XSB0664 : unnamed protein product [Macaca fascicularis]
[ CaMP Format ]
This entry is computationally expanded from SB0006
* Basic Information
| Organism | Macaca fascicularis (crab-eating macaque) |
| Protein Names | Testis cDNA clone: QtsA-11811, similar to human calcium/calmodulin-dependent protein kinase IV (CAMK4),mRNA, RefSeq: NM_001744.3 |
| Gene Names | Not available |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
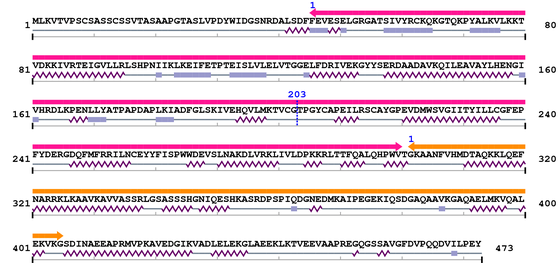
Length: 473 aa
Average Mass: 52.070 kDa
Monoisotopic Mass: 52.037 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Pkinase 1. | 46 | 300 | 400.9 | 2.1e-117 |
| --- cleavage 203 (inside Pkinase 46..300) --- | ||||
| Rrp15p 1. | 302 | 405 | -51.5 | 7.7 |
3. Sequence Information
Fasta Sequence: XSB0664.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] HQVLMKTVCG203-TPGYCAPEIL
Gly203  Thr
Thr
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| His194 | Gln195 | Val196 | Leu197 | Met198 | Lys199 | Thr200 | Val201 | Cys202 | Gly203 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Thr204 | Pro205 | Gly206 | Tyr207 | Cys208 | Ala209 | Pro210 | Glu211 | Ile212 | Leu213 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TPAPDAPLKIADFGLSKIVEHQVLMKTVCGTPGYCAPEILRSCAYGPEVDMWSVGIITYI |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 129.00 | 16 | 1 |
| 2 | Macaca fascicularis | 129.00 | 3 | unnamed protein product |
| 3 | Homo sapiens | 128.00 | 19 | calcium/calmodulin-dependent protein kinase IV |
| 4 | Mus musculus | 128.00 | 18 | unnamed protein product |
| 5 | synthetic construct | 128.00 | 8 | calcium/calmodulin-dependent protein kinase IV |
| 6 | Rattus norvegicus | 128.00 | 8 | calcium/calmodulin-dependent protein kinase IV |
| 7 | Macaca mulatta | 128.00 | 5 | PREDICTED: calcium/calmodulin-dependent protein ki |
| 8 | Pan troglodytes | 128.00 | 5 | PREDICTED: calcium/calmodulin-dependent protein ki |
| 9 | Rattus sp. | 128.00 | 1 | Ca2+/calmodulin-dependent protein kinase IV beta p |
| 10 | Xenopus laevis | 121.00 | 8 | calcium/calmodulin-dependent protein kinase IV |
| 11 | Gallus gallus | 121.00 | 4 | calcium/calmodulin-dependent protein kinase IV |
| 12 | Danio rerio | 118.00 | 14 | calcium/calmodulin-dependent protein kinase IV |
| 13 | Monodelphis domestica | 118.00 | 7 | PREDICTED: similar to calcium/calmodulin-dependent |
| 14 | Tetraodon nigroviridis | 93.60 | 5 | unnamed protein product |
| 15 | Strongylocentrotus purpuratus | 89.00 | 7 | PREDICTED: hypothetical protein |
| 16 | Ciona intestinalis | 75.90 | 1 | calmodulin-dependent protein kinase homologue |
| 17 | Dictyostelium discoideum AX4 | 75.50 | 7 | myosin light chain kinase |
| 18 | Paramecium tetraurelia | 74.70 | 6 | hypothetical protein |
| 19 | Entamoeba histolytica HM-1:IMSS | 72.40 | 3 | protein kinase |
| 20 | Yarrowia lipolytica | 72.00 | 2 | hypothetical protein |
| 21 | Aedes aegypti | 71.60 | 3 | calcium/calmodulin-dependent protein kinase type 1 |
| 22 | Xenopus tropicalis | 71.20 | 3 | Unknown (protein for MGC:122933) |
| 23 | Tetrahymena thermophila SB210 | 71.20 | 2 | Protein kinase domain containing protein |
| 24 | Phaeosphaeria nodorum | 71.20 | 2 | Ca/Cm-dependent protein kinase B |
| 25 | Drosophila pseudoobscura | 70.90 | 1 | GA13377-PA |
| 26 | Drosophila melanogaster | 70.50 | 3 | Calcium/calmodulin-dependent protein kinase I CG14 |
| 27 | Tribolium castaneum | 70.50 | 2 | PREDICTED: similar to CG1495-PG, isoform G |
| 28 | Apis mellifera | 70.10 | 3 | PREDICTED: similar to Calcium/calmodulin-dependent |
| 29 | Aspergillus terreus NIH2624 | 70.10 | 2 | calcium/calmodulin-dependent protein kinase type I |
| 30 | Aspergillus clavatus NRRL 1 | 70.10 | 2 | calcium/calmodulin dependent protein kinase, putat |
| 31 | Aspergillus oryzae | 70.10 | 2 | unnamed protein product |
| 32 | Aspergillus fumigatus Af293 | 70.10 | 2 | calcium/calmodulin dependent protein kinase, putat |
| 33 | Aspergillus niger | 70.10 | 2 | hypothetical protein An16g03050 |
| 34 | Neosartorya fischeri NRRL 181 | 70.10 | 2 | calcium/calmodulin dependent protein kinase, putat |
| 35 | Glossina morsitans morsitans | 70.10 | 1 | calcium/ calmodulin-dependent protein kinase 1 |
| 36 | Coccidioides immitis RS | 70.10 | 1 | calcium/calmodulin-dependent protein kinase |
| 37 | Cryptococcus neoformans var. neoformans JEC21 | 70.10 | 1 | calmodulin-dependent protein kinase |
| 38 | Gibberella zeae PH-1 | 69.70 | 2 | hypothetical protein FG00337.1 |
| 39 | Arthrobotrys dactyloides | 69.70 | 1 | calmodulin-binding protein kinase |
| 40 | Aspergillus nidulans FGSC A4 | 69.70 | 1 | hypothetical protein AN3065.2 |
| 41 | Glomerella cingulata | 69.30 | 2 | hypothetical protein ( (AF034963) calmodulin-depen |
| 42 | Magnaporthe grisea 70-15 | 69.30 | 1 | hypothetical protein MGG_00925 |
| 43 | Emericella nidulans | 69.30 | 1 | AF156027_1 calcium/calmodulin dependent protein ki |
| 44 | Bos taurus | 68.90 | 6 | hypothetical protein LOC511504 |
| 45 | Canis familiaris | 68.90 | 5 | PREDICTED: similar to calcium/calmodulin-dependent |
| 46 | Anopheles gambiae str. PEST | 68.90 | 3 | ENSANGP00000019618 |
| 47 | Bombyx mori | 68.90 | 1 | cell cycle checkpoint kinase 2 |
| 48 | Ustilago maydis 521 | 68.60 | 1 | hypothetical protein UM05544.1 |
| 49 | Caenorhabditis elegans | 68.20 | 3 | CaM Kinase family member (cmk-1) |
| 50 | Caenorhabditis briggsae | 68.20 | 1 | Hypothetical protein CBG08406 |
| 51 | Schizophyllum commune | 68.20 | 1 | putative serine/threonine protein kinase |
| 52 | Oncorhynchus mykiss | 67.00 | 1 | calcium/calmodulin-dependent protein kinase I |
| 53 | Schizosaccharomyces pombe | 65.90 | 2 | calmodulin kinase I homolog |
| 54 | Schizosaccharomyces pombe 972h- | 65.90 | 2 | hypothetical protein SPAC25D11.02C |
| 55 | Cochliobolus carbonum | 65.90 | 1 | AF159253_1 serine threonine protein kinase SNF1p |
| 56 | Arabidopsis thaliana | 65.10 | 5 | SnRK1.3 (SNF1-RELATED PROTEIN KINASE 1.3); kinase |
| 57 | Guillardia theta | 64.70 | 1 | SNF-related kinase |
| 58 | Trichomonas vaginalis G3 | 64.70 | 1 | CAMK family protein kinase |
| 59 | Candida albicans SC5314 | 64.30 | 2 | putative protein kinase |
| 60 | Sclerotinia sclerotiorum | 64.30 | 1 | serine threonine protein kinase |
| 61 | Triticum aestivum | 63.90 | 2 | calcium-dependent protein kinase |
| 62 | Phaeosphaeria nodorum SN15 | 63.90 | 1 | hypothetical protein SNOG_05495 |
| 63 | Candida glabrata CBS138 | 63.90 | 1 | hypothetical protein CAGL0M08910g |
| 64 | Medicago truncatula | 63.90 | 1 | Protein kinase |
| 65 | Oryza sativa (japonica cultivar-group) | 63.50 | 3 | putative CBL-interacting protein kinase |
| 66 | Kluyveromyces lactis | 63.50 | 2 | unnamed protein product |
| 67 | Chaetomium globosum CBS 148.51 | 63.50 | 2 | conserved hypothetical protein |
| 68 | Debaryomyces hansenii CBS767 | 63.50 | 1 | hypothetical protein DEHA0E15741g |
| 69 | Pichia stipitis CBS 6054 | 63.50 | 1 | carbon catabolite derepressing ser/thr protein kin |
| 70 | Lodderomyces elongisporus NRRL YB-4239 | 63.50 | 1 | carbon catabolite derepressing protein kinase |
| 71 | Glycine max | 63.50 | 1 | AF128443_1 SNF-1-like serine/threonine protein kin |
| 72 | Candida tropicalis | 63.50 | 1 | SNF1_CANTR Carbon catabolite derepressing protein |
| 73 | Pichia guilliermondii ATCC 6260 | 63.50 | 1 | conserved hypothetical protein |
| 74 | Oryza sativa (indica cultivar-group) | 63.50 | 1 | hypothetical protein OsI_028364 |
| 75 | Neurospora crassa | 63.50 | 1 | probable serine/threonine protein kinase (SNF1) |
| 76 | Ashbya gossypii ATCC 10895 | 63.50 | 1 | AEL230Wp |
| 77 | Fusarium oxysporum | 63.50 | 1 | AF420488_1 protein kinase SNF1 |
| 78 | Vitis vinifera | 63.20 | 1 | hypothetical protein |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 |
| Macaca fascicularis | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 |
| Homo sapiens | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||||||||| Sbjct 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 233 |
| Mus musculus | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||||||||| Sbjct 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |
| synthetic construct | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||||||||| Sbjct 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 233 |
| Rattus norvegicus | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||||||||| Sbjct 170 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 229 |
| Macaca mulatta | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||||||||| Sbjct 314 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 373 |
| Pan troglodytes | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||||||||| Sbjct 304 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 363 |
| Rattus sp. | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 ||||||||||||||||||||||||||||||#||||||||||| |||||||||||||||||| Sbjct 198 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 257 |
| Xenopus laevis | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 |||||||||||||||||||+ || ||||||#||||||||||| |||||||||||||||||| Sbjct 178 TPAPDAPLKIADFGLSKIVDDQVTMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGIITYI 237 |
| Gallus gallus | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 |||||||||||||||||||| || ||||||#||||||||||| |||||||||||+|||||| Sbjct 159 TPAPDAPLKIADFGLSKIVEDQVTMKTVCG#TPGYCAPEILRGCAYGPEVDMWSLGIITYI 218 |
| Danio rerio | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 | |||||||||||||||||+ || ||||||#||||||||||| |||||||||||||+|||| Sbjct 157 TSAPDAPLKIADFGLSKIVDDQVTMKTVCG#TPGYCAPEILRGCAYGPEVDMWSVGVITYI 216 |
| Monodelphis domestica | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 || ||||||||||||||||| || |||+||#||||||||||| |||||||||||+|+|||| Sbjct 239 TPDPDAPLKIADFGLSKIVEDQVTMKTICG#TPGYCAPEILRGCAYGPEVDMWSLGVITYI 298 |
| Tetraodon nigroviridis | Query 184 ADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 ||||||||++ || ||||||#||||||||||| ||||||||||||+| || Sbjct 1 ADFGLSKIIDDQVTMKTVCG#TPGYCAPEILRGNAYGPEVDMWSVGVILYI 50 |
| Strongylocentrotus purpuratus | Query 178 DAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 +||||+||||||||++ | ||||||#||||||||+| | |||||||+|||| Sbjct 173 EAPLKLADFGLSKIIDQDVSMKTVCG#TPGYCAPEVLCGKPYCKAVDMWSVGVITYI 228 |
| Ciona intestinalis | Query 176 APDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 | |+ + |+||||||+ ++|| ||#|||| |||+|+ || |||+||+|+| || Sbjct 147 AEDSKIMISDFGLSKVEVEGQMLKTACG#TPGYVAPEVLKQKPYGKEVDVWSIGVIAYI 204 |
| Dictyostelium discoideum AX4 | Query 183 IADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 |||||||||+ ++|+| ||#|| | |||+| + | ||||||+|+|||| Sbjct 149 IADFGLSKIIGQTLVMQTACG#TPSYVAPEVLNATGYDKEVDMWSIGVITYI 199 |
| Paramecium tetraurelia | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 || ||| +||+|||++|++ + || | ||#|||| ||||| | || ||+|+| |+ Sbjct 146 TPDPDATVKISDFGVAKVISDE-LMLTACG#TPGYVAPEILTGVGYDMAVDYWSIGVILYV 204 |
| Entamoeba histolytica HM-1:IMSS | Query 174 TPAPDAPLKIADFGLSKIV-EHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITY 232 +| | ++||||| ||++ | ++ | ||#|| | |||+| + || ||||||+|+||| Sbjct 138 SPDDDTDVRIADFGFSKMITEDAQILLTACG#TPVYVAPEVLNAKGYGMEVDMWSIGVITY 197 Query 233 I 233 + Sbjct 198 V 198 |
| Yarrowia lipolytica | Query 174 TPAPDAPLKIADFGLSKIVEHQVL--MKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 | | |+ | |||||||+|++ + +|| ||#|||| |||| +| ||||++|+|| Sbjct 143 TAAEDSDLLIADFGLSRIIDEERFRTLKTTCG#TPGYMAPEIFTKAGHGKPVDMWALGVIT 202 Query 232 Y 232 | Query 232 Y 232 |
| Aedes aegypti | Query 175 PAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 || |+ + |+||||||+ | | | ||#|||| |||+| || ||+||+|+|+|| Sbjct 176 PAEDSKIMISDFGLSKM-EDSGFMATACG#TPGYVAPEVLAQKPYGKAVDVWSIGVISYI 233 |
| Xenopus tropicalis | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 || |+ + |+||||||| | +| | ||#|||| |||+| || ||+|++|+|+|| Sbjct 149 TPFEDSKIMISDFGLSKI-EDGGMMATACG#TPGYVAPELLEQKPYGKAVDVWAIGVISYI 207 |
| Tetrahymena thermophila SB210 | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 +| ||| +||+||||+|++ + || | ||#|| | ||||| | || ||+|+| |+ Sbjct 214 SPDPDATIKISDFGLAKVISDE-LMTTACG#TPSYIAPEILEGKGYTFAVDYWSIGVILYV 272 |
| Phaeosphaeria nodorum | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| | +| ||+|++|+|| Sbjct 156 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFRKTGHGKPVDIWAIGVIT 215 Query 232 Y 232 | Query 232 Y 232 |
| Drosophila pseudoobscura | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 +| |+ + |+||||||+ | +| | ||#|||| |||+| || ||+||+|+|+|| Sbjct 170 SPEDDSKIMISDFGLSKM-EDSGIMATACG#TPGYVAPEVLAQKPYGKAVDVWSIGVISYI 228 |
| Drosophila melanogaster | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 +| |+ + |+||||||+ | +| | ||#|||| |||+| || ||+||+|+|+|| Sbjct 178 SPDDDSKIMISDFGLSKM-EDSGIMATACG#TPGYVAPEVLAQKPYGKAVDVWSIGVISYI 236 |
| Tribolium castaneum | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 +| |+ + |+||||||+ | +| | ||#|||| |||+| || ||+||+|+|+|| Sbjct 174 SPDEDSKIMISDFGLSKM-EDSGIMATACG#TPGYVAPEVLAQKPYGKAVDVWSIGVISYI 232 |
| Apis mellifera | Query 175 PAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 | |+ + |+||||||+ | +| | ||#|||| |||+| || ||+||+|+|+|| Sbjct 162 PDEDSKIMISDFGLSKM-EDSGIMATACG#TPGYVAPEVLAQKPYGKAVDVWSIGVISYI 219 |
| Aspergillus terreus NIH2624 | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 151 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKSGHGKPVDIWAIGVIT 210 Query 232 Y 232 | Query 232 Y 232 |
| Aspergillus clavatus NRRL 1 | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 151 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKSGHGKPVDIWAIGVIT 210 Query 232 Y 232 | Query 232 Y 232 |
| Aspergillus oryzae | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 177 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKSGHGKPVDIWAIGVIT 236 Query 232 Y 232 | Query 232 Y 232 |
| Aspergillus fumigatus Af293 | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 151 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKSGHGKPVDIWAIGVIT 210 Query 232 Y 232 | Query 232 Y 232 |
| Aspergillus niger | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 151 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKSGHGKPVDIWAIGVIT 210 Query 232 Y 232 | Query 232 Y 232 |
| Neosartorya fischeri NRRL 181 | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 151 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKSGHGKPVDIWAIGVIT 210 Query 232 Y 232 | Query 232 Y 232 |
| Glossina morsitans morsitans | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 +| || + |+|||||| | | | ||#|||| |||+| || ||+||+|+|+|| Sbjct 179 SPDDDAKIMISDFGLSK-TEDSGTMATACG#TPGYVAPEVLAQKPYGKAVDVWSIGVISYI 237 |
| Coccidioides immitis RS | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 151 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKTGHGKPVDIWAIGVIT 210 Query 232 Y 232 | Query 232 Y 232 |
| Cryptococcus neoformans var. neoformans JEC21 | Query 176 APDAPLKIADFGLSKIVEHQ--VLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITY 232 | || | +|||||||+++ ++ | ||#|||| |||| + +| ||+|++|+||| Sbjct 145 AEDADLMLADFGLSKVLDDDKFAILTTTCG#TPGYMAPEIFKKAGHGKPVDIWAIGVITY 203 |
| Gibberella zeae PH-1 | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 149 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKTGHGKPVDLWALGVIT 208 Query 232 Y 232 | Query 232 Y 232 |
| Arthrobotrys dactyloides | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 160 TPEDNADLLIADFGLSRIMDEERFHVLTTTCG#TPGYMAPEIFKKTGHGKPVDIWAIGVIT 219 Query 232 Y 232 | Query 232 Y 232 |
| Aspergillus nidulans FGSC A4 | Query 174 TPAPDAPLKIADFGLSKIVEHQVL--MKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + | + | ||#|||| |||| +| ||+|++|+|| Sbjct 151 TPEDNADLLIADFGLSRIMDEEQLHVLTTTCG#TPGYMAPEIFDKSGHGKPVDIWAIGVIT 210 Query 232 Y 232 | Query 232 Y 232 |
| Glomerella cingulata | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 149 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKTGHGKPVDIWALGVIT 208 Query 232 Y 232 | Query 232 Y 232 |
| Magnaporthe grisea 70-15 | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++|+|| Sbjct 149 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKTGHGKPVDIWALGVIT 208 Query 232 Y 232 | Query 232 Y 232 |
| Emericella nidulans | Query 174 TPAPDAPLKIADFGLSKIVEHQVL--MKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 || +| | |||||||+|++ + | + | ||#|||| |||| +| ||+|++|+|| Sbjct 151 TPEDNADLLIADFGLSRIMDEEQLHVLTTTCG#TPGYMAPEIFDKSGHGKPVDIWAIGLIT 210 Query 232 Y 232 | Query 232 Y 232 |
| Bos taurus | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 || ++ + | ||||||+ ++ | | | ||#|||| |||+| | || ||+|+|||| Sbjct 153 TPEENSKIMITDFGLSKMEQNGV-MSTACG#TPGYVAPEVLAQKPYSKAVDCWSIGVITYI 211 |
| Canis familiaris | Query 174 TPAPDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 || ++ + | ||||||+ | +| | ||#|||| |||+| | || ||+|+|||| Sbjct 186 TPEENSKIMITDFGLSKM-EQSGVMSTACG#TPGYVAPEVLAQKPYSKAVDCWSIGVITYI 244 |
| Anopheles gambiae str. PEST | Query 176 APDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 | |+ + |+||||||+ | | | ||#|||| |||+| || ||+||+|+|+|| Sbjct 170 AEDSKIMISDFGLSKM-EDSGFMATACG#TPGYVAPEVLAQKPYGKAVDVWSIGVISYI 226 |
| Bombyx mori | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRS---CAYGPEVDMWSVGIITYI 233 +|| |||||| | |||+||#|| | |||+||+ ||||||+||+|+| ++ Sbjct 318 VKITDFGLSKFVGEDSFMKTMCG#TPLYLAPEVLRANGQNTYGPEVDVWSLGVIFFV 373 |
| Ustilago maydis 521 | Query 178 DAPLKIADFGLSKIVEHQ--VLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITY 232 ++ | |||||||++|+ + ++ | ||#|||| |||| + +| ||||++|+||| Sbjct 150 ESDLLIADFGLSRVVDDEKITVLSTTCG#TPGYMAPEIFKKTGHGKPVDMWAIGVITY 206 |
| Caenorhabditis elegans | Query 178 DAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 |+ + |+|||||| | +| | ||#|||| |||+|+ || ||+||+|+| || Sbjct 158 DSKIMISDFGLSK-TEDSGVMATACG#TPGYVAPEVLQQKPYGKAVDVWSIGVIAYI 212 |
| Caenorhabditis briggsae | Query 178 DAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 |+ + |+|||||| | +| | ||#|||| |||+|+ || ||+||+|+| || Sbjct 158 DSKIMISDFGLSK-TEDSGVMATACG#TPGYVAPEVLQQKPYGKAVDVWSIGVIAYI 212 |
| Schizophyllum commune | Query 174 TPAPDAPLKIADFGLSKIVEHQVL--MKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIIT 231 | | || + |||||||+++| + | + +||#|||| |||| + + ||+|++|+|| Sbjct 237 TQAEDADIMIADFGLSRVMEEEKLNMLTEICG#TPGYMAPEIFKKTGHSKPVDIWAMGVIT 296 Query 232 Y 232 | Query 232 Y 232 |
| Oncorhynchus mykiss | Query 178 DAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 |+ + |+||||||| +| | ||#|||| |||+| | || ||+|+| || Sbjct 156 DSKIMISDFGLSKIEGSGSVMSTACG#TPGYVAPEVLAQKPYSKAVDCWSIGVIAYI 211 |
| Schizosaccharomyces pombe | Query 177 PDAPLKIADFGLSKIVEHQ--VLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITY 232 |++ | ||||||| | ++ | ||#|| | |||+ | || ||||++|+||| Sbjct 167 PNSDLLIADFGLSHFYEDSQYYMLMTACG#TPEYMAPEVFRRTGYGKPVDMWAIGVITY 224 |
| Schizosaccharomyces pombe 972h- | Query 177 PDAPLKIADFGLSKIVEHQ--VLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITY 232 |++ | ||||||| | ++ | ||#|| | |||+ | || ||||++|+||| Sbjct 167 PNSDLLIADFGLSHFYEDSQYYMLMTACG#TPEYMAPEVFRRTGYGKPVDMWAIGVITY 224 |
| Cochliobolus carbonum | Query 178 DAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 |+ +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 197 DSNVKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILYV 253 |
| Arabidopsis thaliana | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITY 232 +|| ||||| ++ +|| ||#+| | |||++ |||+||+|| |+| | Sbjct 156 IKIVDFGLSNVMHDGHFLKTSCG#SPNYAAPEVISGKPYGPDVDIWSCGVILY 207 |
| Guillardia theta | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITY 232 +|||||||| |++ +|| ||#+| | |||++ +| |||||+|| |+| | Sbjct 149 VKIADFGLSNIMKDGNFLKTSCG#SPNYAAPEVINGKSYLGPEVDVWSCGVIMY 201 |
| Trichomonas vaginalis G3 | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITY 232 +|| ||||| | |+| | +||#|| | |||++++ +| +| ||+||| | Sbjct 87 IKIIDFGLSSIHRHRVKMHKLCG#TPEYIAPEVIQNSSYTDNIDTWSIGIILY 138 |
| Candida albicans SC5314 | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 188 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSAGVILYV 241 |
| Sclerotinia sclerotiorum | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 195 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVINGKLYAGPEVDVWSCGVILYV 248 |
| Triticum aestivum | Query 176 APDAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 | ||||| ||||| + | |#+| | |||+|+ | |||| |+|| |+| || Sbjct 183 AEDAPLKTTDFGLSMFYKPGDKFSDVVG#SPYYVAPEVLQKC-YGPEADVWSAGVILYI 239 |
| Phaeosphaeria nodorum SN15 | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 62 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILYV 115 |
| Candida glabrata CBS138 | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 175 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILYV 228 |
| Medicago truncatula | Query 178 DAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITY 232 || |||||||||+ | ++||||#|| | |||+|+ | + |||||| + + Sbjct 161 DAVLKIADFGLSRTVRPGEYVETVCG#TPSYMAPEVLQFQRYDHKADMWSVGAMLF 215 |
| Oryza sativa (japonica cultivar-group) | Query 181 LKIADFGLSKIVEH---QVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 ||+ ||||| + +| |+ |+||#|||| |||+|| | | + |+|| |+| |+ Sbjct 160 LKVVDFGLSALADHARADGLLHTLCG#TPGYAAPEVLRDKGYDGAKADLWSCGVILYV 216 |
| Kluyveromyces lactis | Query 178 DAPLKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGIITYI 233 | +|||||||+| |+||#|| | |||+| | ||||| |++ |+ Sbjct 259 DIQVKIADFGLAKFTGEMKFTNTLCG#TPSYVAPEVLVKTGYTSRVDMWSAGVLLYV 314 |
| Chaetomium globosum CBS 148.51 | Query 174 TPAPDAPLKIADFGLSKIVEHQV--LMKTVCG#TPGYCAPEILRSCAYGPEVDMWSVGII 230 || +| | |||||||+|++ + ++ | ||#|||| |||| + +| ||+|++| | Sbjct 112 TPEDNADLLIADFGLSRIMDEEQFHVLTTTCG#TPGYMAPEIFKKTGHGKPVDLWALGAI 170 |
| Debaryomyces hansenii CBS767 | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 191 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSSGVILYV 244 |
| Pichia stipitis CBS 6054 | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 174 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSSGVILYV 227 |
| Lodderomyces elongisporus NRRL YB-4239 | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 210 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSSGVILYV 263 |
| Glycine max | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITY 232 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| | Sbjct 157 IKIADFGLSNIMRDGHFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSCGVILY 209 |
| Candida tropicalis | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 188 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSSGVILYV 241 |
| Pichia guilliermondii ATCC 6260 | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 176 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSSGVILYV 229 |
| Oryza sativa (indica cultivar-group) | Query 181 LKIADFGLSKIVEH---QVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 ||+ ||||| + +| |+ |+||#|||| |||+|| | | + |+|| |+| |+ Sbjct 159 LKVVDFGLSALADHARADGLLHTLCG#TPGYAAPEVLRDKGYDGAKADLWSCGVILYV 215 |
| Neurospora crassa | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 212 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVIGGKLYAGPEVDVWSCGVILYV 265 |
| Ashbya gossypii ATCC 10895 | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 175 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVISGKLYAGPEVDVWSSGVILYV 228 |
| Fusarium oxysporum | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITYI 233 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| |+ Sbjct 199 VKIADFGLSNIMTDGNFLKTSCG#SPNYAAPEVIGGKLYAGPEVDVWSCGVILYV 252 |
| Vitis vinifera | Query 181 LKIADFGLSKIVEHQVLMKTVCG#TPGYCAPEILRSCAY-GPEVDMWSVGIITY 232 +|||||||| |+ +|| ||#+| | |||++ | |||||+|| |+| | Sbjct 143 VKIADFGLSNIMRDGHFLKTSCG#SPNYAAPEVISGRLYAGPEVDVWSCGVILY 195 |
* References
[PubMed ID: 15944441] Osada N, Hirata M, Tanuma R, Kusuda J, Hida M, Suzuki Y, Sugano S, Gojobori T, Shen CK, Wu CI, Hashimoto K, Substitution rate and structural divergence of 5'UTR evolution: comparative analysis between human and cynomolgus monkey cDNAs. Mol Biol Evol. 2005 Oct;22(10):1976-82. Epub 2005 Jun 8.