XSB0416 : ITGB2 protein [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0002
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Integrin beta; ITGB2 protein |
| Gene Names | ITGB2 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Description: The leukocyte cell adhesion molecule belongs to the class of cell membrane glycoproteins known as integrins, which are alpha-beta heterodimers. The alpha subunits vary in size from 120 to 180 kD and each is noncovalently associated with a beta subunit (90 to 110 kD). There are 8 known beta subunits and 14 known alpha subunits. Although the alpha and beta subunits could in theory associate to give more than 100 integrin heterodimers, the diversity is restricted and different combinations are associated with different cell types (Hynes, 1992).
Function: By quantitative fluorescence flow cytometric analysis, Taylor et al. (1988) showed that the expression of CD18 was increased in lymphoblastoid cells from persons with Down syndrome, consistent with the location of the gene on chromosome 21.
Function: Bianchi et al. (2000) showed that JAB1 (OMIM:604850) interacts with the cytoplasmic domain of the beta-2 subunit of the alpha-L/beta-2 integrin LFA-1. They demonstrated that a fraction of JAB1 colocalizes with LFA-1 at the cell membrane and that LFA-1 engagement is followed by an increase of the nuclear pool of JAB1, paralleled by enhanced binding of c-Jun-containing AP1 complexes to their DNA consensus site and increased transactivation of an AP1-dependent promoter. Bianchi et al. (2000) suggested that signaling through the LFA-1 integrin may affect c-Jun-driven transcription by regulating JAB1 nuclear localization. This represented a new pathway for integrin-dependent modulation of gene expression.
Function: By yeast 2-hybrid analysis and leukocyte adhesion assays, Ostermann et al. (2002) demonstrated that under both static and physiologic flow conditions, JAM1 (OMIM:605721), through its membrane-proximal domain 2, is a ligand of the LFA-1 integrin that contributes to the LFA-1-dependent transendothelial migration of CD45RO (OMIM:151460)-positive memory T cells expressing the CXCR4 (OMIM:162643) chemokine receptor and of neutrophils. These interactions also facilitated LFA-1-mediated arrest of T cells. Activation of endothelium with inflammatory cytokines enhanced memory T-cell transmigration. Ostermann et al. (2002) suggested that a complex interplay of heterophilic binding of LFA-1 to JAM1 and homophilic trans-interactions of JAM1 may provide a molecular 'zipper' for leukocyte transmigration.
Function: Lu and Cyster (2002) studied the mechanisms that control localization of marginal zone B cells. They demonstrated that marginal zone B cells express elevated levels of the integrins LFA-1 and alpha-4-beta-1 (see OMIM:192975 and OMIM:135630), and that the marginal zone B cells bind to the ligands ICAM1 (OMIM:147840) and VCAM1 (OMIM:192225). These ligands are expressed within the marginal zone in a lymphotoxin-dependent manner. Combined inhibition of LFA-1 and alpha-4-beta-1 causes a rapid and selective release of B cells from the marginal zone. Furthermore, lipopolysaccharide-triggered marginal zone B cell relocalization involves downregulation of integrin-mediated adhesion. Lu and Cyster (2002) concluded that their studies identified key requirements for marginal zone B cell localization and established a role for integrins in peripheral lymphoid tissue compartmentalization.
Function: In a patient with features of Glanzmann thrombasthenia (see OMIM:173470) and leukocyte adhesion deficiency-1 (LAD1; OMIM:116920), McDowall et al. (2003) identified a novel form of integrin dysfunction involving ITGB1 (OMIM:135630), ITGB2, and ITGB3 (OMIM:173470). ITGB2 and ITGB3 were constitutively clustered. Although all 3 integrins were expressed on the cell surface at normal levels and were capable of function following extracellular stimulation, they could not be activated via the 'inside-out' signaling pathways.
Function: Kim et al. (2003) investigated cytoplasmic conformational changes in the integrin LFA1 (alpha-L, OMIM:153370; beta-2) in living cells by measuring fluorescence resonance energy transfer between cyan fluorescent protein-fused and yellow fluorescent protein-fused alpha-L and beta-2 cytoplasmic domains. In the resting state these domains were close to each other, but underwent significant spatial separation upon either intracellular activation of integrin adhesiveness (inside-out signaling) or ligand binding (outside-in signaling). Thus, bidirectional integrin signaling is accomplished by coupling extracellular conformational changes to an unclasping and separation of the alpha and beta cytoplasmic domains, which Kim et al. (2003) noted as a distinctive mechanism for transmitting information across the plasma membrane.
Function: Cherry et al. (2004) generated T-cell clones expressing less than half the wildtype amount of RHOH (OMIM:602037), a leukocyte-specific inhibitory Rho family member. Resting cells expressed constitutively adhesive LFA1 and bound spontaneously to ICAM1, ICAM2 (OMIM:146630), and ICAM3 (OMIM:146631). Reconstituting RHOH mRNA levels reverted the adhesion phenotype to that of relatively nonadhesive wildtype cells. Treatment of peripheral blood lymphocytes with RHOH RNA interference altered the nonadhesive phenotype. Cherry et al. (2004) concluded that RHOH is required for maintenance of lymphocyte LFA1 in a nonadhesive state.
Function: Lammermann et al. (2008) studied the interplay between adhesive, contractile, and protrusive forces during interstitial leukocyte chemotaxis in vivo and in vitro. The authors ablated genes encoding integrin heterodimeric partners ITGA5 (OMIM:135620), ITGB1 (OMIM:135630), ITGB2, and ITGB7 (OMIM:147559) from murine leukocytes and demonstrated that functional integrins do not contribute to migration in 3-dimensional environments. Instead, these cells migrate by the sole force of actin network expansion, which promotes protrusive flowing of the leading edge. Myosin II-dependent contraction is required only on passage through narrow gaps, where a squeezing contraction of the trailing edge propels the rigid nucleus.
* Structure Information
1. Primary Information
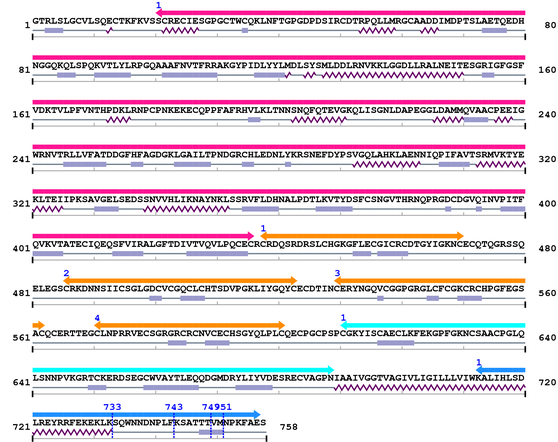
Length: 758 aa
Average Mass: 83.660 kDa
Monoisotopic Mass: 83.605 kDa
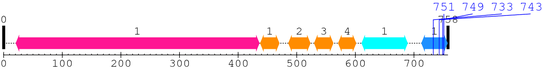
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Integrin_beta 1. | 21 | 436 | 1053.4 | 0 |
| EGF_2 1. | 438 | 470 | 18.7 | 0.02 |
| EGF_2 2. | 486 | 523 | 11.8 | 0.13 |
| EGF_2 3. | 530 | 562 | 16.0 | 0.042 |
| EGF_2 4. | 571 | 601 | 25.7 | 0.00019 |
| Integrin_B_tail 1. | 611 | 689 | 132.0 | 1.8e-36 |
| Integrin_b_cyt 1. | 713 | 757 | 87.8 | 3.9e-23 |
| --- cleavage 751 (inside Integrin_b_cyt 713..757) --- | ||||
| --- cleavage 749 (inside Integrin_b_cyt 713..757) --- | ||||
| --- cleavage 733 (inside Integrin_b_cyt 713..757) --- | ||||
| --- cleavage 743 (inside Integrin_b_cyt 713..757) --- | ||||
3. Sequence Information
Fasta Sequence: XSB0416.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
4 [sites]
Cleavage sites (±10aa)
[Site 1] LFKSATTTVM751-NPKFAES
Met751  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu742 | Phe743 | Lys744 | Ser745 | Ala746 | Thr747 | Thr748 | Thr749 | Val750 | Met751 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn752 | Pro753 | Lys754 | Phe755 | Ala756 | Glu757 | Ser758 | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| REYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 80.50 | 20 | A |
| 2 | Homo sapiens | 80.50 | 14 | ITGB2 protein |
| 3 | Mus musculus | 80.50 | 10 | integrin beta 2 |
| 4 | Pan troglodytes | 80.50 | 6 | integrin, beta 2 |
| 5 | synthetic construct | 80.50 | 3 | integrin, beta 2 (antigen CD18 (p95), lymphocyte f |
| 6 | Rattus norvegicus | 78.60 | 8 | PREDICTED: similar to Integrin beta-2 precursor (C |
| 7 | Bos taurus | 78.60 | 6 | integrin, beta 2 |
| 8 | Ovis canadensis | 78.60 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 9 | Sigmodon hispidus | 78.60 | 1 | AF445415_1 integrin beta-2 precursor |
| 10 | Capra hircus | 78.60 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 11 | Ovis aries | 78.60 | 1 | antigen CD18 |
| 12 | Bubalus bubalis | 78.60 | 1 | integrin beta-2 |
| 13 | Sus scrofa | 77.80 | 4 | CD18 leukocyte adhesion molecule |
| 14 | Canis familiaris | 75.50 | 4 | AF181965_1 B-2 integrin |
| 15 | Gallus gallus | 62.40 | 5 | integrin, beta 2 |
| 16 | Monodelphis domestica | 61.20 | 4 | PREDICTED: similar to Stromal interaction molecule |
| 17 | Xenopus laevis | 48.90 | 5 | hypothetical protein LOC379708 |
| 18 | Mus sp. | 48.90 | 1 | beta 7 integrin |
| 19 | Macaca mulatta | 48.50 | 2 | PREDICTED: integrin, beta 7 |
| 20 | Danio rerio | 48.10 | 12 | integrin, beta 1b |
| 21 | Tetraodon nigroviridis | 48.10 | 5 | unnamed protein product |
| 22 | Oryctolagus cuniculus | 48.10 | 2 | integrin beta 1 |
| 23 | Ictalurus punctatus | 48.10 | 2 | beta-1 integrin |
| 24 | Xenopus tropicalis | 48.10 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 25 | Felis catus | 48.10 | 1 | integrin beta 1 |
| 26 | Pongo pygmaeus | 48.10 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 27 | Oncorhynchus mykiss | 47.00 | 1 | CD18 protein |
| 28 | Anopheles gambiae | 44.70 | 1 | integrin beta subunit |
| 29 | Anopheles gambiae str. PEST | 44.70 | 1 | ENSANGP00000021373 |
| 30 | Aedes aegypti | 44.70 | 1 | integrin beta subunit |
| 31 | Pacifastacus leniusculus | 44.30 | 1 | integrin |
| 32 | Cyprinus carpio | 43.90 | 2 | CD18 type 2 |
| 33 | Drosophila melanogaster | 43.50 | 1 | myospheroid CG1560-PA |
| 34 | Tribolium castaneum | 43.10 | 2 | PREDICTED: similar to CG1560-PA |
| 35 | Pseudoplusia includens | 43.10 | 1 | integrin beta 1 |
| 36 | Ostrinia furnacalis | 43.10 | 1 | integrin beta 1 precursor |
| 37 | Caenorhabditis briggsae | 42.00 | 1 | Hypothetical protein CBG03601 |
| 38 | Apis mellifera | 42.00 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 39 | Strongylocentrotus purpuratus | 41.20 | 5 | integrin beta L subunit |
| 40 | Caenorhabditis elegans | 40.80 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 41 | Crassostrea gigas | 40.40 | 1 | integrin beta cgh |
| 42 | Acropora millepora | 40.40 | 1 | integrin subunit betaCn1 |
| 43 | Biomphalaria glabrata | 39.30 | 1 | beta integrin subunit |
| 44 | Podocoryne carnea | 38.10 | 1 | AF308652_1 integrin beta chain |
| 45 | Lytechinus variegatus | 35.00 | 1 | beta-C integrin subunit |
| 46 | mice, Peptide Partial, 680 aa | 34.70 | 1 | beta 3 integrin, GPIIIA |
| 47 | rats, Peptide Partial, 723 aa | 34.70 | 1 | beta 3 integrin, GPIIIA |
| 48 | Canis lupus familiaris | 34.70 | 1 | integrin beta chain, beta 3 |
| 49 | Equus caballus | 34.70 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 50 | Schistosoma japonicum | 33.90 | 1 | SJCHGC06221 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||||||||||||||||||||||||||||||#||||||| Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 |
| Homo sapiens | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||||||||||||||||||||||||||||||#||||||| Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 |
| Mus musculus | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||||||||||||||||||||||||||||||#||||||| Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 |
| Pan troglodytes | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||||||||||||||||||||||||||||||#||||||| Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 |
| synthetic construct | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||||||||||||||||||||||||||||||#||||||| Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 |
| Rattus norvegicus | Query 723 EYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 |||||||||||||||||||||||||||||#||||||| Query 723 EYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 |
| Bos taurus | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||| ||||||||||||||||||||||||||#||||||| Sbjct 733 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Ovis canadensis | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||| ||||||||||||||||||||||||||#||||||| Sbjct 734 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Sigmodon hispidus | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 |||| |||||||||||||||||||||||||#||||||| Sbjct 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Capra hircus | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||| ||||||||||||||||||||||||||#||||||| Sbjct 734 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Ovis aries | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||| ||||||||||||||||||||||||||#||||||| Sbjct 734 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Bubalus bubalis | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||| ||||||||||||||||||||||||||#||||||| Sbjct 733 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Sus scrofa | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAE 757 |||+||||||||||||||||||||||||||#|||||| Sbjct 733 REYKRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAE 768 |
| Canis familiaris | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||++|||||||+||||||||||||||||||#||+|||| Sbjct 728 REFKRFEKEKLRSQWNNDNPLFKSATTTVM#NPRFAES 764 |
| Gallus gallus | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 755 |||||||||| |++|| |||||||||||||#||+| Sbjct 735 REYRRFEKEKSKAKWNEADNPLFKSATTTVM#NPRF 769 |
| Monodelphis domestica | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NP 753 |||+||||||||||||||||||| |||++# | Sbjct 1199 REYKRFEKEKLKSQWNNDNPLFKKKTTTII#IP 1230 |
| Xenopus laevis | Query 723 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 756 ||+|||||++ |+|| |||+ +||||| #|| |+ Sbjct 736 EYQRFEKERMNSKWNPGHNPLYHNATTTVQ#NPNFS 770 |
| Mus sp. | Query 722 REYRRFEKEKLKSQWNND-NPLFKSATTTVM#NPKF 755 |||||||||+ + | | |||+||| || +#||+| Sbjct 755 REYRRFEKEQQQLNWKQDNNPLYKSAITTTV#NPRF 789 |
| Macaca mulatta | Query 722 REYRRFEKEKLKSQWNND-NPLFKSATTTVM#NPKFAES 758 ||| |||||+ + | | |||+||| || +#||+| |+ Sbjct 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTTI#NPRFQEA 669 |
| Danio rerio | Query 722 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 755 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 758 REFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKY 792 |
| Tetraodon nigroviridis | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 755 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 754 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 788 |
| Oryctolagus cuniculus | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 755 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 26 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 60 |
| Ictalurus punctatus | Query 722 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 755 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 770 REFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKY 804 |
| Xenopus tropicalis | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 755 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Felis catus | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 755 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Pongo pygmaeus | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 755 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Oncorhynchus mykiss | Query 722 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 755 +|+|||| |+ | +|+ ||||| +||||| #|| | Sbjct 740 KEFRRFENEQKKGKWSPGDNPLFMNATTTVA#NPTF 774 |
| Anopheles gambiae | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 756 ||+ |||||++ ++|+ +||++| |||| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYA 835 |
| Anopheles gambiae str. PEST | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 756 ||+ |||||++ ++|+ +||++| |||| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYA 835 |
| Aedes aegypti | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 756 ||+ |||||++ ++|+ +||++| |||| #|| +| Sbjct 798 REFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYA 833 |
| Pacifastacus leniusculus | Query 722 REYRRFEKE-KLKSQWNNDNPLFKSATTTVM#NPKFAES 758 ||| +|||| || | +|||+||| || #|| ||+| Sbjct 763 REYAKFEKERKLPSGKRAENPLYKSAKTTFQ#NPAFAQS 800 |
| Cyprinus carpio | Query 722 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFA 756 ||++||||++ + + ||||++||||| #|| |+ Sbjct 730 REWKRFEKDRKHEKTSGTNPLFQNATTTVQ#NPTFS 764 |
| Drosophila melanogaster | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 756 ||+ |||||++ ++|+ +||++| ||+| #|| +| Sbjct 809 REFARFEKERMNAKWDTGENPIYKQATSTFK#NPMYA 844 |
| Tribolium castaneum | Query 722 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 755 +|| +|| |+ | ||+ |||||++ ||+| #|| + Sbjct 717 QEYAKFENERQKMQWHRNDNPLYRQATSTFK#NPVY 751 |
| Pseudoplusia includens | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 756 ||+ |||||++ ++|+ +||++| ||+| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYA 835 |
| Ostrinia furnacalis | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 756 ||+ |||||++ ++|+ +||++| ||+| #|| +| Sbjct 794 REFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYA 829 |
| Caenorhabditis briggsae | Query 723 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 756 || +| |+| ++|+ |+||++| |||| #|| +| Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTFK#NPVYA 806 |
| Apis mellifera | Query 722 REYRRFEKEK-LKSQWNN-DNPLFKSATTTVM#NPKF 755 ||+++||+|+ | ++|| ||||+| ||+| #|| | Sbjct 778 REFKKFERERILANKWNRRDNPLYKEATSTFK#NPTF 813 |
| Strongylocentrotus purpuratus | Query 722 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFAE 757 ||| ++| + |+||+ +|||++||+||| #|| + + Sbjct 766 REYAQWENDCKKAQWDQSDNPIYKSSTTTFK#NPTYGK 802 |
| Caenorhabditis elegans | Query 723 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 756 || | |+| ++|+ |+||++| |||| #|| +| Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTFK#NPVYA 805 |
| Crassostrea gigas | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFAE 757 || ||||| + ++|+ +||++| ||+| +#|| + + Sbjct 763 RELARFEKEAMNARWDTGENPIYKQATSTFV#NPTYRQ 799 |
| Acropora millepora | Query 723 EYRRFEKEKLKSQWNND-NPLFKSATTTVM#NPKFA 756 ||++||+|++ |+| + |||+++| || #|| +| Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTFE#NPTYA 788 |
| Biomphalaria glabrata | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 755 ||+ +||||+ ++|+ +||++| ||+| #|| + Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATSTFK#NPTY 785 |
| Podocoryne carnea | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 755 ||+ +|||++ +|++ +||++| ||+| #|| + Sbjct 744 REFAKFEKDRQNPKWDSGENPIYKKATSTFQ#NPMY 778 |
| Lytechinus variegatus | Query 722 REYRRFEKEKLKSQW-NNDNPLFKSATTTVM#NPKF 755 ||++ ||||+ + | +||++| +|+ #|| + Sbjct 769 REFQNFEKERANATWEGGENPIYKPSTSVFK#NPTY 803 |
| mice, Peptide Partial, 680 aa | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 752 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 647 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 678 |
| rats, Peptide Partial, 723 aa | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 752 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 686 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 717 |
| Canis lupus familiaris | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 752 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 778 |
| Equus caballus | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 752 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 778 |
| Schistosoma japonicum | Query 722 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFAES 758 || |+++ +| +||+|+| || |+#|| | |+ Sbjct 249 RELANFKQQGENMRWEMAENPIFESPTTNVL#NPTFEEN 286 |
[Site 2] NPLFKSATTT749-VMNPKFAES
Thr749  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn740 | Pro741 | Leu742 | Phe743 | Lys744 | Ser745 | Ala746 | Thr747 | Thr748 | Thr749 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val750 | Met751 | Asn752 | Pro753 | Lys754 | Phe755 | Ala756 | Glu757 | Ser758 | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DLREYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 84.30 | 20 | A |
| 2 | Homo sapiens | 84.30 | 14 | ITGB2 protein |
| 3 | Mus musculus | 84.30 | 10 | integrin beta 2 |
| 4 | Pan troglodytes | 84.30 | 6 | integrin, beta 2 |
| 5 | synthetic construct | 84.30 | 3 | integrin, beta 2 (antigen CD18 (p95), lymphocyte f |
| 6 | Rattus norvegicus | 82.40 | 8 | PREDICTED: similar to Integrin beta-2 precursor (C |
| 7 | Bos taurus | 82.40 | 6 | integrin, beta 2 |
| 8 | Sigmodon hispidus | 82.40 | 1 | AF445415_1 integrin beta-2 precursor |
| 9 | Capra hircus | 82.40 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 10 | Bubalus bubalis | 82.40 | 1 | integrin beta-2 |
| 11 | Ovis aries | 82.40 | 1 | antigen CD18 |
| 12 | Ovis canadensis | 82.40 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 13 | Sus scrofa | 81.60 | 4 | CD18 leukocyte adhesion molecule |
| 14 | Canis familiaris | 79.30 | 4 | AF181965_1 B-2 integrin |
| 15 | Monodelphis domestica | 65.10 | 4 | PREDICTED: similar to Stromal interaction molecule |
| 16 | Gallus gallus | 63.90 | 5 | integrin, beta 2 |
| 17 | Xenopus laevis | 50.40 | 5 | hypothetical protein LOC379708 |
| 18 | Mus sp. | 50.40 | 1 | beta 7 integrin |
| 19 | Macaca mulatta | 50.10 | 2 | PREDICTED: integrin, beta 7 |
| 20 | Felis catus | 50.10 | 1 | integrin beta 1 |
| 21 | Danio rerio | 49.70 | 12 | integrin, beta 1b |
| 22 | Tetraodon nigroviridis | 49.70 | 6 | unnamed protein product |
| 23 | Oryctolagus cuniculus | 49.70 | 2 | integrin beta 1 |
| 24 | Ictalurus punctatus | 49.70 | 2 | beta-1 integrin |
| 25 | Oncorhynchus mykiss | 49.70 | 1 | CD18 protein |
| 26 | Pongo pygmaeus | 49.70 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 27 | Xenopus tropicalis | 49.70 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 28 | Cyprinus carpio | 47.80 | 2 | CD18 type 2 |
| 29 | Anopheles gambiae str. PEST | 46.20 | 1 | ENSANGP00000021373 |
| 30 | Anopheles gambiae | 46.20 | 1 | integrin beta subunit |
| 31 | Aedes aegypti | 46.20 | 1 | integrin beta subunit |
| 32 | Pacifastacus leniusculus | 45.80 | 1 | integrin |
| 33 | Drosophila melanogaster | 45.10 | 1 | myospheroid CG1560-PA |
| 34 | Tribolium castaneum | 44.70 | 3 | PREDICTED: similar to CG1560-PA |
| 35 | Pseudoplusia includens | 44.70 | 1 | integrin beta 1 |
| 36 | Ostrinia furnacalis | 44.70 | 1 | integrin beta 1 precursor |
| 37 | Apis mellifera | 43.10 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 38 | Caenorhabditis briggsae | 43.10 | 1 | Hypothetical protein CBG03601 |
| 39 | Strongylocentrotus purpuratus | 42.70 | 5 | integrin beta L subunit |
| 40 | Caenorhabditis elegans | 42.00 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 41 | Crassostrea gigas | 42.00 | 1 | integrin beta cgh |
| 42 | Biomphalaria glabrata | 41.20 | 1 | beta integrin subunit |
| 43 | Acropora millepora | 40.80 | 1 | integrin subunit betaCn1 |
| 44 | Podocoryne carnea | 39.70 | 1 | AF308652_1 integrin beta chain |
| 45 | Lytechinus variegatus | 36.60 | 1 | beta-C integrin subunit |
| 46 | Equus caballus | 36.20 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 47 | Canis lupus familiaris | 36.20 | 1 | integrin beta chain, beta 3 |
| 48 | rats, Peptide Partial, 723 aa | 36.20 | 1 | beta 3 integrin, GPIIIA |
| 49 | mice, Peptide Partial, 680 aa | 36.20 | 1 | beta 3 integrin, GPIIIA |
| 50 | Schistosoma japonicum | 35.40 | 1 | SJCHGC06221 protein |
| 51 | Ophlitaspongia tenuis | 33.50 | 1 | integrin subunit betaPo1 |
| 52 | Geodia cydonium | 33.10 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 ||||||||||||||||||||||||||||||#||||||||| Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 |
| Homo sapiens | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 ||||||||||||||||||||||||||||||#||||||||| Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 |
| Mus musculus | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 ||||||||||||||||||||||||||||||#||||||||| Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 |
| Pan troglodytes | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 ||||||||||||||||||||||||||||||#||||||||| Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 |
| synthetic construct | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 ||||||||||||||||||||||||||||||#||||||||| Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 |
| Rattus norvegicus | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 || |||||||||||||||||||||||||||#||||||||| Sbjct 730 DLNEYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 768 |
| Bos taurus | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 731 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Sigmodon hispidus | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 |||||| |||||||||||||||||||||||#||||||||| Sbjct 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Capra hircus | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 732 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Bubalus bubalis | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 731 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Ovis aries | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 732 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Ovis canadensis | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 732 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Sus scrofa | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAE 757 |||||+||||||||||||||||||||||||#|||||||| Sbjct 731 DLREYKRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAE 768 |
| Canis familiaris | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 758 ||||++|||||||+||||||||||||||||#||||+|||| Sbjct 726 DLREFKRFEKEKLRSQWNNDNPLFKSATTT#VMNPRFAES 764 |
| Monodelphis domestica | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNP 753 |||||+||||||||||||||||||| |||#++ | Sbjct 1197 DLREYKRFEKEKLKSQWNNDNPLFKKKTTT#IIIP 1230 |
| Gallus gallus | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 755 | |||||||||| |++|| |||||||||||#||||+| Sbjct 733 DRREYRRFEKEKSKAKWNEADNPLFKSATTT#VMNPRF 769 |
| Xenopus laevis | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 756 | ||+|||||++ |+|| |||+ +||||#| || |+ Sbjct 733 DRNEYQRFEKERMNSKWNPGHNPLYHNATTT#VQNPNFS 770 |
| Mus sp. | Query 720 DLREYRRFEKEKLKSQWNND-NPLFKSATTT#VMNPKF 755 | |||||||||+ + | | |||+||| ||# +||+| Sbjct 753 DRREYRRFEKEQQQLNWKQDNNPLYKSAITT#TVNPRF 789 |
| Macaca mulatta | Query 720 DLREYRRFEKEKLKSQWNND-NPLFKSATTT#VMNPKFAES 758 | ||| |||||+ + | | |||+||| ||# +||+| |+ Sbjct 630 DRREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEA 669 |
| Felis catus | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 755 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DTREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Danio rerio | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 755 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 756 DRREFAKFEKEKMNAKWDAGENPIYKSAVTT#VVNPKY 792 |
| Tetraodon nigroviridis | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 755 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 752 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 788 |
| Oryctolagus cuniculus | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 755 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 24 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 60 |
| Ictalurus punctatus | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 755 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 768 DRREFAKFEKEKMNAKWDAGENPIYKSAVTT#VVNPKY 804 |
| Oncorhynchus mykiss | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 755 |++|+|||| |+ | +|+ ||||| +||||#| || | Sbjct 738 DVKEFRRFENEQKKGKWSPGDNPLFMNATTT#VANPTF 774 |
| Pongo pygmaeus | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 755 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Xenopus tropicalis | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 755 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Cyprinus carpio | Query 720 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFA 756 ||||++||||++ + + ||||++||||#| || |+ Sbjct 728 DLREWKRFEKDRKHEKTSGTNPLFQNATTT#VQNPTFS 764 |
| Anopheles gambiae str. PEST | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 756 | ||+ |||||++ ++|+ +||++| ||||# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYA 835 |
| Anopheles gambiae | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 756 | ||+ |||||++ ++|+ +||++| ||||# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYA 835 |
| Aedes aegypti | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 756 | ||+ |||||++ ++|+ +||++| ||||# || +| Sbjct 796 DRREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYA 833 |
| Pacifastacus leniusculus | Query 720 DLREYRRFEKE-KLKSQWNNDNPLFKSATTT#VMNPKFAES 758 | ||| +|||| || | +|||+||| ||# || ||+| Sbjct 761 DRREYAKFEKERKLPSGKRAENPLYKSAKTT#FQNPAFAQS 800 |
| Drosophila melanogaster | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 756 | ||+ |||||++ ++|+ +||++| ||+|# || +| Sbjct 807 DRREFARFEKERMNAKWDTGENPIYKQATST#FKNPMYA 844 |
| Tribolium castaneum | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 755 | +|| +|| |+ | ||+ |||||++ ||+|# || + Sbjct 715 DRQEYAKFENERQKMQWHRNDNPLYRQATST#FKNPVY 751 |
| Pseudoplusia includens | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 756 | ||+ |||||++ ++|+ +||++| ||+|# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYA 835 |
| Ostrinia furnacalis | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 756 | ||+ |||||++ ++|+ +||++| ||+|# || +| Sbjct 792 DRREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYA 829 |
| Apis mellifera | Query 720 DLREYRRFEKEK-LKSQWNN-DNPLFKSATTT#VMNPKF 755 | ||+++||+|+ | ++|| ||||+| ||+|# || | Sbjct 776 DNREFKKFERERILANKWNRRDNPLYKEATST#FKNPTF 813 |
| Caenorhabditis briggsae | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 756 | || +| |+| ++|+ |+||++| ||||# || +| Sbjct 769 DRAEYAKFNNERLMAKWDTNENPIYKQATTT#FKNPVYA 806 |
| Strongylocentrotus purpuratus | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFAE 757 | ||| ++| + |+||+ +|||++||+|||# || + + Sbjct 764 DKREYAQWENDCKKAQWDQSDNPIYKSSTTT#FKNPTYGK 802 |
| Caenorhabditis elegans | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 756 | || | |+| ++|+ |+||++| ||||# || +| Sbjct 768 DRSEYATFNNERLMAKWDTNENPIYKQATTT#FKNPVYA 805 |
| Crassostrea gigas | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFAE 757 | || ||||| + ++|+ +||++| ||+|# +|| + + Sbjct 761 DKRELARFEKEAMNARWDTGENPIYKQATST#FVNPTYRQ 799 |
| Biomphalaria glabrata | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 755 | ||+ +||||+ ++|+ +||++| ||+|# || + Sbjct 749 DTREFAKFEKERQNAKWDTGENPIYKQATST#FKNPTY 785 |
| Acropora millepora | Query 720 DLREYRRFEKEKLKSQWNND-NPLFKSATTT#VMNPKFA 756 | ||++||+|++ |+| + |||+++| ||# || +| Sbjct 751 DRIEYQKFERERMHSKWTREKNPLYQAAKTT#FENPTYA 788 |
| Podocoryne carnea | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 755 | ||+ +|||++ +|++ +||++| ||+|# || + Sbjct 742 DRREFAKFEKDRQNPKWDSGENPIYKKATST#FQNPMY 778 |
| Lytechinus variegatus | Query 720 DLREYRRFEKEKLKSQW-NNDNPLFKSATTT#VMNPKF 755 | ||++ ||||+ + | +||++| +|+ # || + Sbjct 767 DRREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTY 803 |
| Equus caballus | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 752 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 778 |
| Canis lupus familiaris | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 752 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 778 |
| rats, Peptide Partial, 723 aa | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 752 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 684 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 717 |
| mice, Peptide Partial, 680 aa | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 752 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 645 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 678 |
| Schistosoma japonicum | Query 720 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFAES 758 | || |+++ +| +||+|+| || #|+|| | |+ Sbjct 247 DRRELANFKQQGENMRWEMAENPIFESPTTN#VLNPTFEEN 286 |
| Ophlitaspongia tenuis | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFAE 757 |+ | |+||+| +++ |+|||++||| # || + + Sbjct 800 DVVEVRKFEREIKNAKYTKNENPLYRSATKD#YQNPLYGK 838 |
| Geodia cydonium | Query 720 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFAE 757 | | ++|| | | ++ |+|||+ |||| # || + + Sbjct 840 DYTELKKFENELAKVKYTKNENPLYHSATTE#HKNPIYGQ 878 |
[Site 3] YRRFEKEKLK733-SQWNNDNPLF
Lys733  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Tyr724 | Arg725 | Arg726 | Phe727 | Glu728 | Lys729 | Glu730 | Lys731 | Leu732 | Lys733 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser734 | Gln735 | Trp736 | Asn737 | Asn738 | Asp739 | Asn740 | Pro741 | Leu742 | Phe743 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| IGILLLVIWKALIHLSDLREYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 114.00 | 25 | ITGB2 protein |
| 2 | Pan troglodytes | 114.00 | 14 | integrin, beta 2 |
| 3 | synthetic construct | 114.00 | 5 | integrin, beta 2 (antigen CD18 (p95), lymphocyte f |
| 4 | N/A | 110.00 | 28 | - |
| 5 | Mus musculus | 110.00 | 16 | integrin beta 2 |
| 6 | Bos taurus | 110.00 | 13 | integrin, beta 2 |
| 7 | Capra hircus | 110.00 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 8 | Bubalus bubalis | 110.00 | 1 | integrin beta-2 |
| 9 | Rattus norvegicus | 108.00 | 10 | PREDICTED: similar to Integrin beta-2 precursor (C |
| 10 | Ovis aries | 108.00 | 2 | antigen CD18 |
| 11 | Sigmodon hispidus | 108.00 | 1 | AF445415_1 integrin beta-2 precursor |
| 12 | Ovis canadensis | 108.00 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 13 | Sus scrofa | 106.00 | 6 | CD18 leukocyte adhesion molecule |
| 14 | Canis familiaris | 105.00 | 10 | AF181965_1 B-2 integrin |
| 15 | Monodelphis domestica | 87.80 | 6 | PREDICTED: similar to Stromal interaction molecule |
| 16 | Gallus gallus | 79.00 | 7 | integrin, beta 2 |
| 17 | Felis catus | 66.20 | 1 | integrin beta 1 |
| 18 | Danio rerio | 65.90 | 14 | integrin, beta 1b |
| 19 | Tetraodon nigroviridis | 65.90 | 8 | unnamed protein product |
| 20 | Xenopus laevis | 65.90 | 6 | integrin beta-1 subunit |
| 21 | Pongo pygmaeus | 65.90 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 22 | Oryctolagus cuniculus | 65.90 | 2 | integrin beta 1 |
| 23 | Xenopus tropicalis | 65.90 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 24 | Ictalurus punctatus | 65.10 | 2 | beta-1 integrin |
| 25 | Oncorhynchus mykiss | 65.10 | 1 | CD18 protein |
| 26 | Cyprinus carpio | 63.50 | 2 | CD18 type 2 |
| 27 | Pacifastacus leniusculus | 62.40 | 1 | integrin |
| 28 | Anopheles gambiae | 60.50 | 1 | integrin beta subunit |
| 29 | Caenorhabditis briggsae | 60.50 | 1 | Hypothetical protein CBG03601 |
| 30 | Anopheles gambiae str. PEST | 60.50 | 1 | ENSANGP00000021373 |
| 31 | Aedes aegypti | 60.10 | 1 | integrin beta subunit |
| 32 | Caenorhabditis elegans | 59.30 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 33 | Drosophila melanogaster | 58.90 | 1 | myospheroid CG1560-PA |
| 34 | Crassostrea gigas | 58.50 | 1 | integrin beta cgh |
| 35 | Ostrinia furnacalis | 58.20 | 1 | integrin beta 1 precursor |
| 36 | Tribolium castaneum | 56.60 | 3 | PREDICTED: similar to CG1560-PA |
| 37 | Mus sp. | 56.60 | 1 | beta 7 integrin |
| 38 | Biomphalaria glabrata | 56.60 | 1 | beta integrin subunit |
| 39 | Macaca mulatta | 56.20 | 13 | PREDICTED: integrin, beta 7 |
| 40 | Strongylocentrotus purpuratus | 56.20 | 6 | integrin beta G subunit |
| 41 | Pseudoplusia includens | 55.80 | 1 | integrin beta 1 |
| 42 | Podocoryne carnea | 55.80 | 1 | AF308652_1 integrin beta chain |
| 43 | Apis mellifera | 55.50 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 44 | Acropora millepora | 52.40 | 1 | integrin subunit betaCn1 |
| 45 | Lytechinus variegatus | 52.40 | 1 | beta-C integrin subunit |
| 46 | Canis lupus familiaris | 52.00 | 1 | integrin beta chain, beta 3 |
| 47 | mice, Peptide Partial, 680 aa | 52.00 | 1 | beta 3 integrin, GPIIIA |
| 48 | Equus caballus | 52.00 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 49 | rats, Peptide Partial, 723 aa | 52.00 | 1 | beta 3 integrin, GPIIIA |
| 50 | Schistosoma japonicum | 48.50 | 1 | SJCHGC06221 protein |
| 51 | Halocynthia roretzi | 48.10 | 2 | integrin beta Hr2 precursor |
| 52 | Ophlitaspongia tenuis | 45.80 | 1 | integrin subunit betaPo1 |
| 53 | Geodia cydonium | 44.30 | 1 | integrin beta subunit |
| 54 | Spodoptera frugiperda | 43.90 | 1 | integrin beta 1 |
| 55 | Manduca sexta | 41.60 | 1 | plasmatocyte-specific integrin beta 1 |
| 56 | Suberites domuncula | 40.00 | 1 | integrin beta subunit |
| 57 | Drosophila pseudoobscura | 36.60 | 1 | GA14581-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 |
| Pan troglodytes | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 |
| synthetic construct | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 |
| N/A | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 ||+||||||||| ||+||||||||||||||#||||||||||||||||||||||||| Sbjct 717 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 771 |
| Mus musculus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 ||+||||||||| ||+||||||||||||||#||||||||||||||||||||||||| Sbjct 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| Bos taurus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 +||||||||||| |||||||| ||||||||#||||||||||||||||||||||||| Sbjct 715 VGILLLVIWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Capra hircus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 +||||||||||| |||||||| ||||||||#||||||||||||||||||||||||| Sbjct 716 VGILLLVIWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| Bubalus bubalis | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 +||||||||||| |||||||| ||||||||#||||||||||||||||||||||||| Sbjct 715 VGILLLVIWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Rattus norvegicus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 ||+||||||||| ||+|| |||||||||||#||||||||||||||||||||||||| Sbjct 714 IGVLLLVIWKALTHLTDLNEYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 768 |
| Ovis aries | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 +||||| ||||| |||||||| ||||||||#||||||||||||||||||||||||| Sbjct 716 VGILLLAIWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| Sigmodon hispidus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 ||+||||||||| ||+|||||| |||||||#||||||||||||||||||||||||| Sbjct 715 IGVLLLVIWKALTHLTDLREYRHFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Ovis canadensis | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 +||||| ||||| |||||||| ||||||||#||||||||||||||||||||||||| Sbjct 716 VGILLLAIWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| Sus scrofa | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAE 757 +|| |||||| | ||||||||+||||||||#|||||||||||||||||||||||| Sbjct 715 VGIFLLVIWKVLTHLSDLREYKRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAE 768 |
| Canis familiaris | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 758 |||||| ||||| |||||||++|||||||+#||||||||||||||||||||+|||| Sbjct 710 IGILLLAIWKALTHLSDLREFKRFEKEKLR#SQWNNDNPLFKSATTTVMNPRFAES 764 |
| Monodelphis domestica | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNP 753 ||+||| +|| | | ||||||+||||||||#||||||||||| |||++ | Sbjct 1181 IGVLLLAVWKVLTHFSDLREYKRFEKEKLK#SQWNNDNPLFKKKTTTIIIP 1230 |
| Gallus gallus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKF 755 ||+|||+ |+ | + | |||||||||| |#++|| |||||||||||||||+| Sbjct 717 IGLLLLLTWRLLTEIFDRREYRRFEKEKSK#AKWNEADNPLFKSATTTVMNPRF 769 |
| Felis catus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 ||+ ||+||| |+ + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 743 IGLALLLIWKLLMIIHDTREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 795 |
| Danio rerio | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 ||+ ||+||| |+ + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 740 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDAGENPIYKSAVTTVVNPKY 792 |
| Tetraodon nigroviridis | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 ||+ ||+||| |+ + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 736 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 788 |
| Xenopus laevis | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 ||+ ||+||| |+ + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 795 |
| Pongo pygmaeus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 ||+ ||+||| |+ + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 795 |
| Oryctolagus cuniculus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 ||+ ||+||| |+ + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 8 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 60 |
| Xenopus tropicalis | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 ||+ ||+||| |+ + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 795 |
| Ictalurus punctatus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 ||+ || ||| |+ + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 752 IGLALLFIWKLLMIIHDRREFAKFEKEKMN#AKWDAGENPIYKSAVTTVVNPKY 804 |
| Oncorhynchus mykiss | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 ||+|+|+| ||++++ |++|+|||| |+ |# +|+ ||||| +||||| || | Sbjct 722 IGLLILLIIKAILYMRDVKEFRRFENEQKK#GKWSPGDNPLFMNATTTVANPTF 774 |
| Cyprinus carpio | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFA 756 ||+|+|+| ||+++ |||||++||||++ # + + ||||++||||| || |+ Sbjct 712 IGLLILLIIKAVLYASDLREWKRFEKDRKH#EKTSGTNPLFQNATTTVQNPTFS 764 |
| Pacifastacus leniusculus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKE-KLK#SQWNNDNPLFKSATTTVMNPKFAES 758 ||+| |+||| | | | ||| +|||| || #| +|||+||| || || ||+| Sbjct 745 IGLLTLLIWKLLTTLHDRREYAKFEKERKLP#SGKRAENPLYKSAKTTFQNPAFAQS 800 |
| Anopheles gambiae | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 756 ||+ +|++|| | + | ||+ |||||++ #++|+ +||++| |||| || +| Sbjct 782 IGMAVLLLWKVLTSIHDRREFARFEKERMM#AKWDTGENPIYKQATTTFKNPTYA 835 |
| Caenorhabditis briggsae | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 756 +|||||++|| | | | || +| |+| #++|+ |+||++| |||| || +| Sbjct 753 LGILLLLLWKLLTVLHDRAEYAKFNNERLM#AKWDTNENPIYKQATTTFKNPVYA 806 |
| Anopheles gambiae str. PEST | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 756 ||+ +|++|| | + | ||+ |||||++ #++|+ +||++| |||| || +| Sbjct 782 IGMAVLLLWKVLTSIHDRREFARFEKERMM#AKWDTGENPIYKQATTTFKNPTYA 835 |
| Aedes aegypti | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 756 ||+ +|++|| | + | ||+ |||||++ #++|+ +||++| |||| || +| Sbjct 780 IGLAVLLLWKLLTTIHDRREFARFEKERMM#AKWDTGENPIYKQATTTFKNPTYA 833 |
| Caenorhabditis elegans | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 756 +|||||++|| | | | || | |+| #++|+ |+||++| |||| || +| Sbjct 752 LGILLLLLWKLLTVLHDRSEYATFNNERLM#AKWDTNENPIYKQATTTFKNPVYA 805 |
| Drosophila melanogaster | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 756 +|+ +|++|| | + | ||+ |||||++ #++|+ +||++| ||+| || +| Sbjct 791 VGLAILLLWKLLTTIHDRREFARFEKERMN#AKWDTGENPIYKQATSTFKNPMYA 844 |
| Crassostrea gigas | Query 705 GILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFAE 757 ||+||+||| +|| || ||||| + #++|+ +||++| ||+| +|| + + Sbjct 746 GIILLLIWKLFTTISDKRELARFEKEAMN#ARWDTGENPIYKQATSTFVNPTYRQ 799 |
| Ostrinia furnacalis | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 756 +|+ ||++|| + + | ||+ |||||++ #++|+ +||++| ||+| || +| Sbjct 776 VGLALLMLWKMVTTIHDRREFARFEKERMM#AKWDTGENPIYKQATSTFKNPTYA 829 |
| Tribolium castaneum | Query 705 GILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 |++||+ || + | +|| +|| |+ |# ||+ |||||++ ||+| || + Sbjct 700 GVILLLTWKVCTTVHDRQEYAKFENERQK#MQWHRNDNPLYRQATSTFKNPVY 751 |
| Mus sp. | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNND-NPLFKSATTTVMNPKF 755 +|+ |++ ++ + + | |||||||||+ +# | | |||+||| || +||+| Sbjct 737 VGLGLVLAYRLSVEIYDRREYRRFEKEQQQ#LNWKQDNNPLYKSAITTTVNPRF 789 |
| Biomphalaria glabrata | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 +|+ ||++|| | + | ||+ +||||+ #++|+ +||++| ||+| || + Sbjct 733 VGLFLLLLWKLLTFIHDTREFAKFEKERQN#AKWDTGENPIYKQATSTFKNPTY 785 |
| Macaca mulatta | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNND-NPLFKSATTTVMNPKFAES 758 +|+ |++ ++ + + | ||| |||||+ +# | | |||+||| || +||+| |+ Sbjct 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQQ#LNWKQDSNPLYKSAITTTINPRFQEA 669 |
| Strongylocentrotus purpuratus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQW-NNDNPLFKSATTTVMNPKF 755 +|+ ||++|+ |+++ | ||+ ||||+ #+ | |+||++| +|+| || + Sbjct 729 VGLALLLVWRLLVYVQDSREFASFEKERAG#THWGQNENPIYKPSTSTFKNPTY 781 |
| Pseudoplusia includens | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 756 +|+ ||++|| | ||+ |||||++ #++|+ +||++| ||+| || +| Sbjct 782 VGLALLMLWKMATTSHDRREFARFEKERMM#AKWDTGENPIYKQATSTFKNPTYA 835 |
| Podocoryne carnea | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKF 755 ||+ ||+||| | + | ||+ +|||++ # +|++ +||++| ||+| || + Sbjct 726 IGLALLLIWKLLATIQDRREFAKFEKDRQN#PKWDSGENPIYKKATSTFQNPMY 778 |
| Apis mellifera | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEK-LK#SQWN-NDNPLFKSATTTVMNPKF 755 || +|+ || | + | ||+++||+|+ | #++|| ||||+| ||+| || | Sbjct 760 IGFAILLTWKILTMIRDNREFKKFERERILA#NKWNRRDNPLYKEATSTFKNPTF 813 |
| Acropora millepora | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNND-NPLFKSATTTVMNPKFA 756 +|+|+|++ | | + | ||++||+|++ #|+| + |||+++| || || +| Sbjct 735 LGLLILIVIKGLFTMVDRIEYQKFERERMH#SKWTREKNPLYQAAKTTFENPTYA 788 |
| Lytechinus variegatus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQW-NNDNPLFKSATTTVMNPKF 755 +|+ ||+||+ | ++ | ||++ ||||+ #+ | +||++| +|+ || + Sbjct 751 VGLALLLIWRLLTYIHDRREFQNFEKERAN#ATWEGGENPIYKPSTSVFKNPTY 803 |
| Canis lupus familiaris | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 752 ||+ |+||| || + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 778 |
| mice, Peptide Partial, 680 aa | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 752 ||+ |+||| || + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 629 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 678 |
| Equus caballus | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 752 ||+ |+||| || + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 778 |
| rats, Peptide Partial, 723 aa | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 752 ||+ |+||| || + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 668 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 717 |
| Schistosoma japonicum | Query 705 GILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKFAES 758 |++||+|+| +| + | || |+++ # +| +||+|+| || |+|| | |+ Sbjct 232 GLILLLIYKLVITIDDRRELANFKQQGEN#MRWEMAENPIFESPTTNVLNPTFEEN 286 |
| Halocynthia roretzi | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQW-NNDNPLFKSATTTVMNPKF 755 |||+ |+||| + | | ||| +|+||+ # +| +||++ +|++ || | Sbjct 779 IGIIALIIWKVIQTLRDKREYEKFKKEEDL#RKWTKGENPVYVNASSKFDNPMF 831 |
| Ophlitaspongia tenuis | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQW-NNDNPLFKSATTTVMNPKFAE 757 +|+|||++ | |+ | |+ | |+||+| #+++ |+|||++||| || + + Sbjct 784 LGLLLLLLLKGLLLLWDVVEVRKFEREIKN#AKYTKNENPLYRSATKDYQNPLYGK 838 |
| Geodia cydonium | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQW-NNDNPLFKSATTTVMNPKFAE 757 +||| |++ | |+ + | | ++|| | |# ++ |+|||+ |||| || + + Sbjct 824 LGILALILLKILLMVLDYTELKKFENELAK#VKYTKNENPLYHSATTEHKNPIYGQ 878 |
| Spodoptera frugiperda | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 |||| ++||| |+ + | +||++|+ | | #+ ++ + |||++ + || + Sbjct 724 IGILTVIIWKILVDMHDKKEYKKFQDEALA#AGYDVSLNPLYQDPSINFSNPVY 776 |
| Manduca sexta | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 755 ||+| |+ || |+ | | ||| +||+| # ++ + |||++ || + Sbjct 709 IGLLTLIAWKILVDLHDKREYAKFEEESRS#RGFDVSLNPLYQEPEINFSNPVY 761 |
| Suberites domuncula | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQW-NNDNPLFKSATTTVMNPKFAE 757 +|++ |++ | + + |+ | ++|||| |#+++ + |||++|| || + + Sbjct 710 LGVIALLLLKLFLFIIDVIEVKKFEKELAK#TKYPKHQNPLYRSAAKDYQNPIYGQ 764 |
| Drosophila pseudoobscura | Query 704 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPK 754 ||+||+ ++ | | ||| |||+|+ |# + + +|||++ + || Sbjct 710 IGLLLIFLFIWCIRRKDAREYARFEEEQAK#-RVSQENPLYRDPVGRYVVPK 759 |
[Site 4] SQWNNDNPLF743-KSATTTVMNP
Phe743  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser734 | Gln735 | Trp736 | Asn737 | Asn738 | Asp739 | Asn740 | Pro741 | Leu742 | Phe743 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys744 | Ser745 | Ala746 | Thr747 | Thr748 | Thr749 | Val750 | Met751 | Asn752 | Pro753 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ALIHLSDLREYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 95.10 | 16 | ITGB2 protein |
| 2 | Pan troglodytes | 95.10 | 11 | integrin, beta 2 |
| 3 | synthetic construct | 95.10 | 4 | integrin, beta 2 (antigen CD18 (p95), lymphocyte f |
| 4 | N/A | 92.00 | 20 | - |
| 5 | Mus musculus | 92.00 | 11 | integrin beta 2 |
| 6 | Bos taurus | 91.30 | 8 | integrin, beta 2 |
| 7 | Ovis aries | 91.30 | 2 | antigen CD18 |
| 8 | Ovis canadensis | 91.30 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 9 | Capra hircus | 91.30 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 10 | Bubalus bubalis | 91.30 | 1 | integrin beta-2 |
| 11 | Rattus norvegicus | 90.10 | 9 | PREDICTED: similar to Integrin beta-2 precursor (C |
| 12 | Sigmodon hispidus | 90.10 | 1 | AF445415_1 integrin beta-2 precursor |
| 13 | Sus scrofa | 89.00 | 5 | CD18 leukocyte adhesion molecule |
| 14 | Canis familiaris | 88.20 | 9 | AF181965_1 B-2 integrin |
| 15 | Monodelphis domestica | 70.90 | 4 | PREDICTED: similar to Stromal interaction molecule |
| 16 | Gallus gallus | 65.10 | 6 | integrin, beta 2 |
| 17 | Oncorhynchus mykiss | 53.90 | 1 | CD18 protein |
| 18 | Cyprinus carpio | 52.40 | 2 | CD18 type 2 |
| 19 | Danio rerio | 52.00 | 12 | PREDICTED: similar to CD18 type 2 |
| 20 | Ictalurus punctatus | 52.00 | 2 | CD18 |
| 21 | Xenopus laevis | 51.60 | 5 | hypothetical protein LOC379708 |
| 22 | Mus sp. | 51.60 | 1 | beta 7 integrin |
| 23 | Macaca mulatta | 51.20 | 8 | PREDICTED: integrin, beta 7 |
| 24 | Felis catus | 51.20 | 1 | integrin beta 1 |
| 25 | Tetraodon nigroviridis | 50.80 | 6 | unnamed protein product |
| 26 | Oryctolagus cuniculus | 50.80 | 2 | integrin beta 1 |
| 27 | Pongo pygmaeus | 50.80 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 28 | Xenopus tropicalis | 50.80 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 29 | Anopheles gambiae str. PEST | 47.40 | 1 | ENSANGP00000021373 |
| 30 | Pacifastacus leniusculus | 47.40 | 1 | integrin |
| 31 | Anopheles gambiae | 47.40 | 1 | integrin beta subunit |
| 32 | Aedes aegypti | 47.00 | 1 | integrin beta subunit |
| 33 | Drosophila melanogaster | 45.80 | 1 | myospheroid CG1560-PA |
| 34 | Ostrinia furnacalis | 45.10 | 1 | integrin beta 1 precursor |
| 35 | Tribolium castaneum | 44.70 | 3 | PREDICTED: similar to CG1560-PA |
| 36 | Pseudoplusia includens | 44.70 | 1 | integrin beta 1 |
| 37 | Crassostrea gigas | 44.30 | 1 | integrin beta cgh |
| 38 | Caenorhabditis briggsae | 44.30 | 1 | Hypothetical protein CBG03601 |
| 39 | Strongylocentrotus purpuratus | 43.90 | 5 | integrin beta L subunit |
| 40 | Apis mellifera | 43.90 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 41 | Caenorhabditis elegans | 43.10 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 42 | Biomphalaria glabrata | 42.40 | 1 | beta integrin subunit |
| 43 | Acropora millepora | 41.60 | 1 | integrin subunit betaCn1 |
| 44 | Podocoryne carnea | 40.80 | 1 | AF308652_1 integrin beta chain |
| 45 | Canis lupus familiaris | 38.90 | 1 | integrin beta chain, beta 3 |
| 46 | Equus caballus | 38.90 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 47 | Lytechinus variegatus | 38.90 | 1 | beta-C integrin subunit |
| 48 | mice, Peptide Partial, 680 aa | 38.90 | 1 | beta 3 integrin, GPIIIA |
| 49 | rats, Peptide Partial, 723 aa | 38.90 | 1 | beta 3 integrin, GPIIIA |
| 50 | Schistosoma japonicum | 37.40 | 1 | SJCHGC06221 protein |
| 51 | Ophlitaspongia tenuis | 35.00 | 1 | integrin subunit betaPo1 |
| 52 | Geodia cydonium | 34.30 | 1 | integrin beta subunit |
| 53 | Halocynthia roretzi | 33.10 | 1 | integrin beta Hr2 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 ||||||||||||||||||||||||||||||#||||||||||||||| Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 |
| Pan troglodytes | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 ||||||||||||||||||||||||||||||#||||||||||||||| Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 |
| synthetic construct | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 ||||||||||||||||||||||||||||||#||||||||||||||| Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 |
| N/A | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 || ||+||||||||||||||||||||||||#||||||||||||||| Sbjct 727 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 771 |
| Mus musculus | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 || ||+||||||||||||||||||||||||#||||||||||||||| Sbjct 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Bos taurus | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 || |||||||| ||||||||||||||||||#||||||||||||||| Sbjct 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Ovis aries | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 || |||||||| ||||||||||||||||||#||||||||||||||| Sbjct 726 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Ovis canadensis | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 || |||||||| ||||||||||||||||||#||||||||||||||| Sbjct 726 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Capra hircus | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 || |||||||| ||||||||||||||||||#||||||||||||||| Sbjct 726 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Bubalus bubalis | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 || |||||||| ||||||||||||||||||#||||||||||||||| Sbjct 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Rattus norvegicus | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 || ||+|| |||||||||||||||||||||#||||||||||||||| Sbjct 724 ALTHLTDLNEYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 768 |
| Sigmodon hispidus | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 || ||+|||||| |||||||||||||||||#||||||||||||||| Sbjct 725 ALTHLTDLREYRHFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Sus scrofa | Query 715 LIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAE 757 | ||||||||+||||||||||||||||||#|||||||||||||| Sbjct 726 LTHLSDLREYKRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAE 768 |
| Canis familiaris | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 758 || |||||||++|||||||+||||||||||#||||||||||+|||| Sbjct 720 ALTHLSDLREFKRFEKEKLRSQWNNDNPLF#KSATTTVMNPRFAES 764 |
| Monodelphis domestica | Query 715 LIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNP 753 | | ||||||+||||||||||||||||||#| |||++ | Sbjct 1192 LTHFSDLREYKRFEKEKLKSQWNNDNPLF#KKKTTTIIIP 1230 |
| Gallus gallus | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 755 | + | |||||||||| |++|| |||||#||||||||||+| Sbjct 728 LTEIFDRREYRRFEKEKSKAKWNEADNPLF#KSATTTVMNPRF 769 |
| Oncorhynchus mykiss | Query 714 ALIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 755 |++++ |++|+|||| |+ | +|+ |||||# +||||| || | Sbjct 732 AILYMRDVKEFRRFENEQKKGKWSPGDNPLF#MNATTTVANPTF 774 |
| Cyprinus carpio | Query 714 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFA 756 |+++ |||||++||||++ + + ||||#++||||| || |+ Sbjct 722 AVLYASDLREWKRFEKDRKHEKTSGTNPLF#QNATTTVQNPTFS 764 |
| Danio rerio | Query 715 LIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFA 756 +++ |||||+|||||++ + + ||||#++||||| || |+ Sbjct 610 VLYASDLREWRRFEKDRKHEKTSGTNPLF#QNATTTVQNPTFS 651 |
| Ictalurus punctatus | Query 714 ALIHLSDLREYRRFEKEKLKSQW-NNDNPLF#KSATTTVMNPKF 755 || + ||+|+++|||| + || +||||#++||||| || | Sbjct 726 ALFYFKDLKEWKKFEKEAQRRQWAKGENPLF#QNATTTVANPAF 768 |
| Xenopus laevis | Query 715 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 756 + + | ||+|||||++ |+|| |||+# +||||| || |+ Sbjct 728 ITEIKDRNEYQRFEKERMNSKWNPGHNPLY#HNATTTVQNPNFS 770 |
| Mus sp. | Query 716 IHLSDLREYRRFEKEKLKSQWNND-NPLF#KSATTTVMNPKF 755 + + | |||||||||+ + | | |||+#||| || +||+| Sbjct 749 VEIYDRREYRRFEKEQQQLNWKQDNNPLY#KSAITTTVNPRF 789 |
| Macaca mulatta | Query 716 IHLSDLREYRRFEKEKLKSQWNND-NPLF#KSATTTVMNPKFAES 758 + + | ||| |||||+ + | | |||+#||| || +||+| |+ Sbjct 626 VEIYDRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEA 669 |
| Felis catus | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 755 |+ + | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 754 LMIIHDTREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Tetraodon nigroviridis | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 755 |+ + | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 747 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 788 |
| Oryctolagus cuniculus | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 755 |+ + | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 19 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 60 |
| Pongo pygmaeus | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 755 |+ + | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Xenopus tropicalis | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 755 |+ + | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Anopheles gambiae str. PEST | Query 715 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 756 | + | ||+ |||||++ ++|+ +||++#| |||| || +| Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYA 835 |
| Pacifastacus leniusculus | Query 715 LIHLSDLREYRRFEKE-KLKSQWNNDNPLF#KSATTTVMNPKFAES 758 | | | ||| +|||| || | +|||+#||| || || ||+| Sbjct 756 LTTLHDRREYAKFEKERKLPSGKRAENPLY#KSAKTTFQNPAFAQS 800 |
| Anopheles gambiae | Query 715 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 756 | + | ||+ |||||++ ++|+ +||++#| |||| || +| Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYA 835 |
| Aedes aegypti | Query 715 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 756 | + | ||+ |||||++ ++|+ +||++#| |||| || +| Sbjct 791 LTTIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYA 833 |
| Drosophila melanogaster | Query 715 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 756 | + | ||+ |||||++ ++|+ +||++#| ||+| || +| Sbjct 802 LTTIHDRREFARFEKERMNAKWDTGENPIY#KQATSTFKNPMYA 844 |
| Ostrinia furnacalis | Query 718 LSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 756 + | ||+ |||||++ ++|+ +||++#| ||+| || +| Sbjct 790 IHDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYA 829 |
| Tribolium castaneum | Query 720 DLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 755 | +|| +|| |+ | ||+ |||||+#+ ||+| || + Sbjct 715 DRQEYAKFENERQKMQWHRNDNPLY#RQATSTFKNPVY 751 |
| Pseudoplusia includens | Query 720 DLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 756 | ||+ |||||++ ++|+ +||++#| ||+| || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYA 835 |
| Crassostrea gigas | Query 718 LSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 757 +|| || ||||| + ++|+ +||++#| ||+| +|| + + Sbjct 759 ISDKRELARFEKEAMNARWDTGENPIY#KQATSTFVNPTYRQ 799 |
| Caenorhabditis briggsae | Query 718 LSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 756 | | || +| |+| ++|+ |+||++#| |||| || +| Sbjct 767 LHDRAEYAKFNNERLMAKWDTNENPIY#KQATTTFKNPVYA 806 |
| Strongylocentrotus purpuratus | Query 717 HLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 757 ++ | ||| ++| + |+||+ +|||++#||+||| || + + Sbjct 761 YVQDKREYAQWENDCKKAQWDQSDNPIY#KSSTTTFKNPTYGK 802 |
| Apis mellifera | Query 715 LIHLSDLREYRRFEKEK-LKSQWNN-DNPLF#KSATTTVMNPKF 755 | + | ||+++||+|+ | ++|| ||||+#| ||+| || | Sbjct 771 LTMIRDNREFKKFERERILANKWNRRDNPLY#KEATSTFKNPTF 813 |
| Caenorhabditis elegans | Query 718 LSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 756 | | || | |+| ++|+ |+||++#| |||| || +| Sbjct 766 LHDRSEYATFNNERLMAKWDTNENPIY#KQATTTFKNPVYA 805 |
| Biomphalaria glabrata | Query 715 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 755 | + | ||+ +||||+ ++|+ +||++#| ||+| || + Sbjct 744 LTFIHDTREFAKFEKERQNAKWDTGENPIY#KQATSTFKNPTY 785 |
| Acropora millepora | Query 715 LIHLSDLREYRRFEKEKLKSQWNND-NPLF#KSATTTVMNPKFA 756 | + | ||++||+|++ |+| + |||+#++| || || +| Sbjct 746 LFTMVDRIEYQKFERERMHSKWTREKNPLY#QAAKTTFENPTYA 788 |
| Podocoryne carnea | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 755 | + | ||+ +|||++ +|++ +||++#| ||+| || + Sbjct 737 LATIQDRREFAKFEKDRQNPKWDSGENPIY#KKATSTFQNPMY 778 |
| Canis lupus familiaris | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 752 || + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 778 |
| Equus caballus | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 752 || + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 778 |
| Lytechinus variegatus | Query 715 LIHLSDLREYRRFEKEKLKSQW-NNDNPLF#KSATTTVMNPKF 755 | ++ | ||++ ||||+ + | +||++#| +|+ || + Sbjct 762 LTYIHDRREFQNFEKERANATWEGGENPIY#KPSTSVFKNPTY 803 |
| mice, Peptide Partial, 680 aa | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 752 || + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 640 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 678 |
| rats, Peptide Partial, 723 aa | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 752 || + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 679 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 717 |
| Schistosoma japonicum | Query 715 LIHLSDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKFAES 758 +| + | || |+++ +| +||+|#+| || |+|| | |+ Sbjct 242 VITIDDRRELANFKQQGENMRWEMAENPIF#ESPTTNVLNPTFEEN 286 |
| Ophlitaspongia tenuis | Query 715 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 757 |+ | |+ | |+||+| +++ |+|||+#+||| || + + Sbjct 795 LLLLWDVVEVRKFEREIKNAKYTKNENPLY#RSATKDYQNPLYGK 838 |
| Geodia cydonium | Query 715 LIHLSDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 757 |+ + | | ++|| | | ++ |+|||+# |||| || + + Sbjct 835 LLMVLDYTELKKFENELAKVKYTKNENPLY#HSATTEHKNPIYGQ 878 |
| Halocynthia roretzi | Query 718 LSDLREYRRFEKEKLKSQW-NNDNPLF#KSATTTVMNPKF 755 | | ||| +|+||+ +| +||++# +|++ || | Sbjct 793 LRDKREYEKFKKEEDLRKWTKGENPVY#VNASSKFDNPMF 831 |
* References
[PubMed ID: 12477932] Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA, Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002 Dec 24;99(26):16899-903. Epub 2002 Dec 11.