | Lys774 | Trp775 | Asp776 | Thr777 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Gly778 | Glu779 | Asn780 | Pro781 | Ile782 | Tyr783 | Lys784 | Ser785 | Ala786 | Val787 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LLIWKLLMIIHDRREFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKYEGK |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 109.00 | 33 | I |
| 2 | Homo sapiens | 109.00 | 25 | ITB1_HUMAN Integrin beta-1 precursor (Fibronectin |
| 3 | Mus musculus | 109.00 | 18 | integrin beta 1 (fibronectin receptor beta) |
| 4 | Pan troglodytes | 109.00 | 14 | PREDICTED: hypothetical protein |
| 5 | Bos taurus | 109.00 | 13 | integrin beta 1 |
| 6 | Rattus norvegicus | 109.00 | 10 | Integrin beta 1 (fibronectin receptor beta) |
| 7 | Tetraodon nigroviridis | 109.00 | 8 | unnamed protein product |
| 8 | Gallus gallus | 109.00 | 7 | integrin, beta 1 (fibronectin receptor, beta polyp |
| 9 | Monodelphis domestica | 109.00 | 6 | PREDICTED: similar to integrin beta-1 subunit isof |
| 10 | Xenopus laevis | 109.00 | 6 | integrin beta 1 |
| 11 | Sus scrofa | 109.00 | 6 | integrin beta-1 subunit |
| 12 | synthetic construct | 109.00 | 5 | hypothetical protein |
| 13 | Oryctolagus cuniculus | 109.00 | 2 | integrin beta 1 |
| 14 | Pongo pygmaeus | 109.00 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 15 | Xenopus tropicalis | 109.00 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 16 | Danio rerio | 108.00 | 14 | integrin beta1 subunit-like protein 1 |
| 17 | Felis catus | 107.00 | 1 | integrin beta 1 |
| 18 | Ictalurus punctatus | 105.00 | 2 | beta-1 integrin |
| 19 | Drosophila melanogaster | 88.60 | 1 | myospheroid CG1560-PA |
| 20 | Canis familiaris | 88.20 | 10 | PREDICTED: similar to integrin beta 1 isoform 1D p |
| 21 | Aedes aegypti | 87.00 | 1 | integrin beta subunit |
| 22 | Anopheles gambiae | 85.50 | 2 | integrin beta subunit |
| 23 | Anopheles gambiae str. PEST | 85.50 | 2 | ENSANGP00000021373 |
| 24 | Tribolium castaneum | 82.80 | 3 | PREDICTED: similar to CG1560-PA |
| 25 | Ostrinia furnacalis | 82.80 | 1 | integrin beta 1 precursor |
| 26 | Biomphalaria glabrata | 82.00 | 1 | beta integrin subunit |
| 27 | Pseudoplusia includens | 79.70 | 1 | integrin beta 1 |
| 28 | Podocoryne carnea | 78.20 | 1 | AF308652_1 integrin beta chain |
| 29 | Caenorhabditis briggsae | 77.80 | 1 | Hypothetical protein CBG03601 |
| 30 | Crassostrea gigas | 77.00 | 1 | integrin beta cgh |
| 31 | Caenorhabditis elegans | 75.50 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Macaca mulatta | 74.30 | 13 | PREDICTED: similar to integrin beta chain, beta 3 |
| 33 | Canis lupus familiaris | 74.30 | 1 | integrin beta chain, beta 3 |
| 34 | Equus caballus | 74.30 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 35 | rats, Peptide Partial, 723 aa | 74.30 | 1 | beta 3 integrin, GPIIIA |
| 36 | mice, Peptide Partial, 680 aa | 70.90 | 1 | beta 3 integrin, GPIIIA |
| 37 | Strongylocentrotus purpuratus | 69.70 | 5 | integrin beta-C subunit |
| 38 | Lytechinus variegatus | 69.70 | 1 | beta-C integrin subunit |
| 39 | Ovis aries | 63.20 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 40 | Pacifastacus leniusculus | 62.80 | 1 | integrin |
| 41 | Mus sp. | 60.80 | 1 | beta 7 integrin |
| 42 | Capra hircus | 58.90 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 43 | Bubalus bubalis | 58.90 | 1 | integrin beta-2 |
| 44 | Ovis canadensis | 58.20 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 45 | Sigmodon hispidus | 57.80 | 1 | AF445415_1 integrin beta-2 precursor |
| 46 | Schistosoma japonicum | 57.00 | 1 | SJCHGC06221 protein |
| 47 | Apis mellifera | 56.20 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 48 | Halocynthia roretzi | 54.70 | 2 | integrin beta Hr2 precursor |
| 49 | Oncorhynchus mykiss | 54.70 | 1 | CD18 protein |
| 50 | Acropora millepora | 52.80 | 1 | integrin subunit betaCn1 |
| 51 | Manduca sexta | 49.70 | 1 | plasmatocyte-specific integrin beta 1 |
| 52 | Spodoptera frugiperda | 47.40 | 1 | integrin beta 1 |
| 53 | Ophlitaspongia tenuis | 44.70 | 1 | integrin subunit betaPo1 |
| 54 | Cyprinus carpio | 44.30 | 2 | CD18 type 2 |
| 55 | Geodia cydonium | 43.10 | 1 | integrin beta subunit |
| 56 | Suberites domuncula | 41.20 | 1 | integrin beta subunit |
| 57 | Drosophila pseudoobscura | 33.10 | 1 | GA14581-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Homo sapiens | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Mus musculus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Pan troglodytes | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Bos taurus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Rattus norvegicus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Tetraodon nigroviridis | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Gallus gallus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Monodelphis domestica | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Xenopus laevis | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Sus scrofa | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| synthetic construct | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Oryctolagus cuniculus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Pongo pygmaeus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Xenopus tropicalis | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#||||||||||||||||||||| Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Danio rerio | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||#|||||||||||||+||||||| Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVINPKYEGK 798 |
| Felis catus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |||||||||||| |||||||||||||||||#||||||||||||||||||||| Sbjct 748 LLIWKLLMIIHDTREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |
| Ictalurus punctatus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 | ||||||||||||||||||||||||||| #||||||||||||||||||||| Sbjct 757 LFIWKLLMIIHDRREFAKFEKEKMNAKWDA#GENPIYKSAVTTVVNPKYEGK 807 |
| Drosophila melanogaster | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||+|||| ||||||||+||||+|||||||#||||||| | +| || | || Sbjct 796 LLLWKLLTTIHDRREFARFEKERMNAKWDT#GENPIYKQATSTFKNPMYAGK 846 |
| Canis familiaris | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||||||||||||||||||||||||||# ||||||| + || | | Sbjct 771 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#QENPIYKSPINNFKNPNYGRK 821 |
| Aedes aegypti | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||+|||| ||||||||+||||+| |||||#||||||| | || || | || Sbjct 785 LLLWKLLTTIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAGK 835 |
| Anopheles gambiae | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||+||+| ||||||||+||||+| |||||#||||||| | || || | || Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAGK 837 |
| Anopheles gambiae str. PEST | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||+||+| ||||||||+||||+| |||||#||||||| | || || | || Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAGK 837 |
| Tribolium castaneum | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||+||| ||||||||||||| | |+|||#||||||| | +| || | || Sbjct 779 LLLWKLFTTIHDRREFAKFEKEAMMARWDT#GENPIYKQATSTFKNPTYAGK 829 |
| Ostrinia furnacalis | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |++||++ ||||||||+||||+| |||||#||||||| | +| || | || Sbjct 781 LMLWKMVTTIHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAGK 831 |
| Biomphalaria glabrata | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 795 ||+|||| ||| |||||||||+ ||||||#||||||| | +| || | Sbjct 738 LLLWKLLTFIHDTREFAKFEKERQNAKWDT#GENPIYKQATSTFKNPTY 785 |
| Pseudoplusia includens | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 |++||+ |||||||+||||+| |||||#||||||| | +| || | || Sbjct 787 LMLWKMATTSHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAGK 837 |
| Podocoryne carnea | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||||| | ||||||||||++ | |||+#||||||| | +| || | | Sbjct 731 LLIWKLLATIQDRREFAKFEKDRQNPKWDS#GENPIYKKATSTFQNPMYGNK 781 |
| Caenorhabditis briggsae | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||+|||| ++||| |+||| |++ |||||# |||||| | || || | || Sbjct 758 LLLWKLLTVLHDRAEYAKFNNERLMAKWDT#NENPIYKQATTTFKNPVYAGK 808 |
| Crassostrea gigas | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 795 |||||| | |+|| |+|||| |||+|||#||||||| | +| ||| | Sbjct 750 LLIWKLFTTISDKRELARFEKEAMNARWDT#GENPIYKQATSTFVNPTY 797 |
| Caenorhabditis elegans | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||+|||| ++||| |+| | |++ |||||# |||||| | || || | || Sbjct 757 LLLWKLLTVLHDRSEYATFNNERLMAKWDT#NENPIYKQATTTFKNPVYAGK 807 |
| Macaca mulatta | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |||||||+ ||||+||||||+|+ |||||# ||+|| | +| | | | Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 787 |
| Canis lupus familiaris | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |||||||+ ||||+||||||+|+ |||||# ||+|| | +| | | | Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 783 |
| Equus caballus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |||||||+ ||||+||||||+|+ |||||# ||+|| | +| | | | Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 783 |
| rats, Peptide Partial, 723 aa | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |||||||+ ||||+||||||+|+ |||||# ||+|| | +| | | | Sbjct 673 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRG 722 |
| mice, Peptide Partial, 680 aa | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVN 792 |||||||+ ||||+||||||+|+ |||||# ||+|| | +| | Sbjct 634 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTN 678 |
| Strongylocentrotus purpuratus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||+|| ||||||| ||||+ || |+ #||||||| + + || | | Sbjct 756 LLIWRLLTYIHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTYNIK 806 |
| Lytechinus variegatus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||||+|| ||||||| ||||+ || |+ #||||||| + + || | | Sbjct 756 LLIWRLLTYIHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTYNIK 806 |
| Ovis aries | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 | |||||+ |||+| |||| |+ ||| |#| ||+|+ + +| | |+ | Sbjct 726 LCIWKLLVSFHDRKEVAKFEAERSKAKWQT#GTNPLYRGSTSTFKNVTYKHK 776 |
| Pacifastacus leniusculus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 795 ||||||| +|||||+||||||+ # |||+|||| || || + Sbjct 750 LLIWKLLTTLHDRREYAKFEKERKLPSGKR#AENPLYKSAKTTFQNPAF 797 |
| Mus sp. | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 +| ++| + |+||||+ +||||+ | # ||+||||+|| |||+++| Sbjct 742 VLAYRLSVEIYDRREYRRFEKEQQQLNWKQ#DNNPLYKSAITTTVNPRFQG 791 |
| Capra hircus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 795 |+||| | + | ||+ +|||||+ ++|+ # +||++||| |||+|||+ Sbjct 721 LVIWKALTHLSDLREYHRFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 767 |
| Bubalus bubalis | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 795 |+||| | + | ||+ +|||||+ ++|+ # +||++||| |||+|||+ Sbjct 720 LVIWKALTHLSDLREYHRFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 766 |
| Ovis canadensis | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 795 | ||| | + | ||+ +|||||+ ++|+ # +||++||| |||+|||+ Sbjct 721 LAIWKALTHLSDLREYHRFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 767 |
| Sigmodon hispidus | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 795 |+||| | + | ||+ |||||+ ++|+ # +||++||| |||+|||+ Sbjct 720 LVIWKALTHLTDLREYRHFEKEKLKSQWN-#NDNPLFKSATTTVMNPKF 766 |
| Schistosoma japonicum | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYE 796 |||+||++ | |||| | |+++ | +|+ # ||||++| | |+|| +| Sbjct 236 LLIYKLVITIDDRRELANFKQQGENMRWEM#AENPIFESPTTNVLNPTFE 284 |
| Apis mellifera | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNA-KWDT#GENPIYKSAVTTVVNPKY 795 || ||+| +| | ||| |||+|++ | ||+ # +||+|| | +| || + Sbjct 765 LLTWKILTMIRDNREFKKFERERILANKWNR#RDNPLYKEATSTFKNPTF 813 |
| Halocynthia roretzi | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |+|||++ + |+||+ ||+||+ || #||||+| +| + || + | Sbjct 784 LIIWKVIQTLRDKREYEKFKKEEDLRKWTK#GENPVYVNASSKFDNPMFGG 833 |
| Oncorhynchus mykiss | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ||| | ++ + | +|| +|| |+ || #|+||++ +| ||| || + |+ Sbjct 727 LLIIKAILYMRDVKEFRRFENEQKKGKWSP#GDNPLFMNATTTVANPTFTGE 777 |
| Acropora millepora | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |++ | | + || |+ |||+|+|++|| # +||+|++| || || | | Sbjct 740 LIVIKGLFTMVDRIEYQKFERERMHSKWTR#EKNPLYQAAKTTFENPTYAG 789 |
| Manduca sexta | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 795 |+ ||+|+ +||+||+||||+| + +| # ||+|+ || | Sbjct 714 LIAWKILVDLHDKREYAKFEEESRSRGFDV#SLNPLYQEPEINFSNPVY 761 |
| Spodoptera frugiperda | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEGK 798 ++|||+|+ +||++|+ ||+ | + | +| # ||+|+ || | + Sbjct 729 VIIWKILVDMHDKKEYKKFQDEALAAGYDV#SLNPLYQDPSINFSNPVYNNQ 779 |
| Ophlitaspongia tenuis | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 795 ||+ | |+++ | | |||+| |||+ # |||+|+|| || | Sbjct 789 LLLLKGLLLLWDVVEVRKFEREIKNAKYTK#NENPLYRSATKDYQNPLY 836 |
| Cyprinus carpio | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 ||| | ++ | ||+ +|||++ + | +#| ||++++| ||| || + | Sbjct 717 LLIIKAVLYASDLREWKRFEKDRKHEK-TS#GTNPLFQNATTTVQNPTFSG 765 |
| Geodia cydonium | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 795 |++ |+|+++ | | ||| | |+ # |||+| || | || | Sbjct 829 LILLKILLMVLDYTELKKFENELAKVKYTK#NENPLYHSATTEHKNPIY 876 |
| Suberites domuncula | Query 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKY 795 ||+ || + | | | ||||| |+ # +||+|+|| || | Sbjct 715 LLLLKLFLFIIDVIEVKKFEKELAKTKYPK#HQNPLYRSAAKDYQNPIY 762 |
| Drosophila pseudoobscura | Query 748 LLIWKLLMIIH--DRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPK 794 |||+ + | | ||+|+||+|+ || +# |||+|+ | | || Sbjct 713 LLIFLFIWCIRRKDAREYARFEEEQ--AKRVS#QENPLYRDPVGRYVVPK 759 |
* References
[PubMed ID: 10349636] Carninci P, Hayashizaki Y, High-efficiency full-length cDNA cloning. Methods Enzymol. 1999;303:19-44.
[PubMed ID: 11042159] ... Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y, Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 2000 Oct;10(10):1617-30.
[PubMed ID: 11076861] ... Shibata K, Itoh M, Aizawa K, Nagaoka S, Sasaki N, Carninci P, Konno H, Akiyama J, Nishi K, Kitsunai T, Tashiro H, Itoh M, Sumi N, Ishii Y, Nakamura S, Hazama M, Nishine T, Harada A, Yamamoto R, Matsumoto H, Sakaguchi S, Ikegami T, Kashiwagi K, Fujiwake S, Inoue K, Togawa Y, RIKEN integrated sequence analysis (RISA) system--384-format sequencing pipeline with 384 multicapillary sequencer. Genome Res. 2000 Nov;10(11):1757-71.
[PubMed ID: 11217851] ... Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, Adachi J, Fukuda S, Aizawa K, Izawa M, Nishi K, Kiyosawa H, Kondo S, Yamanaka I, Saito T, Okazaki Y, Gojobori T, Bono H, Kasukawa T, Saito R, Kadota K, Matsuda H, Ashburner M, Batalov S, Casavant T, Fleischmann W, Gaasterland T, Gissi C, King B, Kochiwa H, Kuehl P, Lewis S, Matsuo Y, Nikaido I, Pesole G, Quackenbush J, Schriml LM, Staubli F, Suzuki R, Tomita M, Wagner L, Washio T, Sakai K, Okido T, Furuno M, Aono H, Baldarelli R, Barsh G, Blake J, Boffelli D, Bojunga N, Carninci P, de Bonaldo MF, Brownstein MJ, Bult C, Fletcher C, Fujita M, Gariboldi M, Gustincich S, Hill D, Hofmann M, Hume DA, Kamiya M, Lee NH, Lyons P, Marchionni L, Mashima J, Mazzarelli J, Mombaerts P, Nordone P, Ring B, Ringwald M, Rodriguez I, Sakamoto N, Sasaki H, Sato K, Schonbach C, Seya T, Shibata Y, Storch KF, Suzuki H, Toyo-oka K, Wang KH, Weitz C, Whittaker C, Wilming L, Wynshaw-Boris A, Yoshida K, Hasegawa Y, Kawaji H, Kohtsuki S, Hayashizaki Y, Functional annotation of a full-length mouse cDNA collection. Nature. 2001 Feb 8;409(6821):685-90.
[PubMed ID: 12466851] ... Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y, Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002 Dec 5;420(6915):563-73.
[PubMed ID: 16141072] ... Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y, The transcriptional landscape of the mammalian genome. Science. 2005 Sep 2;309(5740):1559-63.
[PubMed ID: 16141073] ... Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C, Antisense transcription in the mammalian transcriptome. Science. 2005 Sep 2;309(5740):1564-6.