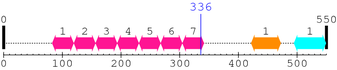
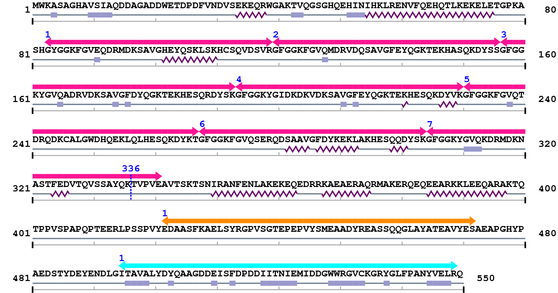
| # |
organism |
max score |
hits |
top seq |
| 1 |
Homo sapiens |
120.00 |
10 |
SRC8_HUMAN Src substrate cortactin (Amplaxin) (Onc |
| 2 |
synthetic construct |
120.00 |
3 |
cortactin |
| 3 |
Macaca mulatta |
120.00 |
2 |
PREDICTED: similar to cortactin isoform a |
| 4 |
Pongo pygmaeus |
118.00 |
1 |
hypothetical protein |
| 5 |
Mus musculus |
113.00 |
5 |
cortactin |
| 6 |
Rattus norvegicus |
113.00 |
4 |
cortactin isoform B |
| 7 |
Bos taurus |
113.00 |
2 |
cortactin |
| 8 |
N/A |
113.00 |
2 |
S |
| 9 |
mice, BALB/c 3T3 cells, Peptide, 546 aa |
113.00 |
1 |
cortactin, p80/p85 |
| 10 |
Canis familiaris |
112.00 |
5 |
PREDICTED: similar to cortactin isoform a isoform |
| 11 |
Macaca fascicularis |
99.40 |
2 |
unnamed protein product |
| 12 |
Gallus gallus |
98.60 |
2 |
cortactin |
| 13 |
Danio rerio |
87.00 |
4 |
cortactin |
| 14 |
Tetraodon nigroviridis |
81.30 |
3 |
unnamed protein product |
| 15 |
Strongylocentrotus purpuratus |
52.40 |
1 |
SRC8 protein |
| 16 |
Drosophila melanogaster |
48.10 |
2 |
Cortactin CG3637-PA |
| 17 |
Drosophila pseudoobscura |
48.10 |
1 |
GA17577-PA |
| 18 |
Pan troglodytes |
43.50 |
5 |
PREDICTED: cortactin isoform 3 |
| 19 |
Tribolium castaneum |
42.70 |
1 |
PREDICTED: similar to Xaa-Pro dipeptidase (X-Pro d |
| 20 |
Monodelphis domestica |
39.70 |
1 |
PREDICTED: hypothetical protein |
| 21 |
Suberites domuncula |
38.10 |
1 |
cortactin |
| 22 |
Anopheles gambiae str. PEST |
33.50 |
1 |
ENSANGP00000029554 |
| 23 |
Trichomonas vaginalis G3 |
33.50 |
1 |
hypothetical protein TVAG_334050 |
| 24 |
Xenopus laevis |
32.70 |
3 |
hypothetical protein LOC398923 |
| 25 |
Xenopus tropicalis |
32.70 |
1 |
cortactin |
| 26 |
Aedes aegypti |
32.30 |
1 |
cortactin |
| organism | matching |
|---|
| Homo sapiens |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|
| synthetic construct |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|
| Macaca mulatta |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|
| Pongo pygmaeus |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
||||||||||||||||||||||||||||||#||||||| ||||||||||||||||||||||
Sbjct 270 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVASKTSNIRANFENLAKEKEQEDR 329
|
| Mus musculus |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|||||||||||||||||||+| || |||||#|||+||||||||||||||||||||+|||||
Sbjct 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366
|
| Rattus norvegicus |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|||||||||||||||||||+| || |||||#|||+||||||||||||||||||||+|||||
Sbjct 270 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 329
|
| Bos taurus |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
||||||||||||||||||||| |||+|||#||||||| ||||||||||||||||||||||
Sbjct 307 GGKYGVQKDRMDKNASTFEDVAAVSSSYQK#TVPVEAVNSKTSNIRANFENLAKEKEQEDR 366
|
| N/A |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|||||||||||||||||||+| || |||||#|||+||||||||||||||||||||+|||||
Sbjct 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366
|
| mice, BALB/c 3T3 cells, Peptide, 546 aa |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|||||||||||||||||||+| || |||||#|||+||||||||||||||||||||+|||||
Sbjct 307 GGKYGVQKDRMDKNASTFEEVVQVPSAYQK#TVPIEAVTSKTSNIRANFENLAKEREQEDR 366
|
| Canis familiaris |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
||||||||||||||||||||| ||+ ||||#||||||| |+||||||||||||||||||||
Sbjct 307 GGKYGVQKDRMDKNASTFEDVAQVAPAYQK#TVPVEAVNSRTSNIRANFENLAKEKEQEDR 366
|
| Macaca fascicularis |
Query 317 MDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 317 MDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|
| Gallus gallus |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|||||||||||||||+||||+ + +| |||#| ||| | +|||+|||| ||||||||||||
Sbjct 316 GGKYGVQKDRMDKNAATFEDIEKPTSTYQK#TKPVERVANKTSSIRANLENLAKEKEQEDR 375
|
| Danio rerio |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|||||||||||||+| |||+| + |+||||#| |||| +| |+|+| |||+||+||+|++
Sbjct 306 GGKYGVQKDRMDKSAGTFEEVQKPSAAYQK#TRPVEAASSSASSIKARFENIAKQKEEEEQ 365
|
| Tetraodon nigroviridis |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|||+||||||||| | |||+| + + +|||#| |||| | +|+| |||+||+|| |||
Sbjct 315 GGKFGVQKDRMDKAAGTFEEVEKPTPSYQK#TKPVEAAGSNAGSIKARFENMAKQKEDEDR 374
|
| Strongylocentrotus purpuratus |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|||+|| |+ | | | |+ |||+|+|#| | | |+| || +|+ |+| |
Sbjct 270 GGKFGVDKNAQDATAGGFGDMQGVSSSYKK#TRPQPPAKSGAGNMRNKFEQMAQAGEEESR 329
|
| Drosophila melanogaster |
Query 307 GGKYGVQKDRMDKNASTFEDVT----QVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKE 362
|||+|||+|| ||+| ++ | |# || +| ||+|| |||||| |
Sbjct 197 GGKFGVQEDRKDKSAVGWDHKEAPQKHASQVDHK#VKPV-IEGAKPSNLRAKFENLAKNSE 255
Query 363 QEDR 366
+| |
Sbjct 256 EESR 259
|
| Drosophila pseudoobscura |
Query 307 GGKYGVQKDRMDKNASTFEDVT----QVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKE 362
|||+|||+|| ||+| ++ | |# || +| ||+|| |||||| |
Sbjct 195 GGKFGVQEDRKDKSAVGWDHKEAPQKHASQVDHK#VKPV-IEGAKPSNLRAKFENLAKNSE 253
Query 363 QEDR 366
+| |
Sbjct 254 EESR 257
|
| Pan troglodytes |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKT 347
|||||||||||||++ + | | ||# || |+ + |
Sbjct 307 GGKYGVQKDRMDKSSPSLGQTIVESPAMQK#GFPVAAIVTTT 347
|
| Tribolium castaneum |
Query 307 GGKYGVQKDRMDKNASTFED-VTQVSSAYQK#TVPVEAVTSKTSNIRANFENL 357
|||+|+| ||+||+|+ | | +| + | |# | + +| ++|| |||+
Sbjct 736 GGKFGLQTDRVDKSAANFNDEPAKVGTNYTK#VKP-DIGDAKPKDLRAKFENM 786
|
| Monodelphis domestica |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTS---KTSNIRANFENLAKEKEQ 363
||||||||||||| | | | |#+ + | | |++ | | + |
Sbjct 270 GGKYGVQKDRMDKRGGTAEPQAQSGRPQTK#SSLLTLATGVPCSISGIKSAFSNHQTPQAQ 329
Query 364 ED 365
|
Sbjct 330 SD 331
|
| Suberites domuncula |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLA 358
||||||++ | +| ++|+ | | ++ # | +| +||+ |||+|
Sbjct 271 GGKYGVEEGNQDSSAGGYDDMQAVKSDHRT#ERGVS--KGQTGSIRSKFENMA 320
|
| Anopheles gambiae str. PEST |
Query 307 GGKYGVQKDRMDKNASTFEDVT-----QVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEK 361
|||+||| || ||+| ++ | + ++ # + + ++| ||+|| ||| |
Sbjct 195 GGKFGVQTDRKDKSAFGWDHVEKPQMHESQLDHKI#VIKPDIGSAKPSNLRAKFENFAATA 254
Query 362 EQEDR 366
|+| |
Sbjct 255 EEEAR 259
Query 307 GGKYGVQKDRMDKNASTFEDVTQV 330
|||+||+||||||+| | + +|
Sbjct 84 GGKFGVEKDRMDKSAVGHEHIEKV 107
Query 307 GGKYGVQKDRMDKNASTFEDVTQV 330
|||+||||||+||+| ++ | +|
Sbjct 121 GGKFGVQKDRVDKSAHGWDHVEKV 144
|
| Trichomonas vaginalis G3 |
Query 317 MDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKE 362
+ | | ||+|| + |+| |# ++ + + |++|| + ||
Sbjct 42 LKKYPSAFENVTMICSSYDK#EQAIDYLMTSIKNVQANLNQIFASKE 87
|
| Xenopus laevis |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
||++||| ||+|| || | ++ ||||+|#| |+||+|| |+|+ |||+|+ |+|++
Sbjct 268 GGRFGVQTDRIDKCASGFGEIETPSSAYEK#TQPLEAMTSGAHNLRSRFENMARTAEEENK 327
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#T 337
|||||+|||| ||+| ++ ++ + +#|
Sbjct 194 GGKYGIQKDRQDKSAHSWSHKEELQTHQSQ#T 224
Query 307 GGKYGVQKDRMDKNASTFEDVTQV 330
||+|||| ||+||+| || |++
Sbjct 157 GGRYGVQTDRVDKSAVGFEYKTEL 180
|
| Xenopus tropicalis |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
||++||| ||+|| || | ++ ||||+|#| |+||+|| |+|+ |||+|+ |+|++
Sbjct 268 GGRFGVQTDRVDKCASGFGEIETPSSAYEK#TQPLEAMTSGAHNLRSRFENMARTAEEENK 327
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|||||||||| ||+| ++ | ++# | ++ | | | +|+++|+
Sbjct 194 GGKYGVQKDRQDKSAHSW-------SHKEE#LQPHQSQTDYTQGFGGKF---GVQKDRQDK 243
Query 307 GGKYGVQKDRMDKNASTFE 325
|||||||||| ||+| |+
Sbjct 120 GGKYGVQKDRTDKSAVGFD 138
Query 307 GGKYGVQKDRMDKNASTFEDVTQV 330
||+|||| ||+||+| || |++
Sbjct 157 GGRYGVQMDRVDKSAVGFEYKTEL 180
|
| Aedes aegypti |
Query 307 GGKYGVQKDRMDKNASTFEDVTQVSSAYQK#TVPVEAVTSKTSNIRANFENLAKEKEQEDR 366
|||+||| |||||+| |++ ++ + | |#| | + ++| ||||| ||| | |+| |
Sbjct 231 GGKFGVQSDRMDKSALGFQEQDKIGTNYTK#TKP-DIGSAKPSNIRARFENFAMAAEEETR 289
Query 307 GGKYGVQKDRMDKNASTFEDVTQV 330
|||+||| ||+|| |+|+| +|
Sbjct 120 GGKFGVQSDRVDKCAATWEHKEKV 143
|
 Thr
Thr

 Sequence conservation (by blast)
Sequence conservation (by blast)