XSB0377 : integrin, beta 2 [Bos taurus]
[ CaMP Format ]
This entry is computationally expanded from SB0002
* Basic Information
| Organism | Bos taurus (cattle) |
| Protein Names | Integrin beta-2; Cell surface adhesion glycoproteins LFA-1/CR3/p150,95 subunit beta; Complement receptor C3 subunit beta; integrin; beta 2; CD18 subunit; Leu-CAM receptor; beta 2 (antigen CD18 subunit (p95); lymphocyte function-associated antigen 1; integrin B2) |
| Gene Names | ITGB2; CD18; integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| Gene Locus | 1; chromosome 1 |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| NP_786975 | NM_175781 | 281877 | P32592 | N/A | N/A | N/A | bta:281877 |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 769 aa
Average Mass: 84.400 kDa
Monoisotopic Mass: 84.345 kDa
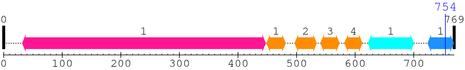
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Integrin_beta 1. | 32 | 447 | 1059.8 | 0 |
| EGF_2 1. | 449 | 481 | 20.0 | 0.01 |
| EGF_2 2. | 497 | 534 | 16.7 | 0.035 |
| EGF_2 3. | 541 | 573 | 11.4 | 0.15 |
| EGF_2 4. | 582 | 612 | 31.0 | 4.8e-06 |
| Integrin_B_tail 1. | 622 | 700 | 125.8 | 1.4e-34 |
| Integrin_b_cyt 1. | 724 | 768 | 86.1 | 1.2e-22 |
| --- cleavage 754 (inside Integrin_b_cyt 724..768) --- | ||||
3. Sequence Information
Fasta Sequence: XSB0377.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] SQWNNDNPLF754-KSATTTVMNP
Phe754  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser745 | Gln746 | Trp747 | Asn748 | Asn749 | Asp750 | Asn751 | Pro752 | Leu753 | Phe754 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys755 | Ser756 | Ala757 | Thr758 | Thr759 | Thr760 | Val761 | Met762 | Asn763 | Pro764 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ALTHLSDLREYHRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Bos taurus | 96.70 | 6 | integrin, beta 2 |
| 2 | Bubalus bubalis | 96.70 | 1 | integrin beta-2 |
| 3 | Capra hircus | 96.70 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 4 | Ovis canadensis | 96.70 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 5 | Ovis aries | 96.70 | 1 | antigen CD18 |
| 6 | N/A | 92.40 | 20 | A |
| 7 | Mus musculus | 92.40 | 10 | integrin beta 2 |
| 8 | Homo sapiens | 91.30 | 14 | integrin beta chain, beta 2 precursor variant |
| 9 | Pan troglodytes | 91.30 | 6 | integrin, beta 2 |
| 10 | synthetic construct | 91.30 | 3 | integrin, beta 2 (antigen CD18 (p95), lymphocyte f |
| 11 | Rattus norvegicus | 90.50 | 8 | PREDICTED: similar to Integrin beta-2 precursor (C |
| 12 | Sigmodon hispidus | 90.50 | 1 | AF445415_1 integrin beta-2 precursor |
| 13 | Sus scrofa | 90.10 | 4 | CD18 leukocyte adhesion molecule |
| 14 | Canis familiaris | 89.40 | 4 | AF181965_1 B-2 integrin |
| 15 | Monodelphis domestica | 72.00 | 4 | PREDICTED: similar to Stromal interaction molecule |
| 16 | Gallus gallus | 65.50 | 6 | integrin, beta 2 |
| 17 | Xenopus laevis | 53.50 | 5 | hypothetical protein LOC379708 |
| 18 | Oncorhynchus mykiss | 50.80 | 1 | CD18 protein |
| 19 | Cyprinus carpio | 50.10 | 2 | CD18 type 2 |
| 20 | Ictalurus punctatus | 50.10 | 2 | CD18 |
| 21 | Macaca mulatta | 50.10 | 2 | PREDICTED: integrin, beta 7 |
| 22 | Felis catus | 50.10 | 1 | integrin beta 1 |
| 23 | Danio rerio | 49.70 | 12 | integrin, beta 1b |
| 24 | Tetraodon nigroviridis | 49.70 | 6 | unnamed protein product |
| 25 | Oryctolagus cuniculus | 49.70 | 2 | integrin beta 1 |
| 26 | Xenopus tropicalis | 49.70 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 27 | Pongo pygmaeus | 49.70 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 28 | Pacifastacus leniusculus | 49.30 | 1 | integrin |
| 29 | Anopheles gambiae | 49.30 | 1 | integrin beta subunit |
| 30 | Anopheles gambiae str. PEST | 49.30 | 1 | ENSANGP00000021373 |
| 31 | Aedes aegypti | 48.90 | 1 | integrin beta subunit |
| 32 | Mus sp. | 48.50 | 1 | beta 7 integrin |
| 33 | Drosophila melanogaster | 47.80 | 1 | myospheroid CG1560-PA |
| 34 | Ostrinia furnacalis | 46.20 | 1 | integrin beta 1 precursor |
| 35 | Caenorhabditis briggsae | 46.20 | 1 | Hypothetical protein CBG03601 |
| 36 | Strongylocentrotus purpuratus | 45.40 | 5 | integrin beta L subunit |
| 37 | Tribolium castaneum | 45.40 | 3 | PREDICTED: similar to CG1560-PA |
| 38 | Caenorhabditis elegans | 45.10 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 39 | Crassostrea gigas | 45.10 | 1 | integrin beta cgh |
| 40 | Apis mellifera | 45.10 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 41 | Biomphalaria glabrata | 44.30 | 1 | beta integrin subunit |
| 42 | Pseudoplusia includens | 44.30 | 1 | integrin beta 1 |
| 43 | Podocoryne carnea | 40.80 | 1 | AF308652_1 integrin beta chain |
| 44 | Lytechinus variegatus | 40.80 | 1 | beta-C integrin subunit |
| 45 | Acropora millepora | 40.40 | 1 | integrin subunit betaCn1 |
| 46 | rats, Peptide Partial, 723 aa | 36.60 | 1 | beta 3 integrin, GPIIIA |
| 47 | Canis lupus familiaris | 36.60 | 1 | integrin beta chain, beta 3 |
| 48 | mice, Peptide Partial, 680 aa | 36.60 | 1 | beta 3 integrin, GPIIIA |
| 49 | Equus caballus | 36.60 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 50 | Schistosoma japonicum | 35.80 | 1 | SJCHGC06221 protein |
| 51 | Trichomonas vaginalis G3 | 35.40 | 1 | hypothetical protein TVAG_480920 |
Top-ranked sequences
| organism | matching |
|---|---|
| Bos taurus | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||| Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Bubalus bubalis | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||| Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Capra hircus | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||| Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Ovis canadensis | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||| Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Ovis aries | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 ||||||||||||||||||||||||||||||#||||||||||||||| Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| N/A | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |||||+||||| ||||||||||||||||||#||||||||||||||| Sbjct 727 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 771 |
| Mus musculus | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |||||+||||| ||||||||||||||||||#||||||||||||||| Sbjct 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Homo sapiens | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || |||||||| ||||||||||||||||||#||||||||||||||| Sbjct 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Pan troglodytes | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || |||||||| ||||||||||||||||||#||||||||||||||| Sbjct 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| synthetic construct | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || |||||||| ||||||||||||||||||#||||||||||||||| Sbjct 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Rattus norvegicus | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |||||+|| || ||||||||||||||||||#||||||||||||||| Sbjct 724 ALTHLTDLNEYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 768 |
| Sigmodon hispidus | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |||||+||||| |||||||||||||||||#||||||||||||||| Sbjct 725 ALTHLTDLREYRHFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Sus scrofa | Query 726 LTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAE 768 |||||||||| ||||||||||||||||||#|||||||||||||| Sbjct 726 LTHLSDLREYKRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAE 768 |
| Canis familiaris | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 ||||||||||+ |||||||+||||||||||#||||||||||+|||| Sbjct 720 ALTHLSDLREFKRFEKEKLRSQWNNDNPLF#KSATTTVMNPRFAES 764 |
| Monodelphis domestica | Query 726 LTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNP 764 ||| |||||| ||||||||||||||||||#| |||++ | Sbjct 1192 LTHFSDLREYKRFEKEKLKSQWNNDNPLF#KKKTTTIIIP 1230 |
| Gallus gallus | Query 726 LTHLSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 || + | ||| |||||| |++|| |||||#||||||||||+| Sbjct 728 LTEIFDRREYRRFEKEKSKAKWNEADNPLF#KSATTTVMNPRF 769 |
| Xenopus laevis | Query 726 LTHLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 +| + | || |||||++ |+|| |||+# +||||| || |+ Sbjct 728 ITEIKDRNEYQRFEKERMNSKWNPGHNPLY#HNATTTVQNPNFS 770 |
| Oncorhynchus mykiss | Query 725 ALTHLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 766 |+ ++ |++|+ ||| |+ | +|+ |||||# +||||| || | Sbjct 732 AILYMRDVKEFRRFENEQKKGKWSPGDNPLF#MNATTTVANPTF 774 |
| Cyprinus carpio | Query 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFA 767 |+ + |||||+ ||||++ + + ||||#++||||| || |+ Sbjct 722 AVLYASDLREWKRFEKDRKHEKTSGTNPLF#QNATTTVQNPTFS 764 |
| Ictalurus punctatus | Query 725 ALTHLSDLREYHRFEKEKLKSQW-NNDNPLF#KSATTTVMNPKF 766 || + ||+|+ +|||| + || +||||#++||||| || | Sbjct 726 ALFYFKDLKEWKKFEKEAQRRQWAKGENPLF#QNATTTVANPAF 768 |
| Macaca mulatta | Query 731 DLREYHRFEKEKLKSQWNND-NPLF#KSATTTVMNPKFAES 769 | ||| |||||+ + | | |||+#||| || +||+| |+ Sbjct 630 DRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEA 669 |
| Felis catus | Query 729 LSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 + | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 757 IHDTREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Danio rerio | Query 729 LSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 766 + | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 754 IHDRREFAKFEKEKMNAKWDAGENPIY#KSAVTTVVNPKY 792 |
| Tetraodon nigroviridis | Query 729 LSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 + | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 750 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 788 |
| Oryctolagus cuniculus | Query 729 LSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 + | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 22 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 60 |
| Xenopus tropicalis | Query 729 LSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 + | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 757 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Pongo pygmaeus | Query 729 LSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 + | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 757 IHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Pacifastacus leniusculus | Query 726 LTHLSDLREYHRFEKE-KLKSQWNNDNPLF#KSATTTVMNPKFAES 769 || | | ||| +|||| || | +|||+#||| || || ||+| Sbjct 756 LTTLHDRREYAKFEKERKLPSGKRAENPLY#KSAKTTFQNPAFAQS 800 |
| Anopheles gambiae | Query 726 LTHLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 || + | ||+ |||||++ ++|+ +||++#| |||| || +| Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYA 835 |
| Anopheles gambiae str. PEST | Query 726 LTHLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 || + | ||+ |||||++ ++|+ +||++#| |||| || +| Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYA 835 |
| Aedes aegypti | Query 726 LTHLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 || + | ||+ |||||++ ++|+ +||++#| |||| || +| Sbjct 791 LTTIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYA 833 |
| Mus sp. | Query 731 DLREYHRFEKEKLKSQWNND-NPLF#KSATTTVMNPKF 766 | ||| |||||+ + | | |||+#||| || +||+| Sbjct 753 DRREYRRFEKEQQQLNWKQDNNPLY#KSAITTTVNPRF 789 |
| Drosophila melanogaster | Query 726 LTHLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 || + | ||+ |||||++ ++|+ +||++#| ||+| || +| Sbjct 802 LTTIHDRREFARFEKERMNAKWDTGENPIY#KQATSTFKNPMYA 844 |
| Ostrinia furnacalis | Query 726 LTHLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 +| + | ||+ |||||++ ++|+ +||++#| ||+| || +| Sbjct 787 VTTIHDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYA 829 |
| Caenorhabditis briggsae | Query 726 LTHLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 || | | || +| |+| ++|+ |+||++#| |||| || +| Sbjct 764 LTVLHDRAEYAKFNNERLMAKWDTNENPIY#KQATTTFKNPVYA 806 |
| Strongylocentrotus purpuratus | Query 727 THLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 768 |++ | ||| ++| + |+||+ +|||++#||+||| || + + Sbjct 760 TYVQDKREYAQWENDCKKAQWDQSDNPIY#KSSTTTFKNPTYGK 802 |
| Tribolium castaneum | Query 727 THLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 766 | + | +|| +|| |+ | ||+ |||||+#+ ||+| || + Sbjct 711 TTVHDRQEYAKFENERQKMQWHRNDNPLY#RQATSTFKNPVY 751 |
| Caenorhabditis elegans | Query 726 LTHLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 || | | || | |+| ++|+ |+||++#| |||| || +| Sbjct 763 LTVLHDRSEYATFNNERLMAKWDTNENPIY#KQATTTFKNPVYA 805 |
| Crassostrea gigas | Query 727 THLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 768 | +|| || ||||| + ++|+ +||++#| ||+| +|| + + Sbjct 757 TTISDKRELARFEKEAMNARWDTGENPIY#KQATSTFVNPTYRQ 799 |
| Apis mellifera | Query 726 LTHLSDLREYHRFEKEK-LKSQWNN-DNPLF#KSATTTVMNPKF 766 || + | ||+ +||+|+ | ++|| ||||+#| ||+| || | Sbjct 771 LTMIRDNREFKKFERERILANKWNRRDNPLY#KEATSTFKNPTF 813 |
| Biomphalaria glabrata | Query 726 LTHLSDLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 766 || + | ||+ +||||+ ++|+ +||++#| ||+| || + Sbjct 744 LTFIHDTREFAKFEKERQNAKWDTGENPIY#KQATSTFKNPTY 785 |
| Pseudoplusia includens | Query 731 DLREYHRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 767 | ||+ |||||++ ++|+ +||++#| ||+| || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYA 835 |
| Podocoryne carnea | Query 726 LTHLSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 766 | + | ||+ +|||++ +|++ +||++#| ||+| || + Sbjct 737 LATIQDRREFAKFEKDRQNPKWDSGENPIY#KKATSTFQNPMY 778 |
| Lytechinus variegatus | Query 726 LTHLSDLREYHRFEKEKLKSQW-NNDNPLF#KSATTTVMNPKF 766 ||++ | ||+ ||||+ + | +||++#| +|+ || + Sbjct 762 LTYIHDRREFQNFEKERANATWEGGENPIY#KPSTSVFKNPTY 803 |
| Acropora millepora | Query 731 DLREYHRFEKEKLKSQWNND-NPLF#KSATTTVMNPKFA 767 | || +||+|++ |+| + |||+#++| || || +| Sbjct 751 DRIEYQKFERERMHSKWTREKNPLY#QAAKTTFENPTYA 788 |
| rats, Peptide Partial, 723 aa | Query 726 LTHLSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 | + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 679 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 717 |
| Canis lupus familiaris | Query 726 LTHLSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 | + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 778 |
| mice, Peptide Partial, 680 aa | Query 726 LTHLSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 | + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 640 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 678 |
| Equus caballus | Query 726 LTHLSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMN 763 | + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 778 |
| Schistosoma japonicum | Query 729 LSDLREYHRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKFAES 769 + | || |+++ +| +||+|#+| || |+|| | |+ Sbjct 245 IDDRRELANFKQQGENMRWEMAENPIF#ESPTTNVLNPTFEEN 286 |
| Trichomonas vaginalis G3 | Query 727 THLSDLREYHRFEKEKLKSQWNNDNP-LF#KSATTTVMNPKFAE 768 ||| ||++ |+ | |+ + | | | +#+ ++ ++| |+ | Sbjct 158 THLEDLKQKHQKELEEFEESWKKDKPKQY#RKPSSALLNMKYQE 200 |
* References
[PubMed ID: 18577362] Lecchi C, Ceciliani F, Bernasconi S, Franciosi F, Bronzo V, Sartorelli P, Bovine alpha-1 acid glycoprotein can reduce the chemotaxis of bovine monocytes and modulate CD18 expression. Vet Res. 2008 Sep-Oct;39(5):50. Epub 2008 Jun 26.
[PubMed ID: 17698568] ... Dassanayake RP, Maheswaran SK, Srikumaran S, Monomeric expression of bovine beta2-integrin subunits reveals their role in Mannheimia haemolytica leukotoxin-induced biological effects. Infect Immun. 2007 Oct;75(10):5004-10. Epub 2007 Aug 13.
[PubMed ID: 17590223] ... Dileepan T, Kannan MS, Walcheck B, Maheswaran SK, Integrin-EGF-3 domain of bovine CD18 is critical for Mannheimia haemolytica leukotoxin species-specific susceptibility. FEMS Microbiol Lett. 2007 Sep;274(1):67-72. Epub 2007 Jun 21.
[PubMed ID: 17039534] ... Pate BJ, White KL, Winger QA, Rickords LF, Aston KI, Sessons BR, Li GP, Campbell KD, Weimer B, Bunch TD, Specific integrin subunits in bovine oocytes, including novel sequences for alpha 6 and beta 3 subunits. Mol Reprod Dev. 2007 May;74(5):600-7.
[PubMed ID: 14984592] ... Nagahata H, Higuchi H, Teraoka H, Takahashi K, Kuwabara M, Inanami O, Decreased apoptosis of beta 2- integrin-deficient bovine neutrophils. Immunol Cell Biol. 2004 Feb;82(1):32-7.
[PubMed ID: 12906050] ... Diez-Fraile A, Meyer E, Duchateau L, Burvenich C, L-selectin and beta2-integrin expression on circulating bovine polymorphonuclear leukocytes during endotoxin mastitis. J Dairy Sci. 2003 Jul;86(7):2334-42.
[PubMed ID: 9361228] ... Kriegesmann B, Jansen S, Baumgartner BG, Brenig B, Partial genomic structure of the bovine CD18 gene and the refinement of test for bovine leukocyte adhesion deficiency. J Dairy Sci. 1997 Oct;80(10):2547-9.
[PubMed ID: 1384046] ... Shuster DE, Kehrli ME Jr, Ackermann MR, Gilbert RO, Identification and prevalence of a genetic defect that causes leukocyte adhesion deficiency in Holstein cattle. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9225-9.
[PubMed ID: 1351021] ... Shuster DE, Bosworth BT, Kehrli ME Jr, Sequence of the bovine CD18-encoding cDNA: comparison with the human and murine glycoproteins. Gene. 1992 May 15;114(2):267-71.