XSB0415 : connexin50 [Mesocricetus auratus]
[ CaMP Format ]
This entry is computationally expanded from SB0016
* Basic Information
| Organism | Mesocricetus auratus (golden hamster) |
| Protein Names | Gap junction protein; connexin50 |
| Gene Names | Cx50 |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein |
Entrez Nucleotide |
Entrez Gene |
UniProt |
OMIM |
HGNC |
HPRD |
KEGG |
| AAS83438 |
N/A |
N/A |
Q6PYT1 |
N/A |
N/A |
N/A |
N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 420 aa
Average Mass: 46.885 kDa
Monoisotopic Mass: 46.855 kDa
2. Domain Information
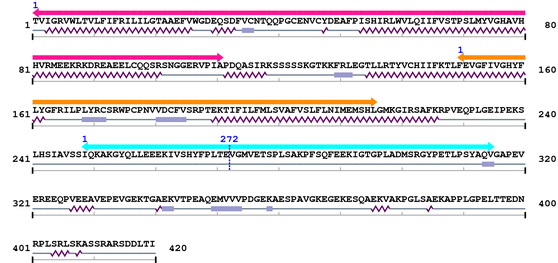
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
| Connexin 1. | 1 | 111 | 246.0 | 9.1e-71 |
| Connexin_CCC 1. | 150 | 216 | 165.8 | 1.3e-46 |
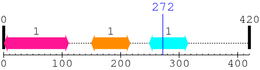
| Connexin50 1. | 249 | 315 | 182.5 | 1.2e-51 |
| --- cleavage 272 (inside Connexin50 249..315) --- |
3. Sequence Information
Fasta Sequence: XSB0415.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] IVSHYFPLTE272-VGMVETSPLS
Glu272  Val
Val
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| Ile263 | Val264 | Ser265 | His266 | Tyr267 | Phe268 | Pro269 | Leu270 | Thr271 | Glu272 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Val273 | Gly274 | Met275 | Val276 | Glu277 | Thr278 | Ser279 | Pro280 | Leu281 | Ser282 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
SIAVSSIQKAKGYQLLEEEKIVSHYFPLTEVGMVETSPLSAKPFSQFEEKIGTGPLADMS
|
Summary
| # |
organism |
max score |
hits |
top seq |
| 1 |
Rattus norvegicus |
119.00 |
1 |
gap junction membrane channel protein alpha 8 |
| 2 |
Mesocricetus auratus |
119.00 |
1 |
connexin50 |
| 3 |
Mus musculus |
119.00 |
1 |
gap junction membrane channel protein alpha 8 |
| 4 |
Cricetulus griseus |
119.00 |
1 |
connexin50 |
| 5 |
Canis familiaris |
115.00 |
6 |
PREDICTED: similar to gap junction membrane channe |
| 6 |
N/A |
112.00 |
1 |
C |
| 7 |
Ovis aries |
112.00 |
1 |
AF177913_1 connexin 49 protein |
| 8 |
Bos taurus |
112.00 |
1 |
PREDICTED: similar to Gap junction alpha-8 protein |
| 9 |
Homo sapiens |
111.00 |
3 |
AAA77062.1 |
| 10 |
Pan troglodytes |
111.00 |
1 |
PREDICTED: gap junction protein, alpha 8, 50kDa (c |
| 11 |
Monodelphis domestica |
105.00 |
1 |
PREDICTED: similar to Gap junction alpha-8 protein |
| 12 |
Macaca mulatta |
97.40 |
1 |
PREDICTED: gap junction protein, alpha 8, 50kDa (c |
| 13 |
Gallus gallus |
82.40 |
1 |
gap junction protein, alpha 8, 50kDa (connexin 50) |
| 14 |
Danio rerio |
63.90 |
3 |
AF288817_1 gap junction protein connexin 44.1 |
| 15 |
Tetraodon nigroviridis |
47.80 |
1 |
unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|
| Rattus norvegicus |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
|
| Mesocricetus auratus |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
|
| Mus musculus |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
|
| Cricetulus griseus |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
|
| Canis familiaris |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#||||||||||||||||||||+||||| |+|
Sbjct 241 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 300
|
| N/A |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#||||| ||||||||||||||+| ||| |+|
Sbjct 261 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVEASPLSAKPFSQFEEKVGPGPLGDLS 320
|
| Ovis aries |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#||||| ||||||||||||||+| ||| |+|
Sbjct 261 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVEASPLSAKPFSQFEEKVGPGPLGDLS 320
|
| Bos taurus |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#||||| ||||||||||||||+| ||| |+|
Sbjct 261 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVEASPLSAKPFSQFEEKVGPGPLGDLS 320
|
| Homo sapiens |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#||||||||| ||||+|||||| |||| |+|
Sbjct 253 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLPAKPFNQFEEKISTGPLGDLS 312
|
| Pan troglodytes |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#||||||||| ||||+|||||| |||| |+|
Sbjct 254 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLPAKPFNQFEEKISTGPLGDLS 313
|
| Monodelphis domestica |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#||+||| ||+| |||+||||| |||| |+|
Sbjct 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGVVETRPLAATPFSRFEEKISTGPLGDLS 319
|
| Macaca mulatta |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
||||||||||||||||||||||||||||||#|||||| ||||||||||| ||| |+|
Sbjct 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVET----AKPFSQFEEKITTGPPEDLS 315
|
| Gallus gallus |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
+|||||| |||||+|||||| |||||||||#|| || ||| + |++|||||| ||| |+|
Sbjct 256 AIAVSSIPKAKGYKLLEEEKPVSHYFPLTE#VG-VEPSPLPS-AFNEFEEKIGMGPLEDLS 313
|
| Danio rerio |
Query 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKI--GTGPLAD 300
||| |||||||||+||||+| ||+|||||#|| +| | | + |||| | |
Sbjct 262 SIAASSIQKAKGYKLLEEDKSTSHFFPLTE#VGGMEAGRLPAS-YEPFEEKSDEAMAPKKD 320
Query 301 MS 302
||
Query 301 MS 302
|
| Tetraodon nigroviridis |
Query 243 SIAVSSIQKAKGYQLLEEEKI--VSHYFPLTE#VGMVETSPLSAKPFSQFEEK 292
|+|| |+|+ |||+|||||| ++| +|| |#||| + + || |||
Sbjct 268 SLAVPSMQRVKGYRLLEEEKAPPITHLYPLAE#VGM--EAGRGSPPFQGLEEK 317
|
* References
[PubMed ID: 16194882] Cruciani V, Heintz KM, Husoy T, Hovig E, Warren DJ, Mikalsen SO, The detection of hamster connexins: a comparison of expression profiles with wild-type mouse and the cancer-prone Min mouse. Cell Commun Adhes. 2004 Sep-Dec;11(5-6):155-71.
 Val
Val

 Sequence conservation (by blast)
Sequence conservation (by blast)