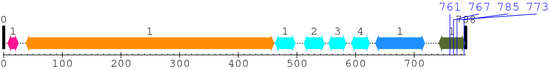
XSB0943 : PREDICTED: similar to integrin beta chain, beta 3 precursor isoform 2 [Macaca mulatta]
[ CaMP Format ]
This entry is computationally expanded from SB0043
* Basic Information
| Organism | Macaca mulatta (rhesus monkey) |
| Protein Names | similar to integrin beta chain; beta 3 precursor isoform 2 |
| Gene Names | LOC717379; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 5 mRNAs, 61 ESTs, 11 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001116013 | XM_001116013 | 717379 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 788 aa
Average Mass: 86.961 kDa
Monoisotopic Mass: 86.903 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| DUF2474 1. | 7 | 25 | -21.9 | 8.3 |
| Integrin_beta 1. | 38 | 461 | 1050.2 | 0 |
| EGF_2 1. | 463 | 497 | 13.2 | 0.09 |
| EGF_2 2. | 512 | 547 | 32.0 | 2.4e-06 |
| EGF_2 3. | 554 | 584 | 22.8 | 0.0014 |
| EGF_2 4. | 593 | 624 | 30.2 | 8.2e-06 |
| Integrin_B_tail 1. | 634 | 718 | 128.0 | 3e-35 |
| Integrin_b_cyt 1. | 742 | 787 | 106.8 | 7.1e-29 |
| --- cleavage 761 (inside Integrin_b_cyt 742..787) --- | ||||
| --- cleavage 767 (inside Integrin_b_cyt 742..787) --- | ||||
| --- cleavage 785 (inside Integrin_b_cyt 742..787) --- | ||||
| --- cleavage 773 (inside Integrin_b_cyt 742..787) --- | ||||
3. Sequence Information
Fasta Sequence: XSB0943.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
4 [sites]
Cleavage sites (±10aa)
[Site 1] EFAKFEEERA761-RAKWDTANNP
Ala761  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu752 | Phe753 | Ala754 | Lys755 | Phe756 | Glu757 | Glu758 | Glu759 | Arg760 | Ala761 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg762 | Ala763 | Lys764 | Trp765 | Asp766 | Thr767 | Ala768 | Asn769 | Asn770 | Pro771 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LIGLATLLIWKLLITIHDRKEFAKFEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 84.70 | 16 | unnamed protein product |
| 2 | Bos taurus | 84.70 | 12 | PREDICTED: similar to platelet glycoprotein IIIa i |
| 3 | Macaca mulatta | 84.70 | 12 | PREDICTED: similar to integrin beta chain, beta 3 |
| 4 | Canis familiaris | 84.70 | 10 | platelet glycoprotein IIIa |
| 5 | Rattus norvegicus | 84.70 | 10 | integrin beta 3 |
| 6 | Sus scrofa | 84.70 | 6 | glycoprotein GPIIIa |
| 7 | synthetic construct | 84.70 | 5 | AF239959_1 hybrid integrin beta 3 subunit precurso |
| 8 | rats, Peptide Partial, 723 aa | 84.70 | 1 | beta 3 integrin, GPIIIA |
| 9 | Canis lupus familiaris | 84.70 | 1 | integrin beta chain, beta 3 |
| 10 | Equus caballus | 84.70 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 11 | N/A | 82.80 | 29 | - |
| 12 | Homo sapiens | 82.80 | 24 | integrin beta chain, beta 3 precursor |
| 13 | Pan troglodytes | 82.80 | 11 | PREDICTED: integrin beta chain, beta 3 isoform 4 |
| 14 | Oryctolagus cuniculus | 80.90 | 2 | glycoprotein IIIa |
| 15 | Monodelphis domestica | 80.50 | 5 | PREDICTED: similar to platelet glycoprotein IIIa |
| 16 | Gallus gallus | 79.30 | 7 | integrin, beta 3 precursor |
| 17 | mice, Peptide Partial, 680 aa | 75.90 | 1 | beta 3 integrin, GPIIIA |
| 18 | Xenopus laevis | 74.70 | 6 | integrin beta-3 subunit |
| 19 | Danio rerio | 70.90 | 13 | hypothetical protein LOC100001836 |
| 20 | Tetraodon nigroviridis | 70.90 | 5 | unnamed protein product |
| 21 | Aedes aegypti | 63.90 | 1 | integrin beta subunit |
| 22 | Drosophila melanogaster | 62.00 | 1 | myospheroid CG1560-PA |
| 23 | Tribolium castaneum | 61.60 | 2 | PREDICTED: similar to CG1560-PA |
| 24 | Ostrinia furnacalis | 61.60 | 1 | integrin beta 1 precursor |
| 25 | Anopheles gambiae | 60.50 | 1 | integrin beta subunit |
| 26 | Anopheles gambiae str. PEST | 60.50 | 1 | ENSANGP00000021373 |
| 27 | Pseudoplusia includens | 58.50 | 1 | integrin beta 1 |
| 28 | Ovis aries | 56.60 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 29 | Pongo pygmaeus | 55.80 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 30 | Felis catus | 55.80 | 1 | integrin beta 1 |
| 31 | Biomphalaria glabrata | 55.80 | 1 | beta integrin subunit |
| 32 | Xenopus tropicalis | 55.80 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 33 | Crassostrea gigas | 55.10 | 1 | integrin beta cgh |
| 34 | Podocoryne carnea | 54.70 | 1 | AF308652_1 integrin beta chain |
| 35 | Caenorhabditis briggsae | 53.90 | 1 | Hypothetical protein CBG03601 |
| 36 | Caenorhabditis elegans | 53.90 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 37 | Ictalurus punctatus | 52.40 | 1 | beta-1 integrin |
| 38 | Strongylocentrotus purpuratus | 50.80 | 4 | integrin beta-C subunit |
| 39 | Lytechinus variegatus | 50.80 | 1 | beta-C integrin subunit |
| 40 | Pacifastacus leniusculus | 46.20 | 1 | integrin |
| 41 | Apis mellifera | 44.70 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 42 | Manduca sexta | 43.10 | 1 | plasmatocyte-specific integrin beta 1 |
| 43 | Spodoptera frugiperda | 42.70 | 1 | integrin beta 1 |
| 44 | Halocynthia roretzi | 38.90 | 1 | integrin beta Hr2 precursor |
| 45 | Mus sp. | 38.10 | 1 | beta 7 integrin |
| 46 | Schistosoma japonicum | 37.00 | 1 | SJCHGC06221 protein |
| 47 | Oncorhynchus mykiss | 36.60 | 1 | CD18 protein |
| 48 | Acropora millepora | 35.80 | 1 | integrin subunit betaCn1 |
| 49 | Bubalus bubalis | 33.50 | 1 | integrin beta-2 |
| 50 | Capra hircus | 33.50 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 51 | Sigmodon hispidus | 33.10 | 1 | AF445415_1 integrin beta-2 precursor |
| 52 | Ovis canadensis | 32.70 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 731 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 787 |
| Bos taurus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| Macaca mulatta | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Canis familiaris | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| Rattus norvegicus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 731 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 787 |
| Sus scrofa | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| synthetic construct | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| rats, Peptide Partial, 723 aa | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 667 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 723 |
| Canis lupus familiaris | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| Equus caballus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| N/A | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 ||||| |||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Homo sapiens | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 ||||| |||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Pan troglodytes | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 ||||| |||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Oryctolagus cuniculus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 ||| | |||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGFALLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Monodelphis domestica | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYR 786 |||||||||||||||||| # |||||||||||||||||||||| Sbjct 707 LIGLATLLIWKLLITIHDRREFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYR 761 |
| Gallus gallus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 ||||| |||||||||||| # ||| ||||||||||||||||||| Sbjct 724 LIGLAALLIWKLLITIHDRREFARFEEEKA#RAKWDTGNNPLYKEATSTFTNITYRG 779 |
| mice, Peptide Partial, 680 aa | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNIT 784 |||||||||||||||||| # |||||||||||||||||||| Sbjct 628 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNIT 680 |
| Xenopus laevis | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 |||| |||||||||||| # ||||+||||| |||||||||||| Sbjct 730 LIGLVALLIWKLLITIHDRREFAKFEEERA#KAKWDTAHNPLYKGATSTFTNITYRG 785 |
| Danio rerio | Query 733 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 +||| |||||||||||| # |+| +||||| |||||||||||| Sbjct 734 LGLAALLIWKLLITIHDRREFAKFEEERA#RAKWETGHNPLYKGATSTFTNITYRG 788 |
| Tetraodon nigroviridis | Query 733 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 +||| |||||||||||| # ||| +||||| |||||||||+|| Sbjct 748 LGLAALLIWKLLITIHDRREFAKFEEERA#RAKWDTGHNPLYKGATSTFTNITFRG 802 |
| Aedes aegypti | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 ||||| ||+|||| |||| # ||| ||+||+||+|| | || | Sbjct 779 LIGLAVLLLWKLLTTIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAG 834 |
| Drosophila melanogaster | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 |+||| ||+|||| |||| # ||| ||+||+||||| | | | Sbjct 790 LVGLAILLLWKLLTTIHDRREFARFEKERM#NAKWDTGENPIYKQATSTFKNPMYAG 845 |
| Tribolium castaneum | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 |+|+| ||+||| |||| # ||| ||+||+||||| | || | Sbjct 773 LLGMAILLLWKLFTTIHDRREFAKFEKEAM#MARWDTGENPIYKQATSTFKNPTYAG 828 |
| Ostrinia furnacalis | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 |+||| |++||++ |||| # ||| ||+||+||||| | || | Sbjct 775 LVGLALLMLWKMVTTIHDRREFARFEKERM#MAKWDTGENPIYKQATSTFKNPTYAG 830 |
| Anopheles gambiae | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 |||+| ||+||+| +||| # ||| ||+||+||+|| | || | Sbjct 781 LIGMAVLLLWKVLTSIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAG 836 |
| Anopheles gambiae str. PEST | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 |||+| ||+||+| +||| # ||| ||+||+||+|| | || | Sbjct 781 LIGMAVLLLWKVLTSIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAG 836 |
| Pseudoplusia includens | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 |+||| |++||+ | || # ||| ||+||+||||| | || | Sbjct 781 LVGLALLMLWKMATTSHDRREFARFEKERM#MAKWDTGENPIYKQATSTFKNPTYAG 836 |
| Ovis aries | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYR 786 |||+| | |||||++ || # | | ||||+ +|||| |+||+ Sbjct 720 LIGVALLCIWKLLVSFHDRKEVAKFEAERS#KAKWQTGTNPLYRGSTSTFKNVTYK 774 |
| Pongo pygmaeus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 ||||| |||||||+ ||| # ||| ||+|| | +| | | | Sbjct 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |
| Felis catus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 ||||| |||||||+ ||| # ||| ||+|| | +| | | | Sbjct 742 LIGLALLLIWKLLMIIHDTREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |
| Biomphalaria glabrata | Query 733 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 785 +|| ||+|||| ||| # ||| ||+||+||||| | || Sbjct 733 VGLFLLLLWKLLTFIHDTREFAKFEKERQ#NAKWDTGENPIYKQATSTFKNPTY 785 |
| Xenopus tropicalis | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 ||||| |||||||+ ||| # ||| ||+|| | +| | | | Sbjct 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |
| Crassostrea gigas | Query 734 GLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYR 786 |+ |||||| || | # ||| ||+||+||||| | ||| Sbjct 746 GIILLLIWKLFTTISDKRELARFEKEAM#NARWDTGENPIYKQATSTFVNPTYR 798 |
| Podocoryne carnea | Query 733 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 785 |||| ||||||| || | # ||+ ||+||+||||| | | Sbjct 726 IGLALLLIWKLLATIQDRREFAKFEKDRQ#NPKWDSGENPIYKKATSTFQNPMY 778 |
| Caenorhabditis briggsae | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 ++|+ ||+|||| +|| # ||| ||+||+||+|| | | | Sbjct 752 ILGILLLLLWKLLTVLHDRAEYAKFNNERL#MAKWDTNENPIYKQATTTFKNPVYAG 807 |
| Caenorhabditis elegans | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 ++|+ ||+|||| +|| # ||| ||+||+||+|| | | | Sbjct 751 ILGILLLLLWKLLTVLHDRSEYATFNNERL#MAKWDTNENPIYKQATTTFKNPVYAG 806 |
| Ictalurus punctatus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 ||||| | |||||+ ||| # || ||+|| | +| | | | Sbjct 751 LIGLALLFIWKLLMIIHDRREFAKFEKEKM#NAKWDAGENPIYKSAVTTVVNPKYEG 806 |
| Strongylocentrotus purpuratus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 785 |+||| ||||+|| ||| # |+ ||+|| +|| | | || Sbjct 750 LVGLALLLIWRLLTYIHDRREFQNFEKERA#NATWEGGENPIYKPSTSVFKNPTY 803 |
| Lytechinus variegatus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 785 |+||| ||||+|| ||| # |+ ||+|| +|| | | || Sbjct 750 LVGLALLLIWRLLTYIHDRREFQNFEKERA#NATWEGGENPIYKPSTSVFKNPTY 803 |
| Pacifastacus leniusculus | Query 733 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTN 782 ||| |||||||| |+|| # | ||||| | +|| | Sbjct 745 IGLLTLLIWKLLTTLHDRREYAKFEKERK#LPSGKRAENPLYKSAKTTFQN 794 |
| Apis mellifera | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXX-WDTANNPLYKEATSTFTNITY 785 +|| | || ||+| | | # |+ +||||||||||| | |+ Sbjct 759 IIGFAILLTWKILTMIRDNREFKKFERERI#LANKWNRRDNPLYKEATSTFKNPTF 813 |
| Manduca sexta | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 785 |||| ||+ ||+|+ +|| # +| + ||||+| |+| | Sbjct 708 LIGLLTLIAWKILVDLHDKREYAKFEEESR#SRGFDVSLNPLYQEPEINFSNPVY 761 |
| Spodoptera frugiperda | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 785 |||+ |++|||+|+ +|| # +| + ||||++ + |+| | Sbjct 723 LIGILTVIIWKILVDMHDKKEYKKFQDEAL#AAGYDVSLNPLYQDPSINFSNPVY 776 |
| Halocynthia roretzi | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 |||+ |+|||++ |+ | # | ||+| |+| | | + | Sbjct 778 LIGIIALIIWKVIQTLRDKREYEKFKKEED#LRKWTKGENPVYVNASSKFDNPMFGG 833 |
| Mus sp. | Query 733 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 788 +|| +| ++| + |+| # | |||||| | +| | ++|| Sbjct 737 VGLGLVLAYRLSVEIYDRREYRRFEKEQQ#QLNWKQDNNPLYKSAITTTVNPRFQGT 792 |
| Schistosoma japonicum | Query 734 GLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 785 || |||+||+||| | # |+ | ||+++ |+ | |+ Sbjct 232 GLILLLIYKLVITIDDRRELANFKQQGE#NMRWEMAENPIFESPTTNVLNPTF 283 |
| Oncorhynchus mykiss | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 |||| ||| | ++ + | # | +|||+ ||+| | |+ | Sbjct 721 LIGLLILLIIKAILYMRDVKEFRRFENEQK#KGKWSPGDNPLFMNATTTVANPTFTG 776 |
| Acropora millepora | Query 733 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 787 +|| |++ | | |+ | # | ||||+ | +|| | || | Sbjct 735 LGLLILIVIKGLFTMVDRIEYQKFERERM#HSKWTREKNPLYQAAKTTFENPTYAG 789 |
| Bubalus bubalis | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTN 782 |+|+ |+||| | + | # |+ +|||+| ||+| | Sbjct 714 LVGILLLVIWKALTHLSDLREYHRFEKEKL#KSQWNN-DNPLFKSATTTVMN 763 |
| Capra hircus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTN 782 |+|+ |+||| | + | # |+ +|||+| ||+| | Sbjct 715 LVGILLLVIWKALTHLSDLREYHRFEKEKL#KSQWNN-DNPLFKSATTTVMN 764 |
| Sigmodon hispidus | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTN 782 |||+ |+||| | + | # |+ +|||+| ||+| | Sbjct 714 LIGVLLLVIWKALTHLTDLREYRHFEKEKL#KSQWNN-DNPLFKSATTTVMN 763 |
| Ovis canadensis | Query 732 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTN 782 |+|+ | ||| | + | # |+ +|||+| ||+| | Sbjct 715 LVGILLLAIWKALTHLSDLREYHRFEKEKL#KSQWNN-DNPLFKSATTTVMN 764 |
[Site 2] EERARAKWDT767-ANNPLYKEAT
Thr767  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu758 | Glu759 | Arg760 | Ala761 | Arg762 | Ala763 | Lys764 | Trp765 | Asp766 | Thr767 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala768 | Asn769 | Asn770 | Pro771 | Leu772 | Tyr773 | Lys774 | Glu775 | Ala776 | Thr777 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LLIWKLLITIHDRKEFAKFEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 74.30 | 21 | - |
| 2 | Mus musculus | 74.30 | 13 | unnamed protein product |
| 3 | Homo sapiens | 74.30 | 12 | integrin beta chain, beta 3 precursor |
| 4 | Bos taurus | 74.30 | 11 | PREDICTED: similar to platelet glycoprotein IIIa i |
| 5 | Macaca mulatta | 74.30 | 10 | PREDICTED: similar to integrin beta chain, beta 3 |
| 6 | Pan troglodytes | 74.30 | 8 | PREDICTED: integrin beta chain, beta 3 isoform 4 |
| 7 | Canis familiaris | 74.30 | 8 | platelet glycoprotein IIIa |
| 8 | Sus scrofa | 74.30 | 5 | AF170527_1 glycoprotein IIIa; GPIIIa |
| 9 | Rattus norvegicus | 74.30 | 5 | integrin beta 3 |
| 10 | synthetic construct | 74.30 | 4 | AF239959_1 hybrid integrin beta 3 subunit precurso |
| 11 | Oryctolagus cuniculus | 74.30 | 2 | glycoprotein IIIa |
| 12 | Equus caballus | 74.30 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 13 | rats, Peptide Partial, 723 aa | 74.30 | 1 | beta 3 integrin, GPIIIA |
| 14 | Canis lupus familiaris | 74.30 | 1 | integrin beta chain, beta 3 |
| 15 | Gallus gallus | 70.90 | 7 | integrin, beta 3 precursor |
| 16 | Monodelphis domestica | 70.10 | 5 | PREDICTED: similar to platelet glycoprotein IIIa |
| 17 | Xenopus laevis | 67.80 | 6 | Unknown (protein for MGC:80757) |
| 18 | mice, Peptide Partial, 680 aa | 65.50 | 1 | beta 3 integrin, GPIIIA |
| 19 | Danio rerio | 64.70 | 10 | hypothetical protein LOC100001836 |
| 20 | Tetraodon nigroviridis | 64.70 | 5 | unnamed protein product |
| 21 | Aedes aegypti | 55.50 | 1 | integrin beta subunit |
| 22 | Tribolium castaneum | 55.10 | 2 | PREDICTED: similar to CG1560-PA |
| 23 | Drosophila melanogaster | 54.30 | 1 | myospheroid CG1560-PA |
| 24 | Ostrinia furnacalis | 53.90 | 1 | integrin beta 1 precursor |
| 25 | Crassostrea gigas | 52.80 | 1 | integrin beta cgh |
| 26 | Anopheles gambiae str. PEST | 52.80 | 1 | ENSANGP00000021373 |
| 27 | Anopheles gambiae | 52.80 | 1 | integrin beta subunit |
| 28 | Biomphalaria glabrata | 52.00 | 1 | beta integrin subunit |
| 29 | Pseudoplusia includens | 50.80 | 1 | integrin beta 1 |
| 30 | Caenorhabditis briggsae | 50.10 | 1 | Hypothetical protein CBG03601 |
| 31 | Caenorhabditis elegans | 50.10 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Pongo pygmaeus | 49.70 | 2 | hypothetical protein |
| 33 | Ovis aries | 49.70 | 1 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 34 | Podocoryne carnea | 48.10 | 1 | AF308652_1 integrin beta chain |
| 35 | Felis catus | 47.80 | 1 | integrin beta 1 |
| 36 | Xenopus tropicalis | 47.80 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 37 | Ictalurus punctatus | 44.30 | 1 | beta-1 integrin |
| 38 | Strongylocentrotus purpuratus | 43.10 | 4 | integrin beta-C subunit |
| 39 | Lytechinus variegatus | 43.10 | 1 | beta-C integrin subunit |
| 40 | Pacifastacus leniusculus | 39.30 | 1 | integrin |
| 41 | Apis mellifera | 38.90 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 42 | Spodoptera frugiperda | 35.00 | 1 | integrin beta 1 |
| 43 | Manduca sexta | 34.70 | 1 | plasmatocyte-specific integrin beta 1 |
| 44 | Schistosoma japonicum | 33.90 | 1 | SJCHGC06221 protein |
| 45 | Mus sp. | 33.50 | 1 | beta 7 integrin |
| 46 | Halocynthia roretzi | 33.10 | 1 | integrin beta Hr2 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Mus musculus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 737 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 787 |
| Homo sapiens | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Bos taurus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 784 |
| Macaca mulatta | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Pan troglodytes | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Canis familiaris | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 784 |
| Sus scrofa | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 784 |
| Rattus norvegicus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 737 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 787 |
| synthetic construct | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Oryctolagus cuniculus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Equus caballus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 784 |
| rats, Peptide Partial, 723 aa | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 673 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 723 |
| Canis lupus familiaris | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 |||||||||||| |||#||||||||||||||||||||| Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 784 |
| Gallus gallus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 |||||||||||| |||# ||||||||||||||||||| Sbjct 730 LLIWKLLITIHDRREFARFEEEKARAKWDT#GNNPLYKEATSTFTNITYRG 779 |
| Monodelphis domestica | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYR 786 |||||||||||| |||#||||||||||||||||||| Sbjct 713 LLIWKLLITIHDRREFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYR 761 |
| Xenopus laevis | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 |||||||||||| |||#|+||||| |||||||||||| Sbjct 736 LLIWKLLITIHDRREFAKFEEERAKAKWDT#AHNPLYKGATSTFTNITYRG 785 |
| mice, Peptide Partial, 680 aa | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNIT 784 |||||||||||| |||#||||||||||||||||| Sbjct 634 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNIT 680 |
| Danio rerio | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 |||||||||||| |+|# +||||| |||||||||||| Sbjct 739 LLIWKLLITIHDRREFAKFEEERARAKWET#GHNPLYKGATSTFTNITYRG 788 |
| Tetraodon nigroviridis | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 |||||||||||| |||# +||||| |||||||||+|| Sbjct 753 LLIWKLLITIHDRREFAKFEEERARAKWDT#GHNPLYKGATSTFTNITFRG 802 |
| Aedes aegypti | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 ||+|||| |||| |||# ||+||+||+|| | || | Sbjct 785 LLLWKLLTTIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAG 834 |
| Tribolium castaneum | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 ||+||| |||| |||# ||+||+||||| | || | Sbjct 779 LLLWKLFTTIHDRREFAKFEKEAMMARWDT#GENPIYKQATSTFKNPTYAG 828 |
| Drosophila melanogaster | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 ||+|||| |||| |||# ||+||+||||| | | | Sbjct 796 LLLWKLLTTIHDRREFARFEKERMNAKWDT#GENPIYKQATSTFKNPMYAG 845 |
| Ostrinia furnacalis | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 |++||++ |||| |||# ||+||+||||| | || | Sbjct 781 LMLWKMVTTIHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAG 830 |
| Crassostrea gigas | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYR 786 |||||| || | |||# ||+||+||||| | ||| Sbjct 750 LLIWKLFTTISDKRELARFEKEAMNARWDT#GENPIYKQATSTFVNPTYR 798 |
| Anopheles gambiae str. PEST | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 ||+||+| +||| |||# ||+||+||+|| | || | Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAG 836 |
| Anopheles gambiae | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 ||+||+| +||| |||# ||+||+||+|| | || | Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAG 836 |
| Biomphalaria glabrata | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 785 ||+|||| ||| |||# ||+||+||||| | || Sbjct 738 LLLWKLLTFIHDTREFAKFEKERQNAKWDT#GENPIYKQATSTFKNPTY 785 |
| Pseudoplusia includens | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 |++||+ | || |||# ||+||+||||| | || | Sbjct 787 LMLWKMATTSHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAG 836 |
| Caenorhabditis briggsae | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 ||+|||| +|| |||# ||+||+||+|| | | | Sbjct 758 LLLWKLLTVLHDRAEYAKFNNERLMAKWDT#NENPIYKQATTTFKNPVYAG 807 |
| Caenorhabditis elegans | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 ||+|||| +|| |||# ||+||+||+|| | | | Sbjct 757 LLLWKLLTVLHDRSEYATFNNERLMAKWDT#NENPIYKQATTTFKNPVYAG 806 |
| Pongo pygmaeus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYR 786 | |||||++ || | |# ||||+ +|||| |+||+ Sbjct 654 LCIWKLLVSFHDRKEVAKFEAERSKVKWQT#GTNPLYRGSTSTFKNVTYK 702 |
| Ovis aries | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYR 786 | |||||++ || | |# ||||+ +|||| |+||+ Sbjct 726 LCIWKLLVSFHDRKEVAKFEAERSKAKWQT#GTNPLYRGSTSTFKNVTYK 774 |
| Podocoryne carnea | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 785 ||||||| || | ||+# ||+||+||||| | | Sbjct 731 LLIWKLLATIQDRREFAKFEKDRQNPKWDS#GENPIYKKATSTFQNPMY 778 |
| Felis catus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 |||||||+ ||| |||# ||+|| | +| | | | Sbjct 748 LLIWKLLMIIHDTREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |
| Xenopus tropicalis | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 |||||||+ ||| |||# ||+|| | +| | | | Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |
| Ictalurus punctatus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 | |||||+ ||| || # ||+|| | +| | | | Sbjct 757 LFIWKLLMIIHDRREFAKFEKEKMNAKWDA#GENPIYKSAVTTVVNPKYEG 806 |
| Strongylocentrotus purpuratus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 785 ||||+|| ||| |+ # ||+|| +|| | | || Sbjct 756 LLIWRLLTYIHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTY 803 |
| Lytechinus variegatus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 785 ||||+|| ||| |+ # ||+|| +|| | | || Sbjct 756 LLIWRLLTYIHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTY 803 |
| Pacifastacus leniusculus | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTN 782 ||||||| |+|| #| ||||| | +|| | Sbjct 750 LLIWKLLTTLHDRREYAKFEKERKLPSGKR#AENPLYKSAKTTFQN 794 |
| Apis mellifera | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXX-WDT#ANNPLYKEATSTFTNITY 785 || ||+| | | |+ # +||||||||||| | |+ Sbjct 765 LLTWKILTMIRDNREFKKFERERILANKWNR#RDNPLYKEATSTFKNPTF 813 |
| Spodoptera frugiperda | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 785 ++|||+|+ +|| +| #+ ||||++ + |+| | Sbjct 729 VIIWKILVDMHDKKEYKKFQDEALAAGYDV#SLNPLYQDPSINFSNPVY 776 |
| Manduca sexta | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 785 |+ ||+|+ +|| +| #+ ||||+| |+| | Sbjct 714 LIAWKILVDLHDKREYAKFEEESRSRGFDV#SLNPLYQEPEINFSNPVY 761 |
| Schistosoma japonicum | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 785 |||+||+||| | |+ #| ||+++ |+ | |+ Sbjct 236 LLIYKLVITIDDRRELANFKQQGENMRWEM#AENPIFESPTTNVLNPTF 283 |
| Mus sp. | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 788 +| ++| + |+| | # |||||| | +| | ++|| Sbjct 742 VLAYRLSVEIYDRREYRRFEKEQQQLNWKQ#DNNPLYKSAITTTVNPRFQGT 792 |
| Halocynthia roretzi | Query 738 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 787 |+|||++ |+ | | # ||+| |+| | | + | Sbjct 784 LIIWKVIQTLRDKREYEKFKKEEDLRKWTK#GENPVYVNASSKFDNPMFGG 833 |
[Site 3] ATSTFTNITY785-RGT
Tyr785  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala776 | Thr777 | Ser778 | Thr779 | Phe780 | Thr781 | Asn782 | Ile783 | Thr784 | Tyr785 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg786 | Gly787 | Thr788 | - | - | - | - | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| FEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 72.80 | 22 | - |
| 2 | Mus musculus | 72.80 | 13 | integrin beta 3 |
| 3 | Homo sapiens | 72.80 | 12 | integrin beta chain, beta 3 precursor |
| 4 | Bos taurus | 72.80 | 11 | PREDICTED: similar to platelet glycoprotein IIIa i |
| 5 | Macaca mulatta | 72.80 | 10 | PREDICTED: similar to integrin beta chain, beta 3 |
| 6 | Pan troglodytes | 72.80 | 8 | PREDICTED: integrin beta chain, beta 3 isoform 4 |
| 7 | Canis familiaris | 72.80 | 8 | platelet glycoprotein IIIa |
| 8 | Rattus norvegicus | 72.80 | 8 | integrin beta 3 |
| 9 | Sus scrofa | 72.80 | 5 | glycoprotein GPIIIa |
| 10 | synthetic construct | 72.80 | 4 | AF239959_1 hybrid integrin beta 3 subunit precurso |
| 11 | Oryctolagus cuniculus | 72.80 | 2 | glycoprotein IIIa |
| 12 | Equus caballus | 72.80 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 13 | rats, Peptide Partial, 723 aa | 72.80 | 1 | beta 3 integrin, GPIIIA |
| 14 | Canis lupus familiaris | 72.80 | 1 | integrin beta chain, beta 3 |
| 15 | Monodelphis domestica | 68.60 | 5 | PREDICTED: similar to platelet glycoprotein IIIa |
| 16 | Gallus gallus | 68.20 | 7 | integrin, beta 3 precursor |
| 17 | Xenopus laevis | 65.10 | 6 | Unknown (protein for MGC:80757) |
| 18 | mice, Peptide Partial, 680 aa | 63.90 | 1 | beta 3 integrin, GPIIIA |
| 19 | Tetraodon nigroviridis | 63.50 | 5 | unnamed protein product |
| 20 | Danio rerio | 63.20 | 8 | hypothetical protein LOC100001836 |
| 21 | Pseudoplusia includens | 52.00 | 1 | integrin beta 1 |
| 22 | Ostrinia furnacalis | 52.00 | 1 | integrin beta 1 precursor |
| 23 | Anopheles gambiae str. PEST | 50.80 | 1 | ENSANGP00000021373 |
| 24 | Anopheles gambiae | 50.80 | 1 | integrin beta subunit |
| 25 | Aedes aegypti | 50.80 | 1 | integrin beta subunit |
| 26 | Biomphalaria glabrata | 50.40 | 1 | beta integrin subunit |
| 27 | Drosophila melanogaster | 50.10 | 1 | myospheroid CG1560-PA |
| 28 | Crassostrea gigas | 49.30 | 1 | integrin beta cgh |
| 29 | Ovis aries | 48.90 | 1 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 30 | Tribolium castaneum | 48.50 | 2 | PREDICTED: similar to CG1560-PA |
| 31 | Pongo pygmaeus | 47.40 | 2 | hypothetical protein |
| 32 | Caenorhabditis briggsae | 45.80 | 1 | Hypothetical protein CBG03601 |
| 33 | Caenorhabditis elegans | 45.80 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 34 | Podocoryne carnea | 43.10 | 1 | AF308652_1 integrin beta chain |
| 35 | Felis catus | 43.10 | 1 | integrin beta 1 |
| 36 | Xenopus tropicalis | 43.10 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 37 | Strongylocentrotus purpuratus | 41.60 | 4 | integrin beta-C subunit |
| 38 | Lytechinus variegatus | 41.60 | 1 | beta-C integrin subunit |
| 39 | Ictalurus punctatus | 41.20 | 2 | beta-1 integrin |
| 40 | Apis mellifera | 40.80 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 41 | Acropora millepora | 40.00 | 1 | integrin subunit betaCn1 |
| 42 | Oncorhynchus mykiss | 36.60 | 1 | CD18 protein |
| 43 | Mus sp. | 36.60 | 1 | beta 7 integrin |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Mus musculus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Homo sapiens | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Bos taurus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Macaca mulatta | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Pan troglodytes | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Canis familiaris | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Rattus norvegicus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Sus scrofa | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| synthetic construct | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Oryctolagus cuniculus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Equus caballus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| rats, Peptide Partial, 723 aa | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Canis lupus familiaris | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||||||||||||||||||||||||||||||#||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 |
| Monodelphis domestica | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#R 786 ||||||||||||||||||||||||||||||#| Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#R 786 |
| Gallus gallus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||||+||||||| |||||||||||||||||#|| Sbjct 748 FEEEKARAKWDTGNNPLYKEATSTFTNITY#RG 779 |
| Xenopus laevis | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||||||+||||||+||||| ||||||||||#|| Sbjct 754 FEEERAKAKWDTAHNPLYKGATSTFTNITY#RG 785 |
| mice, Peptide Partial, 680 aa | Query 756 FEEERARAKWDTANNPLYKEATSTFTNIT 784 ||||||||||||||||||||||||||||| Query 756 FEEERARAKWDTANNPLYKEATSTFTNIT 784 |
| Tetraodon nigroviridis | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 |||||||||||| |||||| |||||||| |#+| Sbjct 720 FEEERARAKWDTGNNPLYKGATSTFTNIAY#KG 751 |
| Danio rerio | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||||||||||+| +||||| ||||||||||#|| Sbjct 757 FEEERARAKWETGHNPLYKGATSTFTNITY#RG 788 |
| Pseudoplusia includens | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|| ||||| ||+||+||||| | ||# | Sbjct 805 FEKERMMAKWDTGENPIYKQATSTFKNPTY#AG 836 |
| Ostrinia furnacalis | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|| ||||| ||+||+||||| | ||# | Sbjct 799 FEKERMMAKWDTGENPIYKQATSTFKNPTY#AG 830 |
| Anopheles gambiae str. PEST | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|| ||||| ||+||+||+|| | ||# | Sbjct 805 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AG 836 |
| Anopheles gambiae | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|| ||||| ||+||+||+|| | ||# | Sbjct 805 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AG 836 |
| Aedes aegypti | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|| ||||| ||+||+||+|| | ||# | Sbjct 803 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AG 834 |
| Biomphalaria glabrata | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 ||+|| ||||| ||+||+||||| | ||# Sbjct 756 FEKERQNAKWDTGENPIYKQATSTFKNPTY# 785 |
| Drosophila melanogaster | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|| ||||| ||+||+||||| | |# | Sbjct 814 FEKERMNAKWDTGENPIYKQATSTFKNPMY#AG 845 |
| Crassostrea gigas | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#R 786 ||+| |+||| ||+||+||||| | ||#| Sbjct 768 FEKEAMNARWDTGENPIYKQATSTFVNPTY#R 798 |
| Ovis aries | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#R 786 || ||++||| | ||||+ +|||| |+||#+ Sbjct 744 FEAERSKAKWQTGTNPLYRGSTSTFKNVTY#K 774 |
| Tribolium castaneum | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+| |+||| ||+||+||||| | ||# | Sbjct 797 FEKEAMMARWDTGENPIYKQATSTFKNPTY#AG 828 |
| Pongo pygmaeus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#R 786 || ||++ || | ||||+ +|||| |+||#+ Sbjct 672 FEAERSKVKWQTGTNPLYRGSTSTFKNVTY#K 702 |
| Caenorhabditis briggsae | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 | || ||||| ||+||+||+|| | |# | Sbjct 776 FNNERLMAKWDTNENPIYKQATTTFKNPVY#AG 807 |
| Caenorhabditis elegans | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 | || ||||| ||+||+||+|| | |# | Sbjct 775 FNNERLMAKWDTNENPIYKQATTTFKNPVY#AG 806 |
| Podocoryne carnea | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 ||++| |||+ ||+||+||||| | |# Sbjct 749 FEKDRQNPKWDSGENPIYKKATSTFQNPMY# 778 |
| Felis catus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|+ ||||| ||+|| | +| | |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EG 797 |
| Xenopus tropicalis | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|+ ||||| ||+|| | +| | |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EG 797 |
| Strongylocentrotus purpuratus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 ||+||| | |+ ||+|| +|| | | ||# Sbjct 774 FEKERANATWEGGENPIYKPSTSVFKNPTY# 803 |
| Lytechinus variegatus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY# 785 ||+||| | |+ ||+|| +|| | | ||# Sbjct 774 FEKERANATWEGGENPIYKPSTSVFKNPTY# 803 |
| Ictalurus punctatus | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 ||+|+ |||| ||+|| | +| | |# | Sbjct 775 FEKEKMNAKWDAGENPIYKSAVTTVVNPKY#EG 806 |
| Apis mellifera | Query 756 FEEERARA-KWDTANNPLYKEATSTFTNITY# 785 || || | ||+ +||||||||||| | |+# Sbjct 783 FERERILANKWNRRDNPLYKEATSTFKNPTF# 813 |
| Acropora millepora | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 || || +|| ||||+ | +|| | ||# | Sbjct 758 FERERMHSKWTREKNPLYQAAKTTFENPTY#AG 789 |
| Oncorhynchus mykiss | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 787 || |+ + || +|||+ ||+| | |+# | Sbjct 745 FENEQKKGKWSPGDNPLFMNATTTVANPTF#TG 776 |
| Mus sp. | Query 756 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 788 ||+|+ + | |||||| | +| | +#+|| Sbjct 760 FEKEQQQLNWKQDNNPLYKSAITTTVNPRF#QGT 792 |
[Site 4] KWDTANNPLY773-KEATSTFTNI
Tyr773  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys764 | Trp765 | Asp766 | Thr767 | Ala768 | Asn769 | Asn770 | Pro771 | Leu772 | Tyr773 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys774 | Glu775 | Ala776 | Thr777 | Ser778 | Thr779 | Phe780 | Thr781 | Asn782 | Ile783 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LITIHDRKEFAKFEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 62.00 | 12 | - |
| 2 | Homo sapiens | 62.00 | 8 | integrin beta chain, beta 3 precursor |
| 3 | Pan troglodytes | 62.00 | 7 | PREDICTED: integrin beta chain, beta 3 isoform 4 |
| 4 | Macaca mulatta | 62.00 | 7 | PREDICTED: similar to integrin beta chain, beta 3 |
| 5 | Mus musculus | 62.00 | 7 | unnamed protein product |
| 6 | Canis familiaris | 62.00 | 6 | platelet glycoprotein IIIa |
| 7 | Bos taurus | 62.00 | 5 | PREDICTED: similar to platelet glycoprotein IIIa i |
| 8 | Sus scrofa | 62.00 | 5 | glycoprotein GPIIIa |
| 9 | Rattus norvegicus | 62.00 | 4 | integrin beta 3 |
| 10 | synthetic construct | 62.00 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurso |
| 11 | Oryctolagus cuniculus | 62.00 | 2 | glycoprotein IIIa |
| 12 | Canis lupus familiaris | 62.00 | 1 | integrin beta chain, beta 3 |
| 13 | Equus caballus | 62.00 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 14 | rats, Peptide Partial, 723 aa | 62.00 | 1 | beta 3 integrin, GPIIIA |
| 15 | Gallus gallus | 58.50 | 3 | integrin, beta 3 precursor |
| 16 | Monodelphis domestica | 57.80 | 3 | PREDICTED: similar to platelet glycoprotein IIIa |
| 17 | Xenopus laevis | 55.50 | 4 | Unknown (protein for MGC:80757) |
| 18 | mice, Peptide Partial, 680 aa | 53.10 | 1 | beta 3 integrin, GPIIIA |
| 19 | Danio rerio | 52.40 | 8 | hypothetical protein LOC100001836 |
| 20 | Tetraodon nigroviridis | 52.40 | 5 | unnamed protein product |
| 21 | Tribolium castaneum | 43.90 | 2 | PREDICTED: similar to CG1560-PA |
| 22 | Ostrinia furnacalis | 43.90 | 1 | integrin beta 1 precursor |
| 23 | Aedes aegypti | 43.90 | 1 | integrin beta subunit |
| 24 | Drosophila melanogaster | 42.70 | 1 | myospheroid CG1560-PA |
| 25 | Anopheles gambiae | 42.40 | 1 | integrin beta subunit |
| 26 | Anopheles gambiae str. PEST | 42.40 | 1 | ENSANGP00000021373 |
| 27 | Pseudoplusia includens | 41.60 | 1 | integrin beta 1 |
| 28 | Crassostrea gigas | 41.20 | 1 | integrin beta cgh |
| 29 | Biomphalaria glabrata | 40.40 | 1 | beta integrin subunit |
| 30 | Pongo pygmaeus | 39.30 | 2 | hypothetical protein |
| 31 | Ovis aries | 39.30 | 1 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 32 | Caenorhabditis briggsae | 38.50 | 1 | Hypothetical protein CBG03601 |
| 33 | Caenorhabditis elegans | 38.50 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 34 | Podocoryne carnea | 35.80 | 1 | AF308652_1 integrin beta chain |
| 35 | Xenopus tropicalis | 35.40 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 36 | Felis catus | 35.40 | 1 | integrin beta 1 |
| 37 | Apis mellifera | 35.00 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 38 | Strongylocentrotus purpuratus | 34.30 | 2 | integrin beta L subunit |
| 39 | Ictalurus punctatus | 33.50 | 1 | beta-1 integrin |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Homo sapiens | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Pan troglodytes | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Macaca mulatta | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Mus musculus | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 743 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 787 |
| Canis familiaris | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 784 |
| Bos taurus | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 784 |
| Sus scrofa | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 784 |
| Rattus norvegicus | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 743 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 787 |
| synthetic construct | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Oryctolagus cuniculus | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Canis lupus familiaris | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 784 |
| Equus caballus | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 784 |
| rats, Peptide Partial, 723 aa | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 788 |||||| |||||||||#||||||||||||||| Sbjct 679 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 723 |
| Gallus gallus | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 |||||| ||| |||||#|||||||||||||| Sbjct 736 LITIHDRREFARFEEEKARAKWDTGNNPLY#KEATSTFTNITYRG 779 |
| Monodelphis domestica | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYR 786 |||||| |||||||||#||||||||||||| Sbjct 719 LITIHDRREFAKFEEERARAKWDTANNPLY#KEATSTFTNITYR 761 |
| Xenopus laevis | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 |||||| ||||+||||#| |||||||||||| Sbjct 742 LITIHDRREFAKFEEERAKAKWDTAHNPLY#KGATSTFTNITYRG 785 |
| mice, Peptide Partial, 680 aa | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNIT 784 |||||| |||||||||#||||||||||| Sbjct 640 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNIT 680 |
| Danio rerio | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 |||||| |+| +||||#| |||||||||||| Sbjct 745 LITIHDRREFAKFEEERARAKWETGHNPLY#KGATSTFTNITYRG 788 |
| Tetraodon nigroviridis | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 |||||| ||| +||||#| |||||||||+|| Sbjct 759 LITIHDRREFAKFEEERARAKWDTGHNPLY#KGATSTFTNITFRG 802 |
| Tribolium castaneum | Query 746 TIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 |||| ||| ||+|#|+||||| | || | Sbjct 787 TIHDRREFAKFEKEAMMARWDTGENPIY#KQATSTFKNPTYAG 828 |
| Ostrinia furnacalis | Query 746 TIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 |||| ||| ||+|#|+||||| | || | Sbjct 789 TIHDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYAG 830 |
| Aedes aegypti | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 | |||| ||| ||+|#|+||+|| | || | Sbjct 791 LTTIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAG 834 |
| Drosophila melanogaster | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 | |||| ||| ||+|#|+||||| | | | Sbjct 802 LTTIHDRREFARFEKERMNAKWDTGENPIY#KQATSTFKNPMYAG 845 |
| Anopheles gambiae | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 | +||| ||| ||+|#|+||+|| | || | Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAG 836 |
| Anopheles gambiae str. PEST | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 | +||| ||| ||+|#|+||+|| | || | Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAG 836 |
| Pseudoplusia includens | Query 746 TIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 | || ||| ||+|#|+||||| | || | Sbjct 795 TSHDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYAG 836 |
| Crassostrea gigas | Query 765 WDTANNPLY#KEATSTFTNITYR 786 ||| ||+|#|+||||| | ||| Sbjct 777 WDTGENPIY#KQATSTFVNPTYR 798 |
| Biomphalaria glabrata | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITY 785 | ||| ||| ||+|#|+||||| | || Sbjct 744 LTFIHDTREFAKFEKERQNAKWDTGENPIY#KQATSTFKNPTY 785 |
| Pongo pygmaeus | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYR 786 |++ || | | ||||#+ +|||| |+||+ Sbjct 660 LVSFHDRKEVAKFEAERSKVKWQTGTNPLY#RGSTSTFKNVTYK 702 |
| Ovis aries | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYR 786 |++ || | | ||||#+ +|||| |+||+ Sbjct 732 LVSFHDRKEVAKFEAERSKAKWQTGTNPLY#RGSTSTFKNVTYK 774 |
| Caenorhabditis briggsae | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 | +|| ||| ||+|#|+||+|| | | | Sbjct 764 LTVLHDRAEYAKFNNERLMAKWDTNENPIY#KQATTTFKNPVYAG 807 |
| Caenorhabditis elegans | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 | +|| ||| ||+|#|+||+|| | | | Sbjct 763 LTVLHDRSEYATFNNERLMAKWDTNENPIY#KQATTTFKNPVYAG 806 |
| Podocoryne carnea | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITY 785 | || | ||+ ||+|#|+||||| | | Sbjct 737 LATIQDRREFAKFEKDRQNPKWDSGENPIY#KKATSTFQNPMY 778 |
| Xenopus tropicalis | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 |+ ||| ||| ||+|#| | +| | | | Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEG 797 |
| Felis catus | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 |+ ||| ||| ||+|#| | +| | | | Sbjct 754 LMIIHDTREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEG 797 |
| Apis mellifera | Query 765 WDTANNPLY#KEATSTFTNITY 785 |+ +||||#||||||| | |+ Sbjct 793 WNRRDNPLY#KEATSTFKNPTF 813 |
| Strongylocentrotus purpuratus | Query 765 WDTANNPLY#KEATSTFTNITY 785 || ++||+|#| +|+|| | || Sbjct 780 WDQSDNPIY#KSSTTTFKNPTY 800 |
| Ictalurus punctatus | Query 744 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 787 |+ ||| || ||+|#| | +| | | | Sbjct 763 LMIIHDRREFAKFEKEKMNAKWDAGENPIY#KSAVTTVVNPKYEG 806 |
* References
None.